Isolation and Characterization of 17 Microsatellite DNA Loci for Odorrana margaretae (Anura: Ranidae)
2015-10-31LiangQIAOWeizhaoYANGHuNIEandJiapanHU
Liang QIAO, Weizhao YANG Hu NIEand Jiapan HU
1Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu 610041, Sichuan, China
2College of Life Science and Technology, Xinxiang Medical University, Xinxiang 453003, Henan, China
3College of Life Sciences, Sichuan University, Chengdu 610041, Sichuan, China
Isolation and Characterization of 17 Microsatellite DNA Loci for Odorrana margaretae (Anura: Ranidae)
Liang QIAO1,2*, Weizhao YANG1, Hu NIE3and Jiapan HU3
1Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu 610041, Sichuan, China
2College of Life Science and Technology, Xinxiang Medical University, Xinxiang 453003, Henan, China
3College of Life Sciences, Sichuan University, Chengdu 610041, Sichuan, China
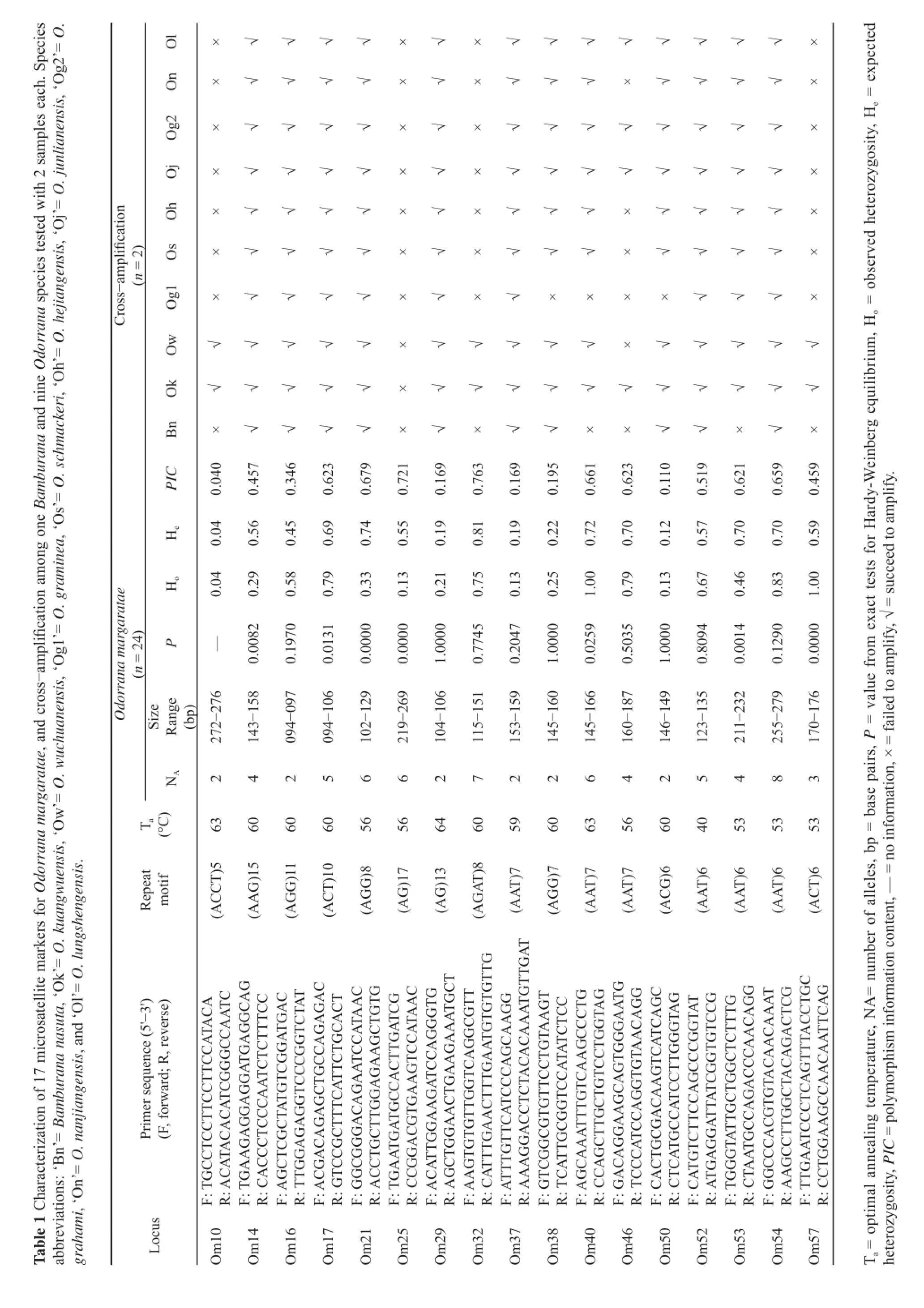
We isolated and characterized 17 microsatellite DNA loci for the odorous frog Odorrana margaretae from its transcriptome sequence data. These loci were screened with 24 individuals from Mt. Emei. All loci were polymorphic,with the number of alleles ranging from 2 to 8. The observed and expected heterozygosity, polymorphism information content, ranged from 0.04 to 1, 0.04 to 0.81, and 0.040 to 0.763, respectively. All loci were in linkage equilibrium and six loci were signifi cantly deviated from Hardy-Weinberg equilibrium after sequential Bonferroni corrections. Crossspecies amplifi cation test was conducted for ten odorous frog species, and 12 loci were amplifi able in most species. With the high cross-species amplifi cation rates, these markers will provide useful molecular tools for conservation genetic and phylogeographic studies on the genus Odorrana and Bamburana.
Microsatellite DNA loci, Transcriptome, Odorrana margaretae, Cross-species amplifi cation test
The green odorous frog Odorrana margaretae is widely distributed in mountain regions of southwestern China. It is a dominant amphibian species in the forest-stream ecosystem, and plays an important role in the function and stability of this ecosystem (Frost, 2014). In addition,the species, as well as most other odorous frogs, are reservoirs for antimicrobial peptides (AMPs) and have recently attracted much attention for the property (Yang et al., 2012).
Microsatellite DNA markers are useful tools for understanding population genetic structure and mating system, as well as for parentage analysis and genetic resource assessment due to their high polymorphism,relatively small size, and well-established analysis protocols (Ellegren, 2004; Jones et al., 2010; Mittal and Dubey, 2009). A major obstacle in developing microsatellite DNA markers, however, is its tedious isolating processes (Guichoux et al., 2011). Recently, with the advances of next generation sequencing technologies,transcriptome sequence data offer an alternative fast,inexpensive, and accurate way to identify microsatellite DNA loci in non-model organisms (Guichoux et al.,2011). We recently reported a transcriptome profile of O. margaretae (Qiao et al., 2013), and subsequently we designed and tested potential useful microsatellite DNA markers for this species based on its trancriptome data. In this paper, we report 17 polymorphic microsatellite DNA loci for O. margaretae and their cross-species amplifi ability for ten closely-related species.
Qiao et al. (2013) identifi ed 2,574 microsatellite DNA loci from the O. margaretae transcriptome sequence data and designed 11,695 pairs of primers using the QDD2 pipeline (Meglecz et al., 2010). In this study, we selected 60 pairs of primers based on their parameter values to test for amplifi ability and polymorphism. A total of 35 pairs of primers were successfully amplifi ed for O. margaretae. PCR amplifications were performed in 15 μl reaction volume containing 0.6 μl of genomic DNA, 0.6 μl of each primer, 5.7 μl ddH2O, and 7.5 μl 2 × EasyTaq SuperMix(TransGen Biotech). The amplification conditions were as following: an initial denaturation at 94°C for 5 min;30 cycles of denaturation at 94°C for 30 s, annealing at temperature Tafor 30 s (Table 1), and extension at 72°C for 30 s; and a fi nal extension at 72°C for 10 min.PCR products were visualized on 8% nondenaturing polyacrylamide gels with silver staining, and their polymorphism was assessed. Ultimately, 17 loci were readily amplifiable and polymorphic, and we proceeded to characterize these loci.
A total of 24 individuals of O. margaretae were collected from Emei Mountain, China (E 103°38918', N 29°56418', 749 m) in June 2012, and used to characterize the 17 microsatellite DNA loci. The forward primers were labeled with a fl uorescent dye (FAM, HEX or TAMRA). The sizes of the PCR products were determined on an ABI3730xl DNA Analyzer (Applied Biosystems)and analyzed using GeneMarker®HID v1.95 analysis software. Each locus was amplified separately and loci with different fluorescent dyes were pooled for size determination. Hardy-Weinberg equilibrium (HWE),linkage disequilibrium (LD), and population genetic parameters were analyzed using Genepop on the web version 4.2 (Raymond and Rousset, 1995). Polymorphism information content (PIC) was calculated by Cervus 3.0.7 (Kalinowski et al., 2007). A sequential Bonferroni correction was applied for multiple tests (Rice, 1989). The null alleles and possible scoring errors were assessed by Micro-Checker 2.2.3 (Van Oosterhout et al., 2004).
As shown in Table 1, the numbers of alleles (NA)and PIC for each locus varied from 2 to 8 (average 4.1)and from 0.040 to 0.763, respectively. The observed heterozygosities (Ho) ranged from 0.04 to 1, and the expected heterozygosities (He) ranged from 0.04 to 0.81. Six loci (Om10, Om14, Om21, Om25, Om52, Om57)showed significantly deviation from HWE (P < 0.01)after the Bonferroni correction, and the micro-checker analysis showed no evidence for scoring error or technical or statistical artifacts. Signifi cant deviation from LD (P <0.01) was not observed. The high number of loci deviated from HWE is likely a consequence of the Wahlund effect,because these 24 individuals were collected from multiple streams in close vicinity, which may not represent a true random mating population.
We tested ten closely-related species for crossspecies amplifiability, including Bamburana nasuta,O. kuangwuensis, O. wuchuanensis, O. graminea, O. schmackeri, O. hejiangensis, O. junlianensis, O.grahami,O. nanjiangensis, and O. lungshengensis. Two individuals of each species were tested. High success rates of crossamplification were obtained, and eight pairs of primers were able to amplify all ten species, and 4 pairs were able to amplify 8 or 9 species (Table 1).
The transcriptome data and next generation sequencing techniques prove to be a much better alternative than conventional methods for isolating microsatellite DNA loci. It is much less time consuming and relatively inexpensive. Furthermore, the primers tested in this study are amplifiable across a large number of species, which will provide useful molecular tools for conservation genetic and phylogeographic studies of odorous frog species.
Acknowledgements This project is supported by the National Natural Science Foundation of China (Grant No. 31172061). Thank Pro. J. FU, Dr. Y. QI and Dr. B. LU for their valuable comments.
Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet, 5(6): 435-445
Frost D. R. 2014. Amphibian Species of the World: an Online Reference. Version 6.0 (10/25/2014). Electronic Database accessible at http://research.amnh.org/herpetology/amphibia/ index.html. American Museum of Natural History, New York,USA
Guichoux E., Lagache L., Wagner S., Chaumeil P., Leger P., Lepais O., Lepoittevin C., Malausa T., Revardel E., Salin F., Petit R. J. 2011. Current trends in microsatellite genotyping. Mol Ecol Resour, 11(4): 591-611
Jones A. G., Small C. M., Paczolt K. A., Ratterman N. L. 2010. A practical guide to methods of parentage analysis. Mol Ecol Resour, 10(1): 6-30
Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol, 16(5):1099-1106
Meglecz E., Costedoat C., Dubut V., Gilles A., Malausa T., Pech N., Martin J. F. 2010. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics, 26(3): 403-404
Mittal N., Dubey A. 2009. Microsatellite markers-A new practice of DNA based markers in molecular genetics. Pharmacognosy Reviews, 3(6): 235-246
Qiao L., Yang W., Fu J., Song Z. 2013. Transcriptome profi le of the green odorous frog (Odorrana margaretae). PLoS ONE,8(9): e75211
Raymond M., Rousset F. 1995. GENEPOP (Version 1.2):Population Genetics Software for Exact Tests and Ecumenicism. Journal of Heredity, 86(3): 248-249
Rice W. R. 1989. Analyzing tables of statistical tests. Evolution,43(1): 223-225
Van Oosterhout C., Hutchinson W. F., Wills D. P. M., Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4(3): 535-538
Yang X., Lee W. H., Zhang Y. 2012. Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J Proteome Res, 11(1): 306-319
Dr. Liang Qiao, from the Department of Herpetology, Chengdu Institute of Biology, Chinese Academy of Sciences, with his research focusing on phylogeography and ecology of amphibians.
E-mail: qiaoliang927@163.com
26 October 2014 Accepted: 19 February 2015
杂志排行
Asian Herpetological Research的其它文章
- Genetic Analysis of Multiple Paternity in an Endangered Ovoviviparous Lizard Shinisaurus crocodilurus
- Is Habitat Preference Associated with Locomotor Performance in Multiocellated Racerunners (Eremias multiocellata) from a Desert Steppe?
- Toxic Effects of Three Heavy Metallic Ions on Rana zhenhaiensis Tadpoles
- Herpetological Diversity of Timor-Leste: Updates and a Review of Species Distributions
