香蕉中4条乙醇脱氢酶基因的克隆及表达分析
2015-07-05贾彩红李建平刘菊华苗红霞张建斌金志强徐碧玉
贾彩红,王 卓,李建平,刘菊华,苗红霞,张建斌,金志强,徐碧玉*
(1 中国热带农业科学院热带生物技术研究所,海口571101;2 中国热带农业科学院海口试验站,海口570101)
Alcohol dehydrogenase(ADH)is one of the most commonly induced enzymes in plant cells.The enzyme activity is induced when the plants are exposed to stress,most importantly,flooding stress.ADH is induced by ethylene and ABA which are involved in the signal transduction pathway in the plant stress response.The main physiological function of ADH is to convert acetaldehyde into ethanol.Under hypoxia,the anaerobic glycolysis pathway produces ATP for plants.ADH plays an important role in embryonic development,seed germination,and plant growth.ADH plays an important role in the growth and development of fruit,as well as in the stress response.For example:in the grape[1]and cantaloupe charentais melon[2],ADH may participate in ethylene signal conduction and modulate the biosynthesis of aromatic substances.ADH also plays an important role in removing puckery in persimmon fruit[3].And in the barley,ADH activity was induced after leaves were infected with Parasitic fungi for 24hours[4].
ADH gene research was originated when two isozyme genes were first cloned from corn[5].The ADH gene has also been cloned from cantaloupe charentais melon(Cucumis melo var.cantalupensis)[2],strawberries (Fragaria spp.)[6],mango(Mangifera indica L.)[7], chrysanthemum(Chrysanthemum zawadskii)[8],soybean(Glycine max)[9],ginseng (Panax ginseng)[10],barley(Hordeum vulgare cv.himalaya)[11],and another plants.There have been many reports on the role of the ADH gene in flooding stress,in addition to chilling injury,wounding injury,pathogenic bacteria,and hormones.The ADH1gene of barley aleurone layer is regulated by the ABA/GA interactions[11].There are six ADH genes in soybean,ADH activity was rose after soybean seedling were waterlog for 2days,and the ADH2gene is a specific gene to the flooding stress response[9].The three ADH genes play different roles in the hormone response in mangoes,suggesting that they have different biochemical reactions[7].The ADH gene can be used as a marker of response to chilling stress in the diploid strawberry[6].In the process of grape fruit ripening,the ADH gene can also be used as a marker of mature fruit[12].ADH activity changes after barley leaves are infected with germs[4].Ginseng has different hormones and a different response to abiotic stress,thus,ethanol dehydrogenase is expressed differentially.ADH plays a role in the growth,development and adaption to stress in ginseng[10].
Our primary interest is in the banana plant(Musa acuminata L.AAA group,cv.cavendish),one of the most important fruit crops in the world in production and consumption[13].During the growth period,banana plantlets are often subjected to various abiotic and biotic stresses,such as drought[14],low temperature[15],high salinity[16],flooding[17]and Panama disease[18].Because ADH plays an important role in plant resistance,growth and development,so we describe the molecular cloning and expression of four different ADH gene members (MaADH1,MaADH2,MaADH3 and MaADH4)from banana plantlets.These results provide theoretical basis for cultivating high quality and resistance of banana varieties.
1 Material and Methods
1.1 Plant materials and treatments
One month old banana(Musa acuminata L.AAA group,cv.cavendish)plantlets were grown in Hoagland solution(0.51g/L KNO3,0.82g/L Ca(NO3)2,0.49g/L MgSO4·7H2O,0.136g/L KH2PO4,0.6 mg/L FeSO4,2.86 mg/L H3BO3,1.81mg/L MnCl2·4H2O,0.08 mg/L CuSO4·5H2O,0.22 mg/L ZnSO4·7H2O,0.09 mg/L H2MoO4·4H2O)(pH 6.0)[19]under greenhouse conditions.The maximum and minimum temperatures in the greenhouse during the experiment were 25℃and 30℃,respectively,while the relative humidity oscillated between 55%and 80%.To investigate the effect of abiotic stress and different signaling molecules on the expression of the four identified ADH genes in banana roots,ten plantlets were cultured with solutions containing 100mol/L abscisic acid (ABA),1% (v/v)ethephon,100 mol/L methyl jasmonate(MeJA),100 mol/L salicylic acid (SA),200 mol/L sodium chloride(NaCl),15% (w/v)PEG 6000and low temperature(plants were shifted to 8℃);control plants were mock-treated with Hoagland solution.The banana plantlet roots were collected 0,6,12,24h later.For infection with Foc TR4,the banana plantlet roots were dipped in a Foc TR4spore suspension of 1.5×106condia/mL in two hour.The entire root system was then harvested at 0,2,4,6d later post infection (DPI)[20].All above experiments were repeated three times.In order to determine expression at a tissue-specific level,samples were collected from roots,pseudostems,corms,leaves,flowers,and fruits of 8 month old plants.All collected materials were flash-frozen in liquid nitrogen and stored at-70℃until use.
1.2 Methods
1.2.1 Cloning of full-length MaADH cDNAsTotal RNA was separately extracted from the roots,pseudostems,corms,leaves,flowers,and fruits of the banana using a modified cetyltrimethylammonium bromide(CTAB)method[21].For cloning of the full-length cDNA,total RNA from the banana roots was isolated using a modified CTAB method[21].First-strand cDNA was synthesized using the SMARTTMPCR cDNA Synthesis Kit and SMARTScribe reverse transcriptase(Clontech,Pa-lo Alto,CA,USA)according to the manufacturer’s instructions.PCR amplification and PCR product sequencing were performed as previously described[22].
Full-length sequences of four ADHs,designated as MaADH1,MaADH2,MaADH3 and MaADH4,were amplified from the banana cDNA library using the 5′-RACE(rapid amplification of cDNA ends).The partial sequences of four ADH genes were obtained from a banana cDNA library by random sequencing[20].To obtain full-length cDNAs,four specific primers were designed based on the fragments and a nested PCR strategy applied for 5′RACE (Table 1).These reverse primers were used in both the cloning and expression analyses.
The obtained ADH sequences were analyzed by the BLASTn algorithm (http://www.ncbi.nlm.nih.gov/blastn),and the deduced amino acid sequences were compared with the banana DH-Pahang(AA group)genome database (http://banana-genome.cirad.fr/greenphyl).The percentages of similarity between MaADHs and ADH proteins from the DH-Pahang genome were calculated using DNAman 6.0 software (Lynnon Biosoft,QC,Canada).The phylogenetic tree was constructed using MEGA4[23].
1.2.2 Semiquantitative RT-PCR analysis of MaADHs expressionTotal RNA was extracted from the roots,pseudostems,corms,leaves,flowers,and fruit tissues(including peel and pulp)using a modified CTAB method.First strand cDNA was synthesized from 2μg poly(A)+RNA from each sample using AMV Reverse Transcriptase(Promega).The primers were designed to analyze the gene expression pattern(Table 1).The PCR conditions were 94℃for 4min,followed by 29cycles of amplification 94℃for 30s,55℃for 30s,and 72℃for 30s,and then 72℃for 10min.MaActin transcripts were amplified using forward (5′-CGAGGCTCAATCAAAGA-3′)and reverse (5′-ACCAGCAAGGTCCAAAC-3′)primers as an internal control.The experiments were repeated at least three times with similar results.
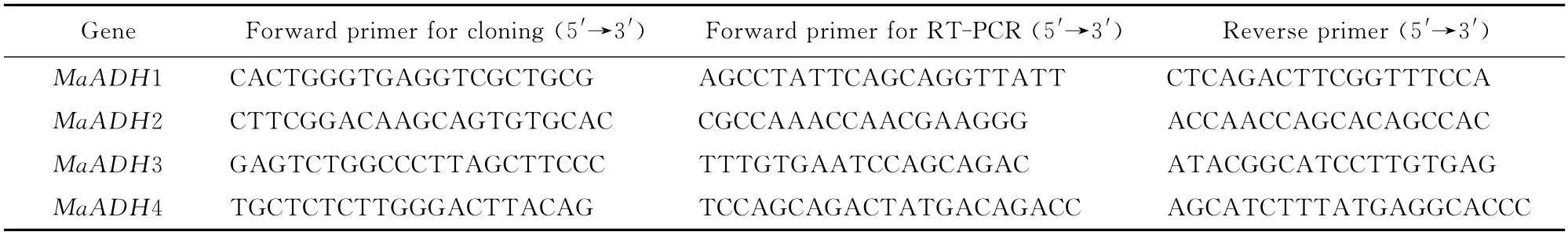
Table 1 PCR primers for cloning and expression analyses of MaADH1,MaADH2,MaADH3,and MaADH4
1.2.3 Real-time RT-PCR analysis of MaADHs expressionTotal RNA was isolated from the banana plantlet roots after treatment.The first strand cDNA was synthesized from 2μg poly(A)+RNA from each sample using AMV Reverse Transcriptase (Promega, Heidelberg, Germany).SYBR?Premix Ex TaqTM(TaKaRa)was used in 25μL reactions with 0.5μL ROX reference dye.Primers(100nmol/L each)were mixed with the equivalent of 100ng reverse-transcribed RNA template per reaction.In all experiments,negative controls containing no template RNA were subjected to the same procedure to exclude or detect any possible contamination.Before proceeding with the actual experiments,a series of template dilutions were performed to determine the optimal template concentration necessary to obtain maximal amplification of the target.
Each quantitative real-time PCR was performed on a Stratagene Mx3000P(Stratagene,CA,USA)machine using SYBR chemistry.The thermal cycling conditions were as follows:94℃for 3 min,followed by 40cycles of 94℃for 7s,55℃for 10s,and 72℃for 15s.Reactions were performed in triplicate,and data were analyzed using Mx-ProTM QPCR software(Stratagene,CA,USA).ACT1was one of the most suitable reference genes used to abiotic stress in banana[24],so MaActin was used as a control.The differences in Ct value between the MaADHs and MaActintranscripts were expressed as fold-changes relative to MaActin.
2 Results and analysis
2.1 Cloning and sequence analysis of ADHs from banana
Four different ADH genes were identified from the banana EST database and their full-length cDNAs cloned by RACE.The cDNA and deduced amino acid sequences of the MaADHsdescribed in this study were deposited in the GenBank under the following accession numbers: MaADH1(KM253748),MaADH2 (KM253749),MaADH3(KM253750)and MaADH4 (KM253753),and MaADH2,MaADH3and MaADH4consisted of 380amino acids,while MaADH1consisted of 381 amino acids.The four MaADHs had the ADH_N domain structure,binding protein and zinc binding sites,and they were then annotated using BLASTp in NCBI and their sequences mapped with respect to the banana DH-Pahang(AA group)genome,which identified four highly similar proteins(Table 2).
2.2 Phylogenetic analysis of MaADHs
To investigate the evolutionary relationships of the MaADHs,aphylogenetic tree was generated from the deduced amino acid sequences of 21 ADHs from various plant species.This phylogenetic tree was analyzed by MEGA4using the Neighbor-Joining method with 1 000bootstrap replicates and the evolutionary distances were calculated by the Poisson correction method.The resulting tree showed that MaADHs could be classified into two different groups,MaADH2,MaADH3and MaADH4belong to the first class,MaADH1does not belong to the first and third classes(Fig.1).MaADH2is a close relative to ADH of persimmon.MaADH3and MaADH4are clustered together and their relation was close,MaADH1is not closely related to various plants.
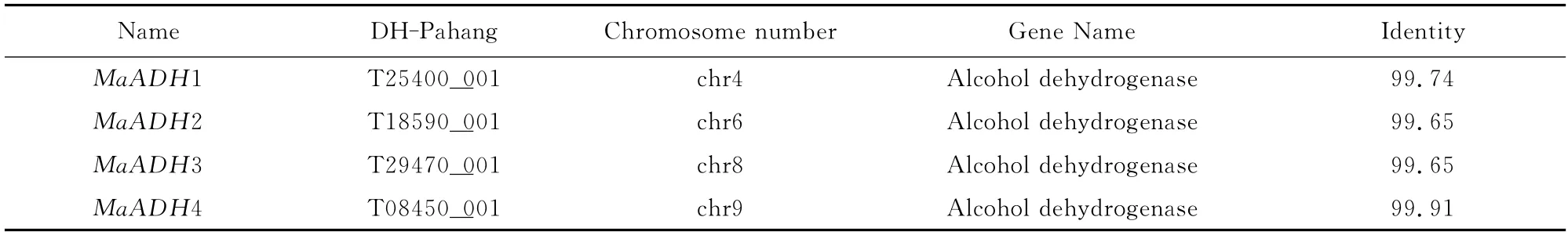
Table 2 Blastp result for four MaADHsin banana A genomics database(DH-Pahang)
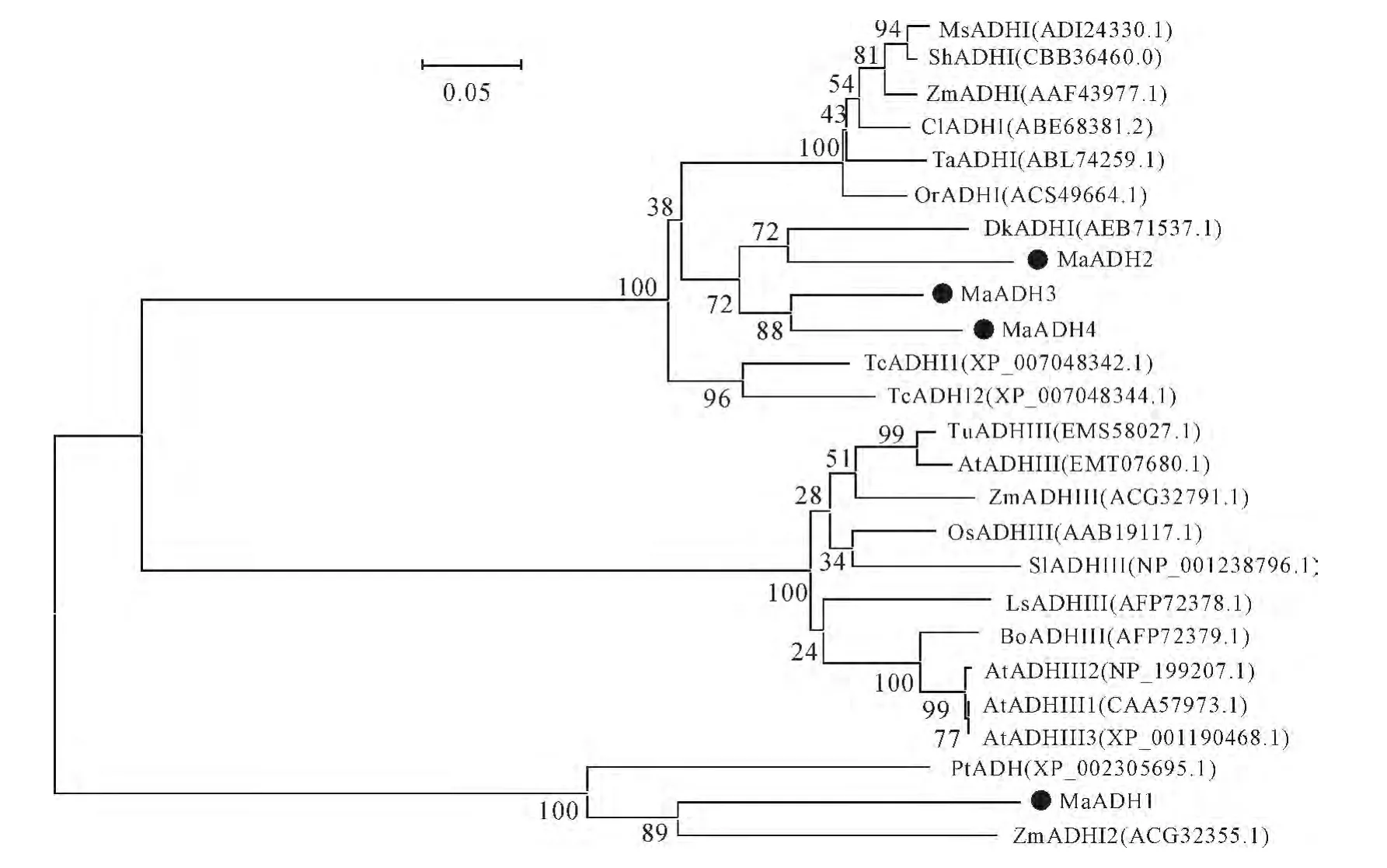
Fig.1 Phylogenetic tree of ADHs from banana and other plant species Values at nodes show the confidence level of bootstrap replication 1 000;the accession No.was given brackets;the circle stand for ADH of banana
2.3 Temporal and spatial transcript accumulation of MaADHs
To clarify the patterns of MaADHs expression in different organs of the banana plant,semi-quantitative RT-PCR was carried out on cDNA from different organs.MaADHs was expressed at different levels in all organs examined:roots,pseudostems,corms,leaves,flowers and fruit.The expression of MaADH1 was the highest in root,corms and pseudostems,and was the lower in the leaf,while in the flower and fruit,the expression was the weakest.MaADH2,MaADH3,and MaADH4 were expressed all organs(Fig.2).The results indicated that alcohol dehydrogenase may play a role in the process of plant growth and development.
2.4 Differential expression of MaADHs in hormone
The expression of alcohol dehydrogenase was analyzed by different hormone treatments.The results showed that MaADH2was strongly induced by ABA,ethylene,JA,and SA,the maximum relative expression is 19.14,428.19,68.21and 61.79 respectively.MaADH3 was induced by ABA,JA,and SA,the maximum relative expression is 16.91,

Fig.2 RT-PCR analysis of MaADH1,MaADH2,MaADH3and MaADH4expression in banana R.root;C.corm;P.pseudostem;L.leaf;F.flower;Fr.fruit 54.71and 52.31respectively.MaADH1and MaADH4 were induced by four hormones,but the expression quantity changed was little(Fig.3).
2.5 Differential expression of MaADHs in abiotic stress
The alcohol dehydrogenase gene expression was analyzed after banana seedlings were treated with different stressors.The MaADH2 was strongly induced by drought and flood,the maximum relative expression is 53.73and 108.43respectively.The MaADH3was strongly induced by NaCl and its maximum relative is 220.27.MaADH1 and MaADH4were induced by four abiotic stress,but the expression quantity changed was little(Fig.4).The MaADH2 may be a marker gene of drought and flood stress and the MaADH3may be a marker gene of salt stress.
2.6 Differential expression of MaADHs in biotic stress
Alcohol dehydrogenase gene expression was analyzed after banana seedlings were infected with Fusarium oxysporum f.specialis(f.sp)cubense Tropical Race 4(Foc TR4,Fig.5).Expression of the four alcohol dehydrogenase genes did not change very much compared to control.

Fig.3 Expression of MaADHs with different treatments
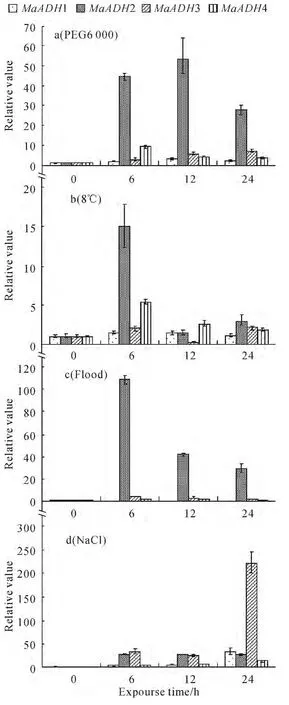
Fig.4 Expression of MaADHs with different treatments
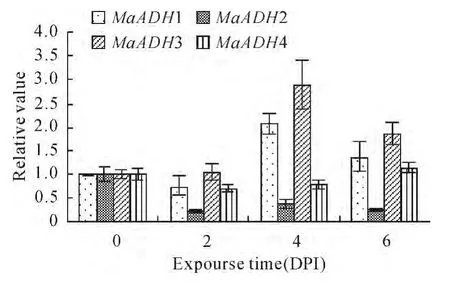
Fig.5 Expression of MaADHs with Foc TR4at day later post infection(DPI)
3 Discussion
The typical features of alcohol dehydrogenase include ADH_N domain structure and NAD binding protein and zinc binding sites[25];the three structure domains are quite conserved in the species.In this study,four ADH genes were cloned from the transcriptome of banana.They were characteried as alcohol dehydrogenase and were named MaADH.Phylogenetic analysis revealed that MaADHs belong to the different category of alcohol dehydrogenase.
Our data indicated that,in 8-month-old plants,the expression of MaADH1,MaADH2,MaADH3 and MaADH4is in all tissues,suggesting that alcohol dehydrogenase exists in various organs of plants and is involved in plant growth and development.MaADH1expression was different compared to the other three alcohol dehydrogenases in the organs of the banana.The MaADH1was alone in a branch in clustering which likely explains why the MaADH1 may have a different effect on the process of growth and development of the banana.
The expression of the four alcohol dehydrogenase genes was analyzed after ABA,ethephon,SA and JA treatment.The results showed that the MaADH2was strongly induced by ABA,ethylene,JA and SA,MaADH3 was strongly induced by ABA,JA,and SA.ADH1and ADH2were induced by ethylene in the mango[7],which is similar to our finding here.We found that a part of the alcohol dehydrogenase gene was induced by ethylene in banana plants,the results demonstrate that different alcohol dehydrogenases play different roles.ADH expression is induced by JA and SA in ginseng[10],which is similar to our results reported here.A part of the ADH was induced by JA and SA in the banana.The expression of ADHis blocked by JA in both ginseng and bananas;however,there are different expression patterns of ADH.In conclusion,alcohol dehydrogenase may be played a role in the growth and development of bananas.
MaADHs were analyzed after banana seedlings were exposed to different abiotic stressors.The MaADH2is sensitive to flood and drought;expression reached levels 110times greater after flood and 60times greater after drought,suggesting that the gene could be a marker gene of flood and drought.The MaADH3 was strongly induced by NaCl and its maximum relative is 220.27,suggesting that the gene could be a marker gene of salt stress.There are six ADHs in soybean,and the ADH2is sensitive to flood and is the specific gene activated in flood stress.The ADH2was strong induced by flood,while it was not induced by penetration,cold or drought[9].The ADH activity of chrysanthemum increased after chrysanthemum was exposed to flood[8].The expression of MaADH2 increased significantly after banana seedlings were exposed to flood.These results found with soybeans and chrysanthemums are similar to our findings of the banana.ADH was induced by flood,which could be a marker gene of flood stress;however,the ADH2gene of soybean was not induced by drought,while the ADH of the banana was induce by drought.
In this study,MaADH1 was expressed at highest levels in the roots under normal condition of development,while the expression level of MaADH1 was lower than other MaADHs after ABA,ethephon,SA,JA and flood and drought treatments,the possible reason is that the organ specificity experiment material is plant with fruit and flower,while the material of stress and hormone treatment is five leaves plant,which may be associated with in different growth periods.
Parasitic fungi susceptibility of the ADH was studied in barley.ADH activity was induced after leaves were infected for 24hours[4].The expression of the four MaADHs was not obviously changed after the banana seedlings had fusarium wilt.ADH activity was induced after 24hours of treatment in barley,while in the banana treatment time was 2,4,and 6days.The results showed that the ADH enzyme may not participate in the process of banana fusarium wilt.It may be the reaction mechanism of different plants caused by different pathogens or may be caused by a different treated time.
Our results indicate that the MaADHs may be played different roles in banana plant growth and development,abiotic stress,biotic stress and hormone.In this study,the MaADH2was found to be apotential marker gene of flood and drought stress in the banana;the MaADH3was found to be a potential marker gene of salt stress in the banana.
[1] TESNIERE C,PRADAL M,El-KEREAMY A,et al.Involvement of ethylene signalling in a non-climacteric fruit:new elements regarding the regulation of ADH expression in grapevine[J].Journal of Experimental Botany,2004,55(406):2 235-2 240.
[2] MANRíQUEZ D,El-SHARKAWY I,FLORES F B,et al.Two highly divergent alcohol dehydrogenases of melon exhibit fruit ripeningspecific expression and distinct biochemical characteristics[J].Plant Molecular Biology,2006,61(4-5):675-685.
[3] MIN T,YIN X,SHI Y,et al.Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon(Diospyros kaki)fruit de-astringency[J].Journal of Experimental Botany,2012,63(18):6 393-6 405.
[4] PATHURI I P,REITBERGER I E,HüCKELHOVEN R,et al.Alcohol dehydrogenase 1of barley modulates susceptibility to the parasitic fungus Blumeria graminis f.sp.hordei[J].Journal of Experimental Botany,2011:err017.
[5] GERLACH W L,PRYOR A J,DENNIS E S,et al.cDNA cloning and induction of the alcohol dehydrogenase gene(Adh1)of maize[J].Proceedings of the National Academy of Sciences,1982,79(9):2 981-2 985.
[6] DAVIK J,KOEHLER G,FROM B,et al.Dehydrin,alcohol dehydrogenase,and central metabolite levels are associated with cold tolerance in diploid strawberry(Fragariaspp.)[J].Planta,2013,237(1):265-277.
[7] SINGH R K,SANE V A,MISRA A,et al.Differential expression of the mango alcohol dehydrogenase gene family during ripening[J].Phytochemistry,2010,71(13):1 485-1 494.
[8] YIN D,NI D,SONG L,et al.Isolation of an alcohol dehydrogenase cDNA from and characterization of its expression in chrysanthemum under waterlogging[J].Plant Science,2013,212:48-54.
[9] KOMATSU S,THIBAUT D,HIRAGA S,et al.Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots[J].Plant Molecular Biology,2011,77(3):309-322.
[10] KIM Y J,SHIM J S,LEE J H,et al.Isolation and characterization of a novel short-chain alcohol dehydrogenase gene fromPanax ginseng[J].BMB Rep,2009,42(10):673-678.
[11] MACNICOL P K,JACOBSEN J V.Regulation of alcohol dehydrogenase gene expression in barley aleurone by gibberellin and abscisic acid[J].Physiologia Plantarum,2001,111(4):533-539.
[12] LABAT E,CALAS M,SAMI-MANCHADO P,et al.Grapevine(Vitis vinifera L.)alcohol dehydrogenase gene:An important marker of the physiological state during fruit ripening[C]//VInternational Symposium on Grapevine Physiology,1997,526:303-310.
[13] AURORE G,PARFAIT B,FAHRASMANE L.Bananas,raw materials for making processed food products[J].Trends in Food Science&Technology,2009,20(2):78-91.
[14] VAN ASTEN P J A,FERMONT A M,TAULYA G.Drought is a major yield loss factor for rainfed East African highland banana[J].Agricultural Water Management,2011,98(4):541-552.
[15] KANG G,WANG Z,XIA K,et al.Protection of ultrastructure in chilling-stressed banana leaves by salicylic acid[J].Journal of Zhejiang University Science B,2007,8(4):277-282.
[16] DENTON G R W,BURDON-JONES C.The influence of temperature and salinity upon the acute toxicity of heavy metals to the banana prawn(Penaeus merguiensis de Man)[J].Chemistry in Ecology,1982,1(2):131-143.
[17] CHAI T T,FADZILLAH N M,KUSNAN M,et al.Water stress-induced oxidative damage and antioxidant responses in micropropagated banana plantlets[J].Biologia Plantarum,2005,49(1):153-156.
[18] O’DONNELL K,KISTLER H C,CIGELNIK E,et al.Multiple evolutionary origins of the fungus causing Panama disease of banana:concordant evidence from nuclear and mitochondrial gene genealogies[J].Proceedings of the National Academy of Sciences,1998,95(5):2 044-2 049.
[19] ARNON D I,HOAGLAND D R.A comparison of water culture and soil as media for crop production[J].Science,1939,89(2 318):512-514.
[20] WANG Z,ZHANG J B,JIA C H,et al.De Novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporumf.sp.Cubense tropical race 4infection[J].BMC Genomics,2012,13(1):650.
[21] ASIF M H,DHAWAN P,NATH P.A simple procedure for the isolation of high quality RNA from ripening banana fruit[J].Plant Molecular Biology Reporter,2000,18(2):109-115.
[22] LI M Y,XU B Y,LIU J H,et al.Identification and expression analysis of four 14-3-3genes during fruit ripening in banana(Musa acuminata L.AAA group,cv.Brazilian)[J].Plant Cell Reports,2012,31(2):369-378.
[23] TAMURA K,DUDLEY J,NEI M,et al.MEGA4:molecular evolutionary genetics analysis(MEGA)software version 4.0[J].Molecular Biology and Evolution,2007,24(8):1 596-1 599.
[24] CHEN L,ZHONG H,KUANG J,et al.Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions[J].Planta,2011,234(2):377-390.
[25] GOTTLIEB L D.Conservation and duplication of isozymes in plants[J].Science,1982,216(4 544):373-380.
