采用QSPR模型预测手性二芳基甲烷衍生物保留因子与分离因子
2015-01-04胡桂香骆成才潘善飞蒋勇军邹建卫浙江大学宁波理工学院生物与化学工程学院浙江宁波3500浙江大学化学系杭州3008
胡桂香 骆成才 潘善飞 蒋勇军 邹建卫(浙江大学宁波理工学院生物与化学工程学院,浙江宁波3500;浙江大学化学系,杭州3008)
采用QSPR模型预测手性二芳基甲烷衍生物保留因子与分离因子
胡桂香1,*骆成才1潘善飞2蒋勇军1邹建卫1
(1浙江大学宁波理工学院生物与化学工程学院,浙江宁波315100;2浙江大学化学系,杭州310028)
对手性化合物的保留因子和分离因子进行定量结构-特征关系(QSPR)研究,对于预测保留因子和分离因子甚至对映体的洗脱顺序都起着重要作用.本文选择手性二芳基甲烷衍生物为研究对象,采用VolSurf程序计算分子结构参数,并分别在其与保留因子以及分离因子间建立模型,采用测试集外部检验、留多法交叉验证和Y随机性检验等方法对分离因子模型的鲁棒性进行了评估,结果令人满意.对变量进行分析显示,分子的球形性,中等能级的亲水区、亲水-亲脂平衡、两亲矩、合适的氢键给体和受体均有利于异构体在手性固定相上的保留;一对对映体的高能级的亲水区、低能级的疏水区、两亲矩、合适的氢键给体和受体以及阴离子区之间大的差异对对映体在手性固定相上的分离是有利的.利用这些模型,可以轻松地预测对映体的保留因子和分离因子,甚至洗脱顺序.
手性;分子建模;VolSurf;二芳基甲烷;偏最小二乘
©Editorial office ofActa Physico-Chimica Sinica
1 Introduction
The derivates possessing diarylmethane moiety often exhibit various biological activities.For example,clemastine((+)-(R,R)-1),1neobenodine((+)-(R)-2),2and chlorpheniramine((+)-(S)-3)3(Fig.1),were used as anticholinergic drugs to treat anaphylactias, such as rhinitis and hives,or used as local anaesthetic and laxative agents.4In recent years,it has been found that this type of compounds shows inhibitory activity against oestrogen synthetase and human immunodeficiency virus(HIV)-1 recombinant reverse transcriptase5,6as well as antitubercular activity7.When two aryl groups differ from each other,the molecule will present chirality and every enantiomer may display different biological activity,e. g.,the antimuscarinic potency of procyclidine((-)-R-4,Fig.1)was about 380 times greater than that of the corresponding(+)-(S) enantiomer.8As a consequence,it is of great significance to study the separation of the chiral diarylmethane derivates,and this has attracted extensive attention in the past decade.
Asymmetric synthesis was usually chosen to prepare diarylmethane derivates catalyzed by lithiation,9,10hemilabile phosphorus ligand11or triphenylborane12.However,it is very difficult to obtain a diarylmethane derivate with 100%enantiomeric purity13through simple separation technique,because two aryl groups in molecule are often close to each other in steric bulkiness.High performance liquid chromatogram(HPLC)using the chiral stationary phase(CSP)is a widely-used technique to separate a chiral compound,14and the cotton effect of the enantiomer can be determined with circular dichroism(CD)spectrum. Nevertheless,the absolute configuration of the isomer cannot be obtained directly.15
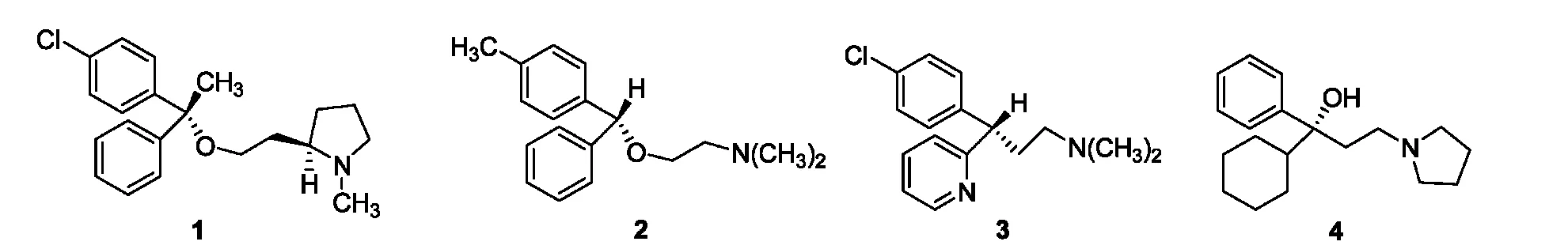
Fig.1 Structures of chiral compounds
Quantitative structure-activity/property relationship(QSAR/ QSPR)is a kind of effective means,by which chemical structure is quantitatively correlated with a well defined activity/property. Since 1990's,it has been applied widely to predict the chromatographic retention factors,16-19separation factors,20-23and chiral resolution ability24,while the chirality and the absolute configurations of the enantiomers were not considered in these studies.As a result,the main reason for the separation of the enantiomers could not be elucidated clearly.Chiral descriptors or codes were presented to build the QSAR25-27or QSPR27-30models.However, these descriptors and codes were not broadly used because of the complicated computational methods.In some studies,31,32however, it has been disclosed that there exists significant differences for some structural descriptors between a pair of enantiomers. Whether can these structural differences be used to build the model to predict the separation of enantiomers as well as judge the absolute configurations and elution order of the enantiomers?For this purpose,we report the QSPR models of chiral diarylmethane derivates.Three-dimensional(3D)structural descriptors derived from VolSurf program were adopted to predict the retention factors and separation factors for 63 samples.
2 Computational methods
2.1 Experimental data
There is one chiral centre in the structure of each diarylmethane derivate.The retention and separation factors of 63 samples as well as the absolute configuration were taken from previous publication by Job et al.33HPLC analyses were performed using brush-type(S,S)-Whelk-O1 CSPat a flow rate of 2.0 mL·min-1at 0°C.33The mobile phase was isopropyl alcohol:hexane(volume ratio,1:99).The data are listed in Table 1.The common logarithm value of retention factors(lgk2,lgk1)and separation factors(lga) were used to build the model.
2.2 VolSurf
VolSurf program is developed for modeling and predicting the physicochemical and pharmacodynamic properties of a compound.34-36It reads or computes 3D molecular interaction fields between molecule and probe and uses image processing methods to convert them into simple molecular descriptors that are easy to understand and interpret.These descriptors quantitatively characterize molecular size,shape,polarity,hydrophilicity,hydrophobicity,and the balance between them and so on.
2.3 Statistical analysis
Chemometrics tool,partial least squares(PLS)analysis,was used to compile the information obtained from VolSurf.Fractional factorial design(FFD)technique was used to remove irrelevant descriptors that did not have any relationship with the property, and,in this way,the predictive ability of the PLS model could be improved.The quality(i.e.,the explanatory power,r2)and the predictive ability(i.e.,q2)of the PLS model were evaluated by leave-one-out cross-validation.
2.4 General procedures
The molecular structures were built in standard geometry and then minimized by means of TRIPOS molecular mechanics force field with Sybyl 6.8.The dielectric constant was set as 2.06 to mimic the mobile phase.All chemicals were modeled in their neutral form.The molecular descriptors were derived by using VolSurf(version 3.07A)/GRID.In our study,3D-maps for water probe OH2,the hydrophobic probe DRY,carbonyl oxygen atom O probe,amide NH group N1 probe,and sp3cationic NH3group N3+probe were used at eight energy levels.And these energy levels could simulate the interaction energy and the distance be-
tween the isomer and CSP.In the meantime,the higher the energy level,the shorter the distance.As a result,102 chemical descriptors were generated.And the value of descriptors of enantiomers for 63 compounds is listed in Table S1 and Table S2(in Supporting Information).These molecular descriptors have clear chemical meanings.Every descriptor is composed of three segments.The first segment is an abbreviation representing the physicochemical property,and the following number(1-8)describes the energy level,while the last segment is probe type.The detailed representation of VolSurf descriptors has been presented by Crucianiet al.37

Table 1 Structures,retention factors,separation factors,and the absolute configuration of MRE

continued Table 1
Two PLS analyses were carried out on VolSurf descriptors in terms of a pair of enantiomers.The whole data set includes 63 compounds and the training set consists of 50 compounds.The training set was selected with the most descriptive compounds (MDC)approach of Hudson et al.38This method privileged a selection scheme that weights the compounds according to their population density.The remaining compounds were used as an external test set to assess the predictive ability of the model.Ultimately,different approaches,including external validation through test set,leave-many-out cross-validation and Y-randomization test,were fully utilized to assess the predictive ability and robustness of the model of separation factors.
3 Results and discussion
3.1 QSPR model for retention factors of the more retained enantiomer
For the whole data set of the more retained enantiomer(MRE), the number of the variables was reduced significantly to 85 after the FFD technique was adopted.With 6 principal components (PCs)used,the model generates an r2value of 0.89 and a q2value of 0.77,and the standard error of calculation(SDEC)is 0.10 and the standard error of prediction(SDEP)is 0.15.
To further test the predictive ability of VolSurf model of MRE, 63 compounds were divided into two sets.By adopting the MDC method,50 compounds were selected as the training set and the remaining compounds as the test set.After using the FFD technique,95 variables were finally introduced into the model for the training set.With 7 PCs,the model produces an r2of 0.91 and a q2of 0.66.And the SDEC is 0.10 and the SDEP is 0.19.When the model is used to predict the retention factors of the compounds in the test set,it yields a predictive r2of 0.67 and SDEPvalue of 0.16 with 7 PCs.This value is very close to that of the training set, which implies,to some degree,that the model possesses good predictive capability.The experimental,calculated and predicted value of lgk2is listed in Table 1,and the relationship between each other is displayed in Fig.2.
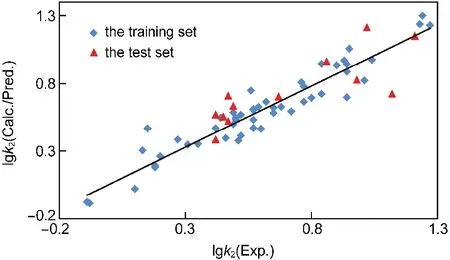
Fig.2 Relationship between the experimental and calculated/ predicted values of lgk2
Fig.3 shows the coefficients of 95 variables,and every barrepresents the contribution of a variable to the retention factors. The longer bar means the stronger correlation between the variable and the retention factors.Variables 1-33 are generated with OH2 probe and these variables are successively V,S,R,G,W1-8OH2,Iw1-8OH2,Cw1-7OH2,Emin1-3OH2,D12OH2, D13OH2,and D23OH2.And variables 34-55,namely D1-8DRY,ID1-8DRY,Emin1-3DRY,D12DRY,D13DRY,and D23DRY,are generated with DRY probe.While variables 56-59, HL1,HL2,A,CP,respectively,belong to the mixed descriptors of OH2 and DRY probe.Variables 60-73,W1-6O,and HB1-8O, which are generated with O probe,variables 74-89,W1-8N1 and HB1−8N1,are with N1 probe,and variables 90-93(W1-4N3+) are with N3+probe.While variables 94-95(POL and MW)are quantities regardless of probes.

Fig.3 PLS coefficient plot of variables for MRE
Among the variables derived from OH2 probe,G is defined as S/Sequiv,where S is the surface area and Sequivis the surface area of a sphere of volume(V).Accordingly,it measures the molecular globularity and its high value indicates the low globularity and high ellipsoid.This descriptor is positively correlated with the retention factors,which means that high ellipsoid is beneficial to the retention of the enantiomer on the CSP.In terms of W type descriptors,they represent the hydrophilic regions.W2-4OH2 variables are positively correlated with the retention factors,while W6-8OH2 negatively,which illustrates that the hydrophilic regions at low energy levels are favourable to the retention.Iw1-3OH2 and Cw5-7OH2 show negative correlation with the retention factors.Like dipole moments,descriptors of Iw type express the imbalance between the centre of mass of a molecule and the barycentre of its hydrophilic regions.Negative correlation of Iw1-3OH2 implies that high imbalance is detrimental to the retention of the enantiomer on the CSP at 1-3 energy levels.Descriptors of Cw,capacity factor,represent the ratio of the hydrophilic surface over the total molecular surface,i.e.,the hydrophilic surface of a surface unit.Negative correlation of Cw5-7OH2 demonstrates that more hydrophilic surface is disadvantageous to the retention at high energy levels.This is consistent with the result of descriptors W6-8OH2.Emin1OH2 represents the best local interaction energy minimum between the water probe and the target molecule.The result shows that high Emin1OH2 value is beneficial to the retention of the enantiomer. D12OH2 is the distance between the first and second local energy minima.Strong negative correlation of this descriptor with retention factors manifests that long distance between the first and second local energy minima is disadvantageous to the retention.
Among the quantities derived from DRY probe,D type descriptors represent the hydrophobic regions whereas ID type descriptors measure the imbalance between the centre of mass of a molecule and the barycentre of the hydrophobic regions.D1-3DRYand ID4-8DRYare negatively correlated with the retention factors,which means that more hydrophobic regions at low energy levels and high imbalance between the centre of mass and the barycentre of the hydrophobic regions at high energy levels are unfavourable to the retention.Besides,descriptor D12DRY also shows strongly negative correlation,which indicates that long distance between the first and second local energy minima for hydrophobic probe is disadvantageous to the retention.D6DRY and ID2DRY are found to be the only two descriptors that are positively correlated with the retention factors.It suggests that more hydrophobic regions at suitable high energy level and high imbalance between the centre of mass and the barycentre of the hydrophobic regions at suitable low energy level are helpful to the retention.
HL1,HL2,A,and CP are mixed descriptors of OH2 and DRY probes.Among them,descriptors of HL refer to the hydrophiliclipophilic balance,which are the ratio between the hydrophilic regions and the hydrophobic regions.The balance describes which effect dominates in the molecule.Positive correlation of HL1 with the retention factors implies that the more hydrophilic regions will be helpful to the retention.Amphiphilic moment(A)is defined as a vector pointing from the centre of the hydrophobic domain to the centre of the hydrophilic domain.Strong positive correlation of this descriptor with the retention factors is found,which signifies that the long vector is conducive to the retention of the enantiomer.
O is hydrogen bond acceptor probe.Descriptors of W show the hydrogen bond donor regions while HB descriptors describe the hydrogen bond acceptor regions.W1-6O variables are positively correlated with the retention factors,which indicates that hydrogen bond donors are beneficial to the retention.HB1O and HB5-8O show negative correlation,which implies that more hydrogen bond acceptors are detrimental to the retention.
Amide NH group N1 probe is a hydrogen bond donor probe. Differing from O probe,here descriptors of W show the hydrogen bond acceptor regions and HB variables describe the hydrogen bond donor regions.W1-4N1 descriptors show weak positive correlation while W6-8N1 for strong negative correlation with the retention factors,which demonstrates that the hydrogen bond acceptor atoms in the molecule have the weak interaction with the CSP,and more hydrogen bond acceptor atoms at high energy levels will be unfavourable to the retention.The descriptors with strong positive correlation are HB2N1 and HB6-7N1.This hints that the suitable hydrogen bond donors and interaction between the enantiomer and the CSP are beneficial to the retention,which is in accord with the results obtained with O probe.
N3+is sp3cationic NH3probe and W descriptors show the anionic regions of a molecule.W1N3+is positively correlated andW2N3+negatively correlated with the retention factors,which demonstrates that the ionic interaction between the enantiomer and the CSP is useful to the retention.
POL and MW are molecular descriptors which are not derived from 3D molecular fields.POL is an estimate of the average molecular polarizability,which is calculated according to the additive method of Miller.39This method is based on the structure of the molecules and is therefore independent of the number and type of probes used.Weak negative correlation with retention factors is found for this descriptor,which indicates that high value is disadvantageous to the retention.MW,the molecular weight,is positively correlated with the retention factors,i.e.,the larger MW of a molecule,the more retained on the CSP.
3.2 QSPR model for retention factors of the less retained enantiomer
Similarly,QSPR models for retention factors of the less retained enantiomer(LRE)have been constructed.For 63 compounds,the number of the variables is 86 after adopting the FFD technique.With 8 PCs used,the model generates an r2value of 0.93 and a q2value of 0.80.The SDEC is 0.08 and the SDEP is 0.14.
Also,63 compounds were divided into two sets.Using the FFD technique,86 variables were finally introduced into the model for the training set,including 50 samples.With 7 PCs,the model produces an r2of 0.94 and a q2of 0.79.The SDEC is 0.09 and the SDEP is 0.16.When the model is used to predict the retention factors of the compounds in the test set,it yields a predictive r2of 0.88 and SDEP value of 0.10 with 7 PCs.This value is lower than the training set,so the model possesses satisfactory predictive capability.The experimental,calculated and predicted value of lgk1is listed in Table 1,and the relationship between them is displayed in Fig.4.
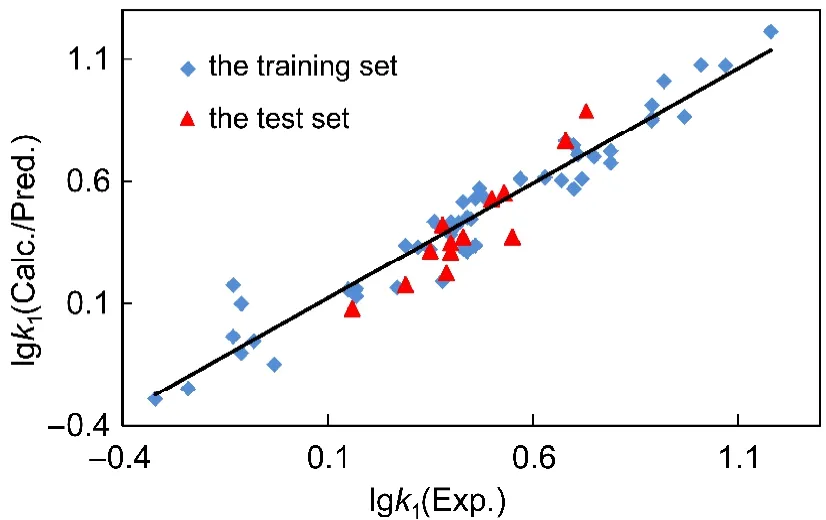
Fig.4 Relationship between the experimental and calculated/predicted values of lgk1
Fig.5 shows the coefficients of 86 variables.These variables are successively V,S,R,G,W1-7OH2,Iw1-8OH2,Cw1-7OH2, Emin1-3OH2,D12OH2,D13OH2,D1-8DRY,ID1-8DRY, Emin2DRY,D12DRY,D23DRY,HL1,HL2,A,CP,W1-6O,HB1-7O,W1-7N1,HB1-5N1,HB7N1,W1-4N3+,POL,and MW.

Fig.5 PLS coefficients plot of variables for LRE
Among the variables derived from OH2 probe,R is the ratio of volume/surface.It scales the molecular wrinkled surface,that is, rugosity,and the small ratio suggests the high rugosity.Strong negative correlation with the retention factors means that the low rugosity provides negative effects on the retention of the enantiomer on the CSP.Positive correlation of descriptor G signifies that high value is advantageous to the retention.W1-4OH2 variables are positive correlation with the retention factors while W5-7OH2 are negative,which is coincident with the result of the MRE.Iw5-8OH2 variables are distinctly positive correlation, which demonstrates that high imbalance is favourable to the retention at high energy levels.Emin3OH2 represents the third best local interaction energy minimum between the water probe and the target molecule.Strong positive correlation implies that high value is beneficial to the retention of the enantiomer.
For DRY probe,D1-5DRY descriptors are significantly negative correlation with the retention factors,which purports that more hydrophobic regions at low energy levels are unfavourable to the retention.Positive correlation of ID5-8DRY descriptors implies that high imbalance between the centre of mass and the barycentre of the hydrophobic regions at high energy levels is helpful to the retention.Negative correlation of D23DRYindicates that long distance between the second and third local energy minimum for hydrophobic probe is disadvantageous to the retention.
Positive correlation of HL1 and A demonstrates that the more hydrophilic regions than the hydrophobic regions and large vector pointing from the centre of the hydrophobic domain to the centre of the hydrophilic domain are beneficial to the retention.
W1-6O descriptors are positive correlation while HB1-7O negative with the retention factors,which signifies that hydrogen bond donors are favourable while hydrogen bond acceptors unfavourable to the retention.W1-3N1 variables are weak positive correlation with the retention factors,while W6-7N1 weak negative,which means that the hydrogen bond acceptor atoms have the weak interaction with the CSP and more hydrogen bond acceptor atoms at high energy levels are detrimental to the retention.Strong positive correlation of HB2N1 and HB7N1 represents the suitable hydrogen bond donors and interaction between the enantiomer and the CSP is beneficial to the retention.Strong negative correlation of W2-4N3+implies that the ionic interaction between the enantiomer and the CSP is disadvantageous to the retention.Positive correlation of POL and MW indicates that high POL value and mass are of benefit to the retention of the enantiomer on the CSP.
Comparing the results of MRE and LRE,it can be found that most of variables possess coincidence with each other.For instance,variable G,W2-4OH2,HL1,A,W1-6O,W1-3N1, HB2N1 and MW are all positively correlated with the retention factors,while variables W6-7OH2,Cw5OH2,D1-3DRY,HB5-7O,W6-7N1,and W2N3+are all negative correlation.However, when the chiral compound is separated with HPLC method,more attention should be paid to the separation factors,instead of the retention factors.Only if LRE is eluted faster than MRE,an effective separation can be obtained.Therefore,we are more concerned about the variables that are negative correlation with LRE and positive correlation with MRE,or weak positive correlation with LRE and strong positive correlation with MRE,or strong negative correlation with LRE and weak negative correlation with MRE.Comparing the results of MRE and LRE,it discloses that the variables matching the described conditions include W3-5OH2,Cw3-4OH2,Emin1OH2,D4-6DRY,ID2-3DRY, D23DRY,A,W2-5O,HB3-5N1,and W1-2N3+.This proves that the hydrophilic and hydrophobic variables with the median energy levels are beneficial to the separation factors.
3.3 QSPR model for the separation factors
The separation factors(a)is defined as the ratio of the retention factors of MRE and LRE,i.e.,a=k2/k1.Accordingly,lga=lgk2-lgk1.
In order to establish the QSPR model for directly predicting the separation factors,it is crucial to seek suitable descriptors that reflect the separation effect of the enantiomers.Instead of using the descriptors of one enantiomer,here the descriptors'difference value of a pair of enantiomers(the value of MRE minus the one of LRE)is used to construct the QSPR model of separation factors.Only 63 variables are used to build the model after some descriptors have been excluded due to the fact that these descriptors have the same or almost the same value.The difference value of 63 variables between the enantiomers is listed inTable S3.
The PLS method was used to generate the linear correlation. Among all samples,compounds27,40,and54were observed to have obvious deviation.The possible explanation is that it is caused by the structures themselves.Comparing compounds27and26,40and41,54and53,it can be seen that only one different substituent can lead to the change of the configuration of the MRE.The change of the substituent brings about the change of the interaction between the enantiomers and the CSP,where the steric interaction is dominating on the base of the compared compounds.After deleting these three outliers,a QSPR model with 7 PCs was obtained.The r2value of the model is 0.92,and the q2value is 0.70.The SDEC is 0.04 and the SDEP is 0.08.The experimental and calculated value of lga is listed in Table 1,and the linear fit of each other is shown in Fig.6(a),from which good consistency can be seen readily.In order to testify the predictive ability,the random 10 compounds(6,12,18,24,31,36,43,48, 51,and60)were chosen as the test set and other compounds (except three outliers)as the training set.The results demonstrate that the r2value of the training set is 0.94,and the q2value is 0.64. The SDEC is 0.03 and the SDEP is 0.09.The model is used to predict the separation factors of the compounds in the test set and it yields a predictive r2of 0.73 and SDEP value of 0.07 with 7 PCs.This states that the model possesses satisfactory predictive capability.The experimental,calculated and predicted value of lga is listed in Table 1,and the relationship between them is displayed in Fig.6(b).

Fig.6 (a)Relationship between the experimental and calculated values of lga;(b)relationship between the experimental and calculated/predicted values of lga
Ten leave-many-out cross-validations were carried out to check the robustness of the model for the separation factors.Fig.7 shows the result of the plot of the q2and the M number.From the figure, it can be seen that the q2is persistently stable and close to the q2of the leave-one-out cross-validation even at M=30,which confirms the robustness of the model.

Fig.7 Plot of the q2and M number for the leave-many-out cross-validation
Y-randomization tests were performed to testify the possible existence of chance correlation of the model.40,41In the tests,the dependent variable(lga)was randomly scrambled and used to build and investigate the PLS model.The result model with the randomized values should have dramatically lower r2and q2than the original one,because the relationship between the structure and the original dependent variable is broken.35 randomization runs were conducted in the present study.Fig.8 shows the results of the Y-randomization tests.From the figure,it can be seen that all randomized models have bad quality when compared with the original model.The r2value is about 0.2-0.5 and all q2values are negative,which means that the models have no predictive ability. The r2and q2value is within the limit recommended in the literature,40which implies that the original model is considered being free of chance correlation.
It is noteworthy that the calculated separation factors are 0.00 for compound44and minus values for compounds9and22(both are-0.03)among whole samples in Table 1.Except these three compounds,all others present positive predictive value,which signifies that the QSPR model predicts qualitatively the elution order very well.The correct rate of prediction on elution order is 95%.And the correct rate of prediction on elution order of the training set to the test set is 100%.

Fig.8 (a)r2and the correlation coefficient between lga and the randomized data;(b)q2and the correlation coefficient between lga and the randomized data
Fig.9 shows the coefficients of 63 descriptors in the QSPR model for the whole samples,which are successively the difference value of V,S,W1-7OH2,Iw8OH2,D12OH2,D13OH2, D23OH2,D1-7DRY,ID1-8DRY,Emin2-3DRY,D12DRY, D13DRY,D23DRY,A,W1-5O,HB1-7O,W1-8N1,HB1-6N1, and W1-3N3+between the enantiomers.
From Fig.9,it can be found that the difference value of V and D23OH2 are negatively correlated with lga,which means that the larger the differences of V and D23OH2,the more difficult the separation of the enantiomers.While the difference value of W5-7OH2 and D13OH2 is remarkably positively correlated with the separation factors,which represents that large differences of these variables are advantageous to the separation.The difference of D3DRY is positive correlation with the separation factors while D5-7DRY and Emin2-3DRY strong negative,which demonstrates that,at low energy level,large differences of hydrophobic regions are beneficial to the separation,while at high energy levels not.Positive correlation of the difference of descriptor A implies that the large difference of the amphiphilic moment between the enantiomers is advantageous to the separation.In addition,the large differences of W3O,HB5-6O,and W3N3+are advantageous to the separation,while the large differences of W5O and W8N1 disadvantageous.

Fig.9 PLS coefficient plot of variables for QSPR model of separation factors
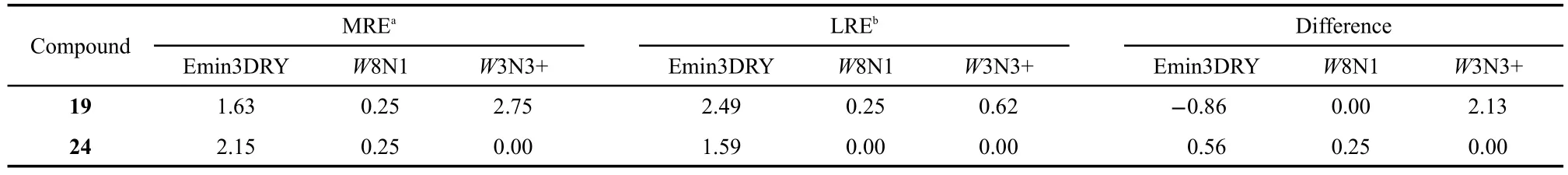
Table 2 Value of variables for MRE,LRE,and their difference of compounds 19 and 24
Among all compounds,compound19has the largest lga value (0.59)while compound24possesses the lowest value(0.01). From Fig.9,it can be seen that the differences of Emin3DRY, W8N1,and W3N3+are the three most strongly correlated descriptors with the separation factors.Table 2 shows the value of descriptor Emin3DRY,W8N1,and W3N3+for MRE,LRE of compounds19and24and their differences between the enan-tiomers.It can be seen from Table 2 that compound19has higher W3N3+and lower Emin3DRY and W8N1 difference value than compound24.W3N3+has a positive correlation with the separation factors.High value is beneficial to the separation,which coincides with the experimental data.Emin3DRY and W8N1 are correlated negatively with the separation.In Table 2,with the higher separation factors,molecule19has much lower Emin3DRY and W8N1 difference value than molecule24.So,it can be concluded that the correlation of the difference of variable is coincident with the separation effect.
4 Conclusions
QSPR models for chiral diarylmethane derivates have been constructed in the present study.For the retention factors,good results were obtained and the models established for the training set had satisfactory predictive capability for the test set.According to analysis of the descriptors introduced in the models,it can be concluded that the molecular globularity,hydrophilic regions at median energy levels,hydrophilic-lipophilic balance,amphiphilic moment,suitable hydrogen bond donors and acceptors are all beneficial factors for the retention of the enantiomer,and large value will result in the long retention time.However,the hydrophilic regions at high energy levels,the hydrophobic regions at low energy levels,more hydrogen bond acceptors at high energy levels and the anion regions are unfavourable factors for the retention of the enantiomer,and also,large value of these descriptors will bring about the short retention time.As for the separation factors,it can be summarized that the differences of the hydrophilic regions at high energy levels,the distance between the first and third energy minima for OH2 probe,the hydrophobic regions at low energy level,amphiphilic moment,suitable hydrogen bond donors and acceptors as well as the anion regions between the enantiomers are favourable factors for the separation,and large difference value will result in the large separation factors.While the differences of the volume,the distance between the second and third energy minima for OH2 probe,the hydrophobic regions at high energy levels,the second and third energy minima for DRY probe,the hydrogen bond donors and acceptors regions at high energy levels are disadvantageous factors to the separation, and large difference value will lead to the small separation factors. The external validation through the test set,leave-many-out crossvalidation and Y-randomization test were carried out and confirmed the robustness and good predictive ability of the model for the separation factors.Using the model,we can predict the retention factors,especially the separation factors and the elution order of the enantiomers of chiral diarylmethane derivates.Also, this study provides a useful guide for studying other chiral compounds.
Supporting Information:The descriptors'value of enantiomers for 63 compounds and the difference value of 63 variables between the enantiomers have been included.This information is available free of charge via the internet at http://www.whxb.pku. edu.cn.
(1) Ebnöther,A.;Weber,H.P.Helv.Chim.Acta1976,59,2462.
(2) Casy,A.F.;Drake,A.F.;Ganellin,C.R.;Mercer,A.D.;Upton, C.Chirality 1992,4,356.
(3) James,M.N.G.;Williams,G.J.B.Can.J.Chem.1974,52,1872.doi:10.1139/v74-267
(4) Sund,R.B.Nor.Pharm.Acta 1983,45,125.
(5) Jones,C.D.;Winter,M.A.;Hirsh,K.S.;Stam,N.;Taylor,H. M.;Holden,H.E.;Davenport,J.D.;Krumkalns,E.V.;Suhr,R. G.J.Med.Chem.1990,33,416.doi:10.1021/jm00163a065
(6) Silvestri,R.;Artico,M.;Martino,G.D.;Ragno,R.;Massa,S.; Loddo,R.;Murgioni,C.;Loi,A.G.;Colla,P.L.;Pani,A.J.Med.Chem.2002,45,1567.doi:10.1021/jm010904a
(7) Panda,G.;Parai,M.K.;Das,S.K.;Shagufta;Sinha,M.; Chaturvedi,V.;Srivastava,A.K.;Manju,Y.S.;Gaikwad,A.N.; Sinha,S.Eur.J.Med.Chem.2007,42,410.doi:10.1016/j. ejmech.2006.09.020
(8) Tacke,R.;Strohmann,C.;Sarge,S.;Cammenga,H.K.; Sehomburg,D.;Mutschler,E.;Lambrecht,G.Liebigs Ann. Chem.1989,No.2,137.
(9) Wilkinson,J.A.;Rossington,S.B.;Ducki,S.;Leonardb,J.; Hussain,N.Tetrahedron2006,62,1833.doi:10.1016/j. tet.2005.11.044
(10) Gao,G.;Gu,F.L.;Jiang,J.X.;Jiang,K.Z.;Sheng,C.Q.;Lai, G.Q.;Xu,L.W.Chem.Eur.J.2011,17,2698.doi:10.1002/ chem.201003111
(11) Arao,T.;Suzuki,K.;Kondo,K.;Aoyama,T.Synthesis2006,No.22,3809.
(12) Rudolph,J.;Schmidt,F.;Bolm,C.Adv.Synth.Catal.2004,346,867.
(13) Stanchev,S.;Rakovska,R.;Berova,N.;Snatzke,G.Tetrahedron-Asymmetry1995,6,183.doi:10.1016/0957-4166 (94)00374-K
(14) Okamoto,Y.;Ikai,T.Chem.Soc.Rev.2008,37,2593.doi: 10.1039/b808881k
(15) Ramillien,M.;Vanthuyne,N.;Jean,M.;Gherase,D.;Giorgi, M.;Naubron,J.V.;Piras,P.;Roussel,C.J.Chromatogr.A2012,1269,82.doi:10.1016/j.chroma.2012.09.025
(16) Héberger,K.J.Chromatogr.A2007,1158,273.doi:10.1016/j. chroma.2007.03.108
(17) Rio,A.D.J.Sep.Sci.2009,32,1566.doi:10.1002/jssc.v32:10
(18) Petric,M.;Crisan,L.;Crisan,M.;Micle,A.;Maranescu,B.; Ilia,G.Heteroatom Chem.2013,24,138.doi:10.1002/ hc.2013.24.issue-2
(19) Durcekova,T.;Boronova,K.;Mocak,J.;Lehotay,J.;Cizmarik, J.J.Pharmaceut.Biomed.2012,59,209.doi:10.1016/j. jpba.2011.09.035
(20) Suzuki,T.;Timofei,S.;Iuoras,B.E.;Uray,G.;Verdino,P.; Fabian,W.M.F.J.Chromatogr.A2001,922,13.doi:10.1016/S0021-9673(01)00921-9
(21) Du,W.;Yang,G.;Wang,X.;Yuan,S.;Zhou,L.;Xu,D.;Liu,C.Talanta2003,29,1187.
(22) Szaleniec,M.;Dudzik,A.;Pawul,M.;Kozik,B.J.Chromatogr.A2009,1216,6224.doi:10.1016/j.chroma.2009.07.002
(23) Dabic,D.;Natic,M.;Dzambaski,Z.;Markovic,R.;Milojkovic-Opsenica,D.;Tesic,Z.J.Sep.Sci.2011,34,2397.doi:10.1002/ jssc.v34.18
(24) Asensi-Bernardi,L.;Escuder-Gilabert,L.;Martin-Biosca,Y.; Medina-Hernandez,M.J.;Sagrado,S.J.Chromatogr.A2013,1308,152.doi:10.1016/j.chroma.2013.08.003
(25) Zhang,Q.Y.;Xu,L.Z.;Li,J.Y.;Zhang,D.D.;Long,H.L.; Leng,J.Y.;Xu,L.J.Chemometrics2012,26,497.doi:10.1002/ cem.v26.10
(26) Chen,G.H.;Xia,Z.N.;Lu,Y.;Liao,L.M.;Shu,M.;Sun,J.Y.; Li,Z.L.Acta Chim.Sin.2008,66,2052.[陈国华,夏之宁,陆 瑶,廖立敏,舒 茂,孙家英,李志良.化学学报,2008,66, 2052.]
(27) Carbonell,P.;Carlsson,L.;Faulon,J.L.J.Chem.Inf.Model.2013,53,887.doi:10.1021/ci300584r
(28) Liu,D.;Zhang,W.J.;Xu,L.Acta Chim.Sin.2009,67,145. [刘 东,章文军,许 禄.化学学报,2009,67,145.]
(29) Zhang,Q.Y.;Hu,W.P.;Hao,J.F.;Liu,X.H.;Xu,L.Acta Chim.Sin.2010,68,883. [张庆友,胡卫平,郝军峰,刘绣华,许 禄.化学学报,2010,68,883.]
(30) Feng,C.J.Acta Phys.-Chim.Sin.2010,26,193. [冯长君.物理化学学报,2010,26,193.]doi:10.3866/PKU. WHXB20100123
(31) Fresqui,M.A.C.;Ferreira,M.M.C.;Trsic,M.Anal.Chim. Acta2013,759,43.doi:10.1016/j.aca.2012.11.004
(32) Hu,G.X.;Zou,J.W.;Zeng,M.;Pan,S.F.;Yu,Q.S.QSAR Comb.Sci.2009,28,1112.doi:10.1002/qsar.v28:10
(33) Job,G.E.;Shvets,A.;Pirkle,W.H.;Kuwahara,S.;Kosaka,M.; Kasai,Y.;Taji,H.;Fujita,K.;Watanabe,M.;Harada,N.J.Chromatogr.A2004,1055,41.doi:10.1016/j. chroma.2004.08.001
(34) Ermondi,G.;Caron,G.J.Chromatogr.A2012,1252,84.doi: 10.1016/j.chroma.2012.06.069
(35) Visentin,S.;Ermondi,G.;Medana,C.;Pedemonte,N.;Galietta, L.;Caron,G.Eur.J.Med.Chem.2012,55,188.doi:10.1016/j. ejmech.2012.07.017
(36) Das,S.;Roy,P.;Islam,M.A.;Saha,A.;Mukherjee,A.Chem. Pharm.Bull.2013,61,125.doi:10.1248/cpb.c12-00475
(37) Cruciani,G.;Crivori,P.;Carrupt,P.A.;Testa,B.J.Mol. Struct.-Theochem2000,503,17.
(38) Hudson,B.D.;Hyde,R.M.;Rahr,E.;Wood,J.;Osman,J.Quant.Struct.-Act.Relat.1996,15,285.doi:10.1002/qsar.v15:4
(39) Miller,K.J.J.Am.Chem.Soc.1990,112,8533.doi:10.1021/ ja00179a044
(40) Kiralj,R.;Ferreira,M.M.C.J.Braz.Chem.Soc.2009,20,770.doi:10.1590/S0103-50532009000400021
(41) Wang,X.J.;Sun,Y.Y.;Wu,L.;Gu,S.J.;Liu,R.N.;Liu,L.; Liu,X.;Xu,J.Chemometr.Intell.Lab.2014,134,1.doi: 10.1016/j.chemolab.2014.03.001
Predicting Retention and Separation Factors of Chiral Diarylmethane Derivates by QSPR Models
HU Gui-Xiang1,*LUO Cheng-Cai1PAN Shan-Fei2JIANG Yong-Jun1ZOU Jian-Wei1
(1School of Biotechnology and Chemical Engineering,Ningbo Institute of Technology,Zhejiang University,Ningbo 315100, Zhejiang Province,P.R.China;2Department of Chemistry,Zhejiang University,Hangzhou 310028,P.R.China)
Quantitative structure-property relationship(QSPR)studies on retention and separation factors of chiral compounds play a key role in predicting the retention and separation factors even the elution order of enantiomers.Chiral diarylmethane derivates were selected for computing molecular structural descriptors using VolSurf program.Models were built between the descriptors and retention as well as separation factors.The robustness of the model with respect to separation factors was assessed by external validation through the test set,leave-many-out cross-validation and Y-randomization test.The results were satisfactory.Analysis on the variables shows that the molecular globularity,hydrophilic regions at median energy levels,hydrophilic-lipophilic balance,amphiphilic moment,suitable hydrogen bond donors and acceptors are beneficial to the retention of enantiomers on the chiral stationary phase.Large differences of the hydrophilic regions at high energy levels, hydrophobic regions at low energy levels,amphiphilic moment,suitable hydrogen bond donors and acceptors, and anion regions between enantiomers are advantageous to the separation of enantiomers on the chiral stationary phase.These models allow the prediction of retention and separation factors,especially the elution order of enantiomers.
Chirality;Molecular modeling;VolSurf;Diarylmethane;Partial least squares
O641
10.3866/PKU.WHXB201410281www.whxb.pku.edu.cn
Received:August 16,2014;Revised:October 27,2014;Published on Web:October 28,2014.
∗Corresponding author.Email:hugx@nit.zju.edu.cn;Tel:+86-574-88130130.
The project was supported by the National Natural Science Foundation of China(21002088,21272211)and Program of Science and Technology of Ningbo,China(2013D1003).
国家自然科学基金(21002088,21272211)和宁波市科技计划(2013D1003)资助项目