MORPHOLOGICAL OBSERVATION AND RBCL SEQUENCE ANALYSIS OF A NEW SPECIES FROM CHINA, GRATELOUPIA BOAOENSIS WANG ET LUAN SP. NOV.(HALYMENIACEAE, RHODOPHYTA)
2014-11-05LIUMiaoWANGHongWeiandLUANRiXiao
LIU Miao, WANG Hong-Wei and LUAN Ri-Xiao
(1. College of Life Sciences, Liaoning Normal University, Dalian 116081, China;2. Dalian Natural History Museum, Dalian 116023, China)
Abstract: During macroalgae investigation, a new Grateloupia species from Wenchang, Hainan Province, South China, was discovered and named Grateloupia boaoensis Wang et Luan sp. nov. By using morphological observation and molecular analysis, we determined that G. boaoensis was purplish red, cartilaginous and slippery in texture, 15—40 cm high, with completely compressed and foliated axes with lateral branches and sword-shaped apex.The transverse sections were 400—800 µm thick consisting of 4—9 layers of cortex cells and solid medulla, and the medullar filaments were longitudinal and intertwined. The new species exhibited typical Grateloupia-type auxiliary cell ampullae. Cystocarps were densely distributed on both axes and lateral branches. The four G. boaoensis sequences in this study were identical and formed a single monophyletic subclade with G. yinggehaiensis embedded in the Grateloupia clade based on ribulose-1, 5-bisphosphate carboxylase/oxygenase gene (rbcL) sequences. G. boaoensis was considered most closely related to G. yinggehaiensis by morphological observation and molecular analysis.
Key words: Rhodophyta; Halymeniaceae; Grateloupia; Grateloupia boaoensis; Morphological observation; rbcL
The genus Grateloupia (Halymeniaceae, Rhodophyta) was established in 1822 with the type species G.filicina (Lamouroux) C. Agardh. Today, more than 80 Grateloupia species have been described and are found distributed worldwide[1—10], 35 of which are endemic to China[11—13]. The genus is notorious in its morphological plasticity due to environmental changes,life periods and intrinsic morphological diversity.Many methods have been applied to the classification of Grateloupia, including morphological and anatomic classification and molecular systematics. Several markers have been applied to red alga taxonomy, and the rbcL gene is considered suitable for Grateloupia due to its comparatively long sequences (more than 1400 bp)and slow variation rate, which allow for the expression of more characters and relatively precise classification.A growing number of new Grateloupia species have been described using rbcL sequencing combined with morphological observation, including G. asiatica Kawaguchi et Wang[3], G. huertana Mateo-Cid, Mendoza-González et Gavio[6], G. capensis De Clerck[7], G.taiwanensis Lin et Liang[10], G. orientalis Lin et Liang[10], G. dalianensis Wang et Zhao[12]and G. yinggehaiensis Wang et Luan[12].
A particular species of Grateloupia was discovered during macroalgae investigation in Hainan province,China. After in-depth study of its outward morphology and internal features, together with rbcL gene analysis,this species was found to differ from other extant species and was named as Grateloupia boaoensis Wang et Luan sp. nov.
1 Materials and methods
1.1 Morphological observations
The study specimens were collected by hand in Longlou, Wenchang, Hainan Province, China, on 8 January, 2009, and voucher herbarium specimens were deposited in the Herbarium of College of Life Sciences, Liaoning Normal University (LNU), Dalian,China.
Grateloupia boaoensis: Longlou, Wenchang, Hainan,China (8 January, 2009, leg. H.W. Wang and R.X.Luan; LNU20092009, LNU2009200901, LNU20092-00902, and LNU 2009200903).
Fresh material, rehydrated herbarium specimens or specimens stored in 10% formalin/seawater were used for morphological observations. Hand sections were made by cryostat microtome, stained with 0.5% (w/v)cotton blue and discolored with 45% acetic acid. Pictures were screened using an Olympus BH2 digital camera (Olympus Beijing Co. Ltd., China) mounted on a Nikon microscope (Nikon Corporation, Japan).
1.2 Molecular analysis
Total DNA were extracted from four samples of G.boaoensis by using a Plant Genomic DNA kit following the manufacturer’s protocols (QIAGEN, Valencia, CA, Beijing). The rbcL PCR amplification and DNA sequencing was conducted as per Wang et al.[1]with three pairs of primers: F8-R646, F481-R1l50 and F765Mix-R1381-ii[1]. No insertion/deletion mutations were detected; therefore, the rbcL sequences were aligned manually.
Forty-one additional Halymeniaceae species sequences downloaded from GenBank were used for sequence alignment. Three species within the family Halymeniaceae, Carpopeltis phyllophora (Hooker et Harvey) Schmitz, Halymenia floresia (Clemente) C.Agardh and Polyopes constrictus (Turner) J. Agardh,were selected to compare with G. boaoensis sequences. Ceramium japonicum Okamura and Gelidium crinale (Hare ex Turner) Gaillon were used as outgroups (Tab. 1).
Tab. 1 List of species for rbcL analysis, collection locations, references and GenBank accession numbers
The rbcL sequence data were aligned and compiled with Clustal X version 1.83[20]. Phylogenetic trees were constructed by Maximum Likelihood (ML),Neighbor Joining (NJ) and Maximum Parsimony (MP)methods. Analyses of base differences and ML and NJ phylogenetic trees (1000 bootstraps) were performed by MEGA 5.0[21]and MP (1000 bootstraps) by MEGA 4.0[22]. The Kimura 2-parameter model was used to analyze the distances matrix[23].
2 Results
2.1 External morphology
Four samples were collected from Longlou, Wenchang, Hainan Province, China, on 8 January, 2009.Thalli clung to rocks by discoid holdfasts in the mid-tolower intertidal. We selected a female gametophyte as the holotype specimen, and labeled it as HN20092009. The other three samples were labeled as HN2009200901,HN2009200902 and HN2009200903.
Holotype: Female gametophyte (LNU20092009 Fig.1A).
Type locality: Longlou in Hainan Province, south China (19.5°N, 108.7°E).
Etymology: Species named for the Boao Forum held in Hainan Province, China.
Thalli were dark purplish red in color, cartilaginous and slippery in texture and arose from discoid holdfasts. The erect axes were completely compressed and foliated, 15—40 cm high and 2—5 cm wide. The 1—3 pinnate lateral branches were also foliated, with constrictions in the bases. The axes were obviously thicker than the lateral branches (Fig. 1A-B). Dense irregular branches were distichous, alternative or secund (Fig. 2A-C). The apices of blades were ensiform.

Fig. 1 External morphology of Grateloupia boaoensis
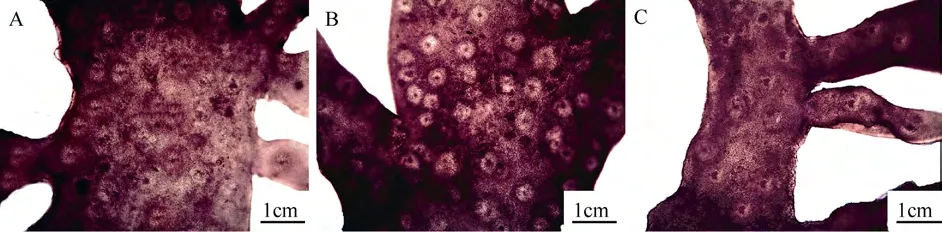
Fig. 2 Branching characteristics of Grateloupia boaoensis and the distribution situation of cystocarps. Showing distichous (Fig. A), alternative (Fig. B) and secund (Fig. C) branch styles
2.2 Vegetative structure
The thalli were 400—800 µm thick consisting of cortex and medulla. The cortex was 100—250 µm thick and consisted of 4—9 layers cells. The outer cortex consisted of 1—2 layers of anticlinally arranged oblong cells, and the inner cortex consisted of 3—6 layers of spherical, oval or irregular cells. The medulla was 200—600 µm thick and consisted of 30—100 µm long and 2—4 µm wide intertwined longitudinal medullar filaments (Fig. 3A-B).
2.3 Reproductive morphology
The reproductive structures were densely distributed on the axes and lateral branches, and obviously bulged from the surface (Fig. 2A-C). Gametophytes were dioecious. Carpogonial branch ampullae and auxiliary cell ampullae both formed from the inner cortex separately (Fig. 4A-B). Carpogonial branch ampullae were composed of a two-celled carpogonial branch and 2—3 secondary filaments (Fig. 4A). Auxiliary cell ampullae consisted of 2—3 secondary filaments (Fig. 4B). The cystocarp development processes were displayed in Fig. 4C-E. The gonimoblasts mingled with medullar filaments and formed a circle at the beginning period (Fig. 4C). Developing cystocarps were embedded in the medullary (Fig. 4D). Mature carpospores were then released from the cystocarp hole (Fig. 4E). Spermatangia were not seen because male gametophytes were not collected. Mature tetraspores were embedded in the inner cortex, cruciately divided, 20—60 μm long and 15—30 μm wide (Fig. 4F).

Fig. 3 Cross-section of the Grateloupia boaoensis thallus, showing cellular cortex and medullar filaments
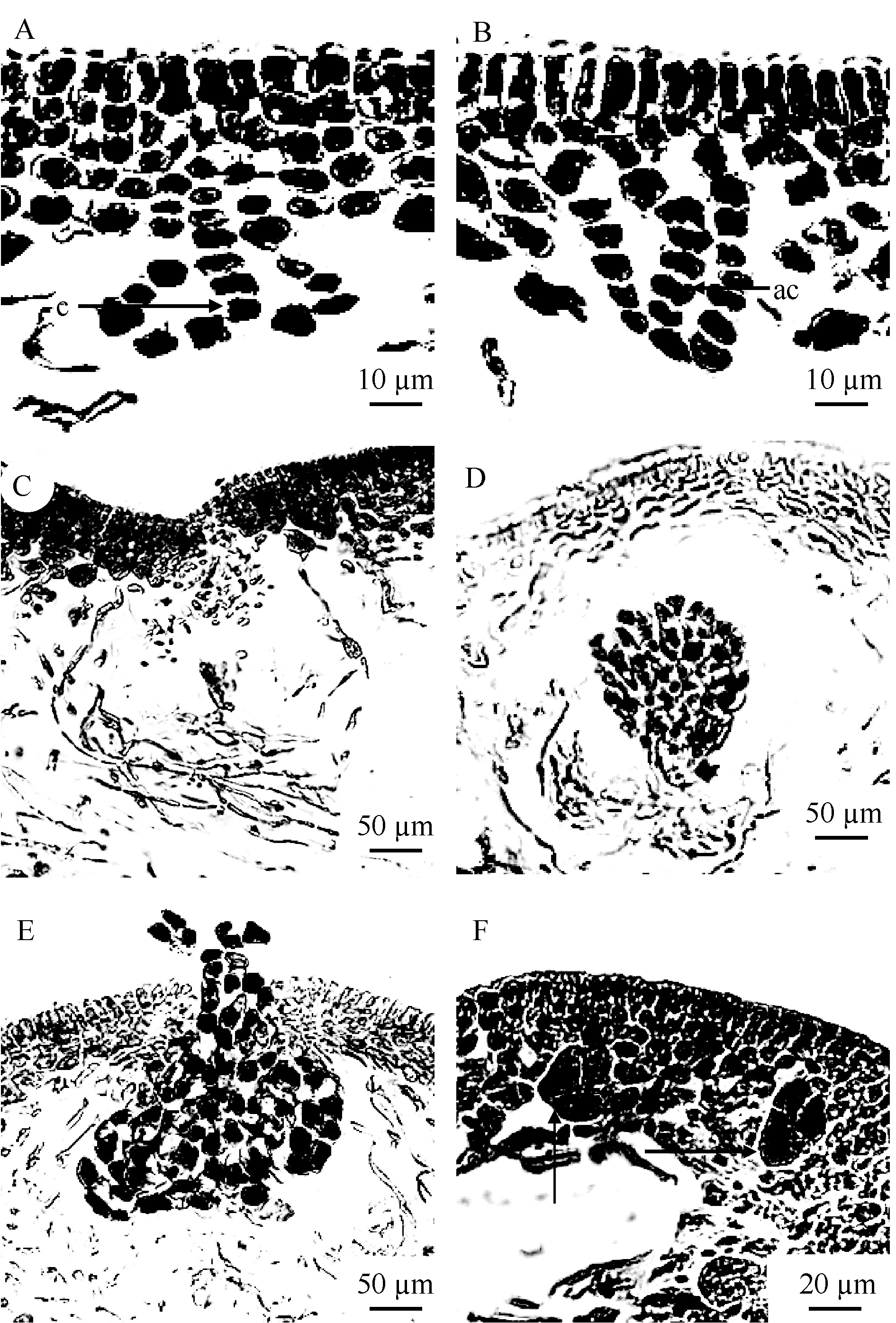
Fig. 4 Reproductive structures of Grateloupia boaoensis
2.4 rbcL analysis
A phylogenetic tree was obtained using Maximum Likelihood (ML), Neighbor-Joining (NJ) and Most Parsimonious (MP) analysis based on rbcL gene sequences (Fig. 5). Numerals at internal nodes are bootstrap values (1000 replicates) inferred from ML (upper), NJ (middle) and MP (lower). The four rbcL sequences of G. boaoensis were identical, which was probably because the specimens were collected from the same place at the same time. The sequence alignment of G. boaoensis consisted of 1298 base pairs (bp),but since many rbcL sequences were incomplete at the 5¢ and 3¢ ends, the first 31 bp and last 17 bp were excluded from the analyses. The phylogenetic tree showed that the pairwise distances between G. boaoensis and the generitype G. filicina from Italy were 69 bp (5.78%). G. boaoensis and G. yinggehaiensis from China formed a single subclade with high bootstrap support within the Grateloupia clade, despite a 22 bp (1.79%) difference between the two species. The pairwise distances between G. boaoensis and other species in the Grateloupia clade ranged from 44 bp(4.13%) to 110 bp (9.56%), and those with other species within family Halymeniaceae, Carpopeltis phyllophora, Halymenia floresia and Polyopes constrictus,ranged from 123—136 bp (10.67%—11.92%). The pairwise distances between G. boaoensis and Ceramium japonicum and Gelidium crinale outgroups were 173 bp and 195 bp (15.32% and 17.59%), respectively.
3 Discussion
The auxiliary cell ampulla is an important feature for discriminating the 20 different genera within Halymeniaceae[2,3,7,24]. Auxiliary cell ampullae types in Halymeniaceae include Aeodes, Cryptonemia, Halymenia, Grateloupia, and Thamnoclonium[16]. A primary fi lament and 2—3 unbranched secondary fi laments constitute the relatively simple Grateloupiatype auxiliary cell ampulla structure[7]. G. boaoensis had a typical Grateloupia-type auxiliary cell ampulla comprised of a primary ampullary filament and two secondary filaments (Fig. 4B).
G. boaoensis had distinct morphological features among Grateloupia species in regards to its completely compressed and foliated axes bearing densely foliated lateral branches. It was morphologically similar to G. yinggehaiensis, though differences were also observed, particularly in relation to the size,thickness, and intensity of the lateral branches (Figs. 1,6) and some internal architecture. Both had multiple ensiform apices, but the thalli of the G. boaoensis were more than twice as large and thick as those of G.yinggehaiensis and had many more lateral branches.The widest point of G. boaoensis was almost 5 cm wide, much wider than the 1 cm of G. yinggehaiensis.The cystocarps were densely distributed on the axis and lateral branches of G. boaoensis, but were scattered over the thallus branches, though not the basal parts and distal ends, of G. yinggehaiensis. Moreover,G. boaoensis had much larger cystocarps than G.yinggehaiensis. The main distinctions between G.boaoensis and G. yinggehaiensis are listed in Tab. 2.Comparison of these features indicated that G. boaoensis was different from G. yinggehaiensis.

Fig. 5 Maximum likelihood (ML) phylogenetic tree based on partial rbcL gene sequences data
Molecular analysis further proved that G. boaoensis was a new species. G. boaoensis and G. yinggehaiensis formed a monophyletic subclade in ML, NJ and MP phylogenetic trees. The pairwise distance between G. boaoensis and G. yinggehaiensis was 22 bp (1.79%).Though the sequence divergence between G. boaoensis and G. yinggehaiensis was small, previous research determining different species has also shown similarly close sequence divergence as demonstrated in this study. The rbcL gene pairwise distances between G.turuturu and G. sparsa was found to be 17 bp (1.37%),indicating they were two different Grate loupia species, though they exhibited similar mor phology[24]. G.asiatica was defined as a new species though only exhibited a 17 bp pairwise distance with G. livida[3].Previous research demonstrated that G. catenata and G. ramosissima had similarities in external characteristics, including axes that were terete below and slightly compressed above, with numerous proliferations, but had a pairwise distance of 28 bp[1,25].

Tab. 2 Comparison of morphological features between Grateloupia boaoensis and G. yinggehaiensis

Fig. 6 External morphology of Grateloupia yinggehaiensis
With the exception of G. yinggehaiensis, the pairwise distances between G. boaoensis and the species clustered in the small subclade with high bootstrap support values,including G. yangjiangensis, G. filiformis, G. orientalis,G. ramosissima, G. catenata and G. filicina, ranged from 44—69 bp (3.62%—5.78%) and with other species in the Grateloupia clade ranged from 80—112 bp (6.75%—9.66%). The pairwise distances between G. boaoensis and species in other genera within family Halymeniaceae were 123—136 bp (10.67%—11.92%), and were 173—195 bp (15.32%—17.59%) with species in other orders.These data help to eliminate G. boaoensis from other families and genera and place it into Grateloupia.
From the morphological features and molecular results analysis, we deduced that the new specimen occupied an independent branch in Grateloupia and was distinguished from other species. Thus, it was defined as a new species in Grateloupia, and named as Grateloupia boaoensis Wang et Luan sp. nov.
Acknowledgements:
The authors are very grateful to Wei Cheng-Xiang,Yu Ling, Li Yazhuo and Guan Yun for their technical assistance and valuable advice.
杂志排行
水生生物学报的其它文章
- GILL MEDIATES IMMUNE RESPONSES AFTER GRASS CARP REOVIRUS CHALLENGE IN GRASS CARP (CTENOPHARYNGODON IDELLA)
- GILL MEDIATES IMMUNE RESPONSES AFTER GRASS CARP REOVIRUS CHALLENGE IN GRASS CARP (CTENOPHARYNGODON IDELLA)
- 晶体氨基酸替代鱼粉蛋白对半滑舌鳎稚鱼消化酶和代谢酶活力的影响
- 生长激素/催乳素家族配体和受体成员在斑马鱼早期胚胎中的表达比较和交叉活性分析
- 青海湖区大型底栖动物群落结构与空间分布格局
- 氮磷比对不同生长时期强壮前沟藻富集2,2’,4,4’-四溴联苯醚的影响
