Multilocus Phylogeny of Lycodon and the Taxonomic Revision of Oligodon multizonatum
2014-07-01JuanLEIXiaoyuSUNKeJIANGGernotVOGELDavidBOOTHandLiDING
Juan LEI, Xiaoyu SUN Ke JIANG, Gernot VOGEL, David T. BOOTHand Li DING*
1Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, Sichuan 610041, China
2College of Life and Sciences, Sichuan University, Chengdu, Sichuan 610064, China
3State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, Yunnan Province, China
4School of Biological Science, The University of Queensland, Brisbane, St Lucia, QLD 4072, Australia
5Society for Southeast Asian Herpetology, Im Sand 3, D-69115 Heidelberg, Germany
Multilocus Phylogeny of Lycodon and the Taxonomic Revision of Oligodon multizonatum
Juan LEI1,2,4, Xiaoyu SUN1, Ke JIANG3, Gernot VOGEL5, David T. BOOTH4and Li DING1*
1Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, Sichuan 610041, China
2College of Life and Sciences, Sichuan University, Chengdu, Sichuan 610064, China
3State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, Yunnan Province, China
4School of Biological Science, The University of Queensland, Brisbane, St Lucia, QLD 4072, Australia
5Society for Southeast Asian Herpetology, Im Sand 3, D-69115 Heidelberg, Germany
Classification of the Asian snake genera Lycodon and Oligodon has proven challenging. We conducted a molecular phylogenetic analysis to estimate the phylogenetic relationships in the genus of Lycodon and clarify the taxonomic status of Oligodon multizonatum using mitochondrial (cyt b, ND4) and nuclear (c-mos) genes. Phylogenetic trees estimated using Maximum Likelihood and Bayesian Inference indicated that O. multizonatum is actually a species of Lycodon. Comparing morphological data from O. multizonatum and its closest relatives also supported this conclusion. Our results imply that a thorough review of the evolutionary relationships in the genus of Lycodon is strong suggested.
Bayesian inference, China, classif i cation, c-mos, cyt b, Lycodon, maximum likelihood, ND4, Oligodon
1. Introduction
The genus Oligodon Fitzinger, 1826 is widespread throughout central and tropical Asia, containing approximately 70 species (Green et al., 2010). Among them, 15 are known to occur in southern China (Zhao et al., 1998). Previous studies aimed at classifying the genus have been based on morphological data and yielded conflicting results (Wall, 1923; Pope, 1935; Smith, 1943; Leviton, 1963; Campden, 1969; Wallach and Bauer, 1996; David et al., 2008; Tillack and Günther, 2009). However, all of these studies were limited to a species group within this complex or a limited geographic area, and no study constructed a phylogenetic tree. Green et al. (2010) produced an updated checklist and key to the entire genus together with a phylogentic tree. The key and checklist were given in his thesis, and the phylogenetic data were later published (Green et al., 2010) and concluded that several uncertainties about the classif i cation still exist. However, no study has included molecular data from Oligodon multizonatum.
Oligodon multizonatum was described by Zhao and Jiang (1981) from Luding County, Sichuan Province, southwest China. The species was classif i ed as a member of the genus Oligodon on the basis of morphological characteristics including a short head that is not distinct from the neck, a large rostral scale that appears protruding when viewed from above, a cylindrical body with paired subcaudals and smooth dorsal scales (Zhao et al., 1998). There have been no published attempts to explore the taxonomic position of the species since it was first described, and no new specimens have been reported. Currently, O. multizonatum is considered an endemicspecies of China, only occurring in Sichuan and Gansu Provinces (Zhao, 2006). A snake specimen (specimen number KIZ01623, Figure 1) was collected in Luding County (29°55′12.58″ N, 102°13′31.07″ E) during a herpetological survey on July in 2009. A detailed comparison with the species description and the holotype specimen (CIB9964, Figure 2) suggested that it was conspecif i c with O. multizonatum.
Recent studies of snakes (Burbrink and Castoe, 2009; Huang et al., 2009), have shown that molecular data are powerful tools for identifying and understanding snake diversity. In view of this, the purpose of the present study was to use molecular methods to clairfy the systematic aff i nities of O. multizonatum. A prior study by us based on molecular analysis with more than three genes and 89 species of Colubridae showed that O. multizonatum clustered within Lycodon. Recently, the genus Lycodon was suggested to include species of the old genus Dinodon (Siler et al., 2013; Guo et al., 2013), suggesting that many relationship within the genus of Lycodon still need to be resolved. For example, Siler et al. (2013) suggested that currently recognized subspecies mayneed to be elevated to species in further studies. Hence, in this study, we sampled species from both Lycodon and Dinodon in order to resolve these issues. For the convenience of our discussion, the historic taxonomic genera Lycodon and Dinodon continue to be used. Additionally, we also compared the morphological data of O. multizonatum with its closest relative as identif i ed by molecular data analysis to verify this conclusion.

Figure 1 Photographs of a new Oligodon multizonatum specimen (specimen number KIZ01623) collected in Luding province. A–C: Whole body; D–F: Head in dorsal, ventral and right lateral views; G: Cloacal region in ventral view. Photo by Mian HOU.

Figure 2 Photographs of the holotype specimen (CIB9964) of Oligodon multizonatum. A and B: Whole body; C: Ventral views; D: Cloacal region in ventral view and hemipenis. Photo by Juan LEI.
2. Materials and methods
2.1 MorphologyMeasurements, except body and tail lengths, were taken with a slide-caliper to the nearest 0.1 mm; all body lengths were made to the nearest millimeter using a tape measure. The number of ventral scales was counted according to Dowling (1951). Divided ventrals were counted as one. The first scale posterior to the cloaca was regarded as the first subcaudal, the terminal scute was not included in the number of subcaudals. The dorsal scale rows were counted at one head length behind the head, at midbody (i.e., at the level of the ventral plate corresponding to half the total number of ventrals), and at one head length before the vent. We considered sublabials being those shields that were completely below a supralabial. Values for paired head characters are given in left/right order.
The hemipenes of O. multizonatum and L. liuchengchaoi were compared. The method for preparing the hemipenes of preserved specimens followed Jiang (2010) and Pesantes (1994). Hemipenial descriptive terminology followed Dowling and Savage (1960), Branch (1986) and Zhang et al. (1984). Drawings were made with the aid of a stereomicroscope.
2.2 Taxon samplingPrevious studies indicated that the systematics of the genera Oligodon, Lycodon and Dinodon are complex and possibly intertwined (Pope, 1935; Smith, 1943; Vogel and Brachtel, 2008; Green et al., 2010; Guo et al., 2013). Therefore, data from seven species in Oligodon, 16 species in Lycodon and one species in Dinodon from GenBank were used along with new data generated during the present study from O. multizonatum, O. formosanus, O. chinensis, L. ruhstrati, L. liuchengchaoi, D. rufozonatum and D. flavozonatum (Table 1). We also selected 10 taxa representing 10 genera of Colubrinae from GenBank. The choice of outgroup taxa (Boa constrictor and Cylindrophis ruffus) was based on Huang et al. (2009). Accession numbers from the Chengdu Institute of Biology (CIB), Kunming Institution of Biology (KIZ) and the laboratory of Ding Li (DL) for all these specimens are provided in Table 1.
2.3 DNA extraction, amplification, and sequencingTissue samples were either skeletal muscle or liver preserved in 95% ethanol at the time of collection and
subsequently stored in either ethanol or frozen at –80°C. All specimens sampled are preserved in the collections of CIB. All tissues were treated by the standard method of proteinase K digestion in lysis buffer followed by a high salt DNA extraction procedure (Sambrook et al., 1989). The mitochondrial cytochrome b (cyt b) gene and the NADH dehydrogenase subunit 4 (ND4) gene, and the nuclear oocyte maturation factor Mos (c-mos) gene were amplif i ed from total DNA extracts using polymerase chain reaction (PCR) with the following primer pairs for cyt b: L14910/H16064 (Burbrink et al., 2000), ND4: ND4/Leu (Arévalo et al., 1994), and c-mos: S77/S78 (Lawson et al., 2005). Amplification was performed in a 20 μl volume reaction with the following settings: initial denaturation step with 4 min at 94°C, 35 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 46°C for cyt b primers and 56°C for ND4 and c-mos, extension for 1 min at 72°C. A final extension at 72°C was conducted for 7 min. Purified PCR products were sequenced in both directions with an ABI automated DNA sequencer (ABI 3700). We conducted a BLAST search of acquired sequences by using the GenBank database to verify that generated sequences were not of pseudogenes. All novel sequences have been deposited in GenBank (Table 1).
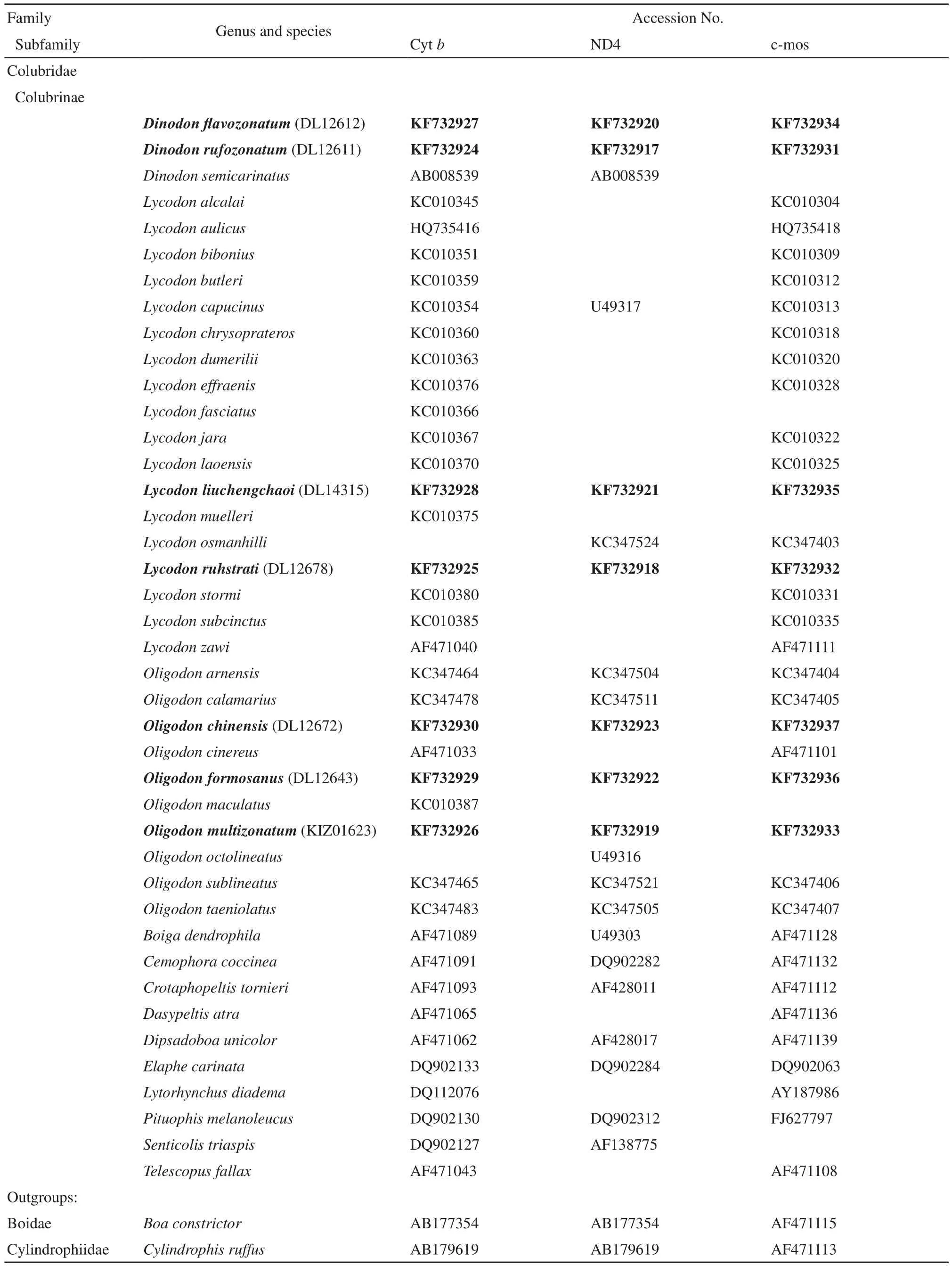
Table 1 The information of sequences retrieved from GenBank and sequenced in this study. New sequences from this study are in bold.
2.4 Phylogenetic analysesThe initial alignments of cyt b, ND4, c-mos were aligned using ClustalX (Thompson et al., 1997) with default parameters, and subsequently verified manually, and translated into amino acid sequences to check for the presence of stop codons. We tested the saturation of 3rdcodon positions of the mitochondrial protein-coding genes. These were highly saturated, therefore, we deleted the 3rdcodon positions of mitochondrial genes (cyt b and ND4). In addition, we also analyzed the phylogeny for each gene independently in order to explore the congruence between different gene data by using likelihood and Bayesian analyses. There was no moderate to highly supported incongruence between cyt b, ND4 and c-mos gene and therefore we used the concatenated and combined data for phylogenetic analyses in this study. Because some of taxa were missing data for cyt b, ND4 and c-mos, we did exploratory analyses of the combined data set of 41 ingroup and two outgroup taxa and found no missing data exhibited identical relationships. Therefore, we chose to use all data (41 taxa) for subsequent analyses of the combined data set.
Phylogenetic analyses were performed using Bayesian Inference (BI) and Maximum Likelihood (ML) methodology. Partitioned Bayesian Inference (BI) approaches were used to reconstruct phylogeny with combined data set of three partial gene sequences, using MrBayes v 3.1 (Huelsenbeck and Ronquist, 2001). Both mitochondrial and nuclear data sets were partitioned by codon position. The best-f i t substitution model (see Table 2) was assigned to each partition using AIC in Modeltest 3.7 (Posada and Crandall, 1998) and PAUP*v4b10 (Swofford, 2003). Two separate runs were performed with four Markov chains. Each run was conducted with 15 000 000 generations and sampled every 1000 generations. When the scores were found to stabilize, a consensus tree was calculated after omitting the f i rst 25% of the trees as burn-in. Node support for the Bayesian consensus tree was determined using posterior probabilities (Erixon et al., 2003). Maximum Likelihood (ML) with the nonpartitioned strategy with the combined data set was used to infer trees and assess nodal support by using RaxML (Stamatakis et al., 2005). The complex model(GTR + Γ) was used for each partition. Support for ML trees was derived from 100 nonparametric bootstrap replicates using RaxML. Each inference was started with a random starting tree, and 100 nonparametric bootstrap pseudoreplicates (Stamatakis et al., 2008) was used to assessed the nodal support. Because of less availability of ND4 gene and the fact that the c-mos gene was highly conserved in this study, average divergence estimation between species was calculated from the two mitochondrial genes using Mega 4.0 (Tamura et al., 2008).
2.5 Topological testThe Bayesian analysis and Maximum Likelihood produced BI and ML trees. The topological structure of phylogenetic trees were slightly different. Results from the Shimodaira-Hasegawa (SH test; Shimodaira and Hasegawa, 1999) and Kishino-Hasegawa tests (KH test; Kishinino and Hasegawa, 1989) indicated that the BI tree was the best f i t. Therefore, the conclusion of our analysis are based mainly on the BI tree topological structure.
3. Results
3.1 MorphologyThe newly collected specimen (KIZ01623) and the specimen of the type series of O. multizonatum Zhao and Jiang, 1981 (CIB9964) were used in this study. A Comparison of the main morphological characters between O. multizonatum (type specimens, CIB9964–9967), O. multizonatum (new specimen, KIZ01623), L. liuchengchaoi (description from Zhang et al. [2011], CWNU867001, CWNU84002, and FMNH15148), L. liuchengchaoi (new specimen, DL14315), and O. joynsoni (description from Jiang et al.[2012], BMNH1946.1.4.23, BMNH 1969.1809, BMNH 1938.8.7.40, BMNH 1969.1808, MNHN 1896.0633, and KIZ09128) are shown in Table 3. Based on morphological examination, our results indicated that the new specimen of O. multizonatum is same as the type specimen of O. multizonatum, but different from the type specimen of L. liuchengchaoi and the new specimen of L. liuchengchaoi. In addition, Zhao and Jiang (1981) reported O. joynsoni is the most similar species to O. multizonatum. Our analysis indicates there are many major differences between O. joynsoni, L. liuchengchaoi and O. multizonatum.
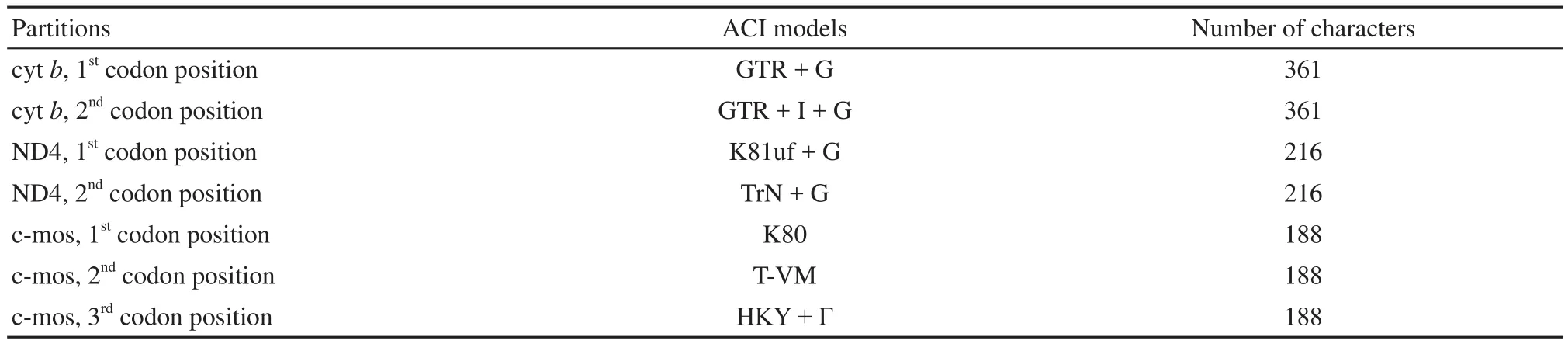
Table 2 Models of evolution selected by AIC and partitions of mitochondrial (cyt b, ND4) and nuclear (c-mos) data applied in model-based analyses.
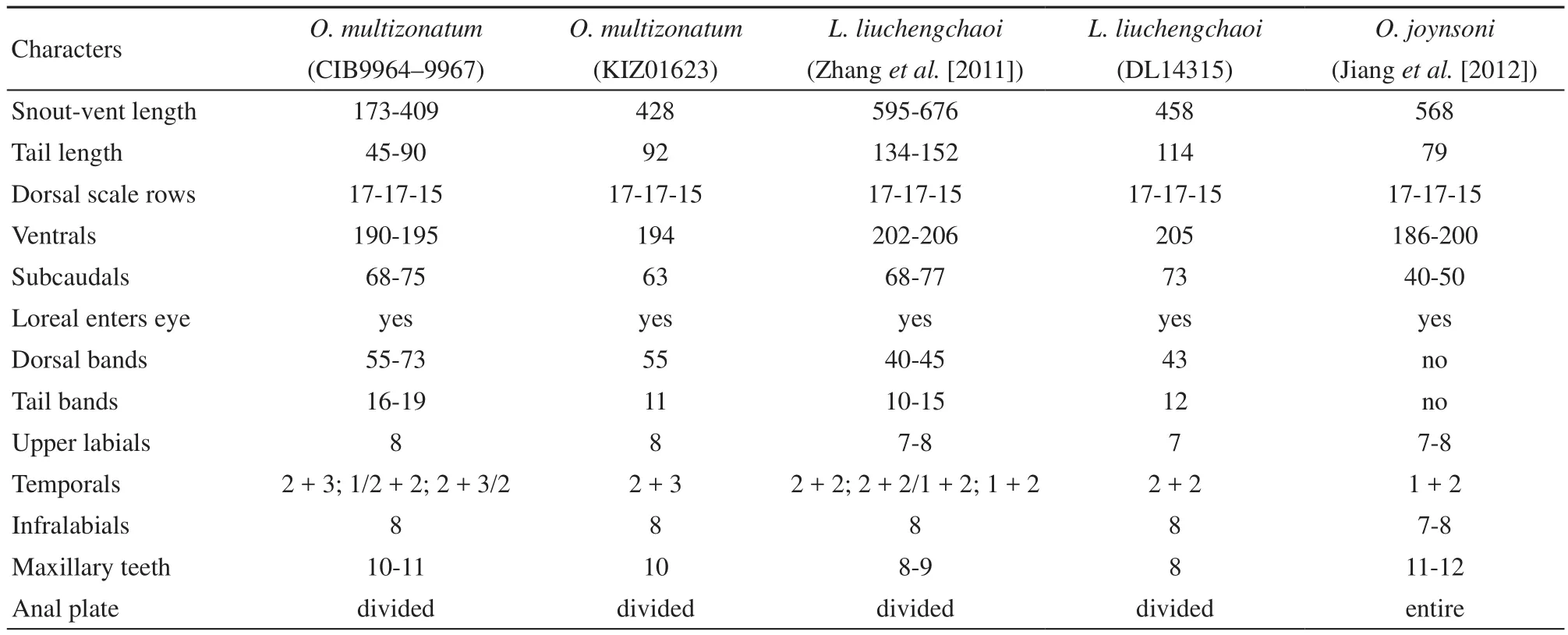
Table 3 A Comparison of the main morphological characters between O. multizonatum (type specimens, CIB9964–9967), O. multizonatum (new specimen, KIZ01623), L. liuchengchaoi (description from Zhang et al. [2011], CWNU867001, CWNU84002, and FMNH15148), L. liuchengchaoi (new specimen, DL14315), and O. joynsoni (description from Jiang et al. [2012], BMNH1946.1.4.23, BMNH 1969.1809, BMNH 1938.8.7.40, BMNH 1969.1808, MNHN 1896.0633, and KIZ09128). Dimensions in mm.
The hemipenis of O. multizonatum (KIZ01623) (Figure 3) is characterized as follows: smaller base, expanding from middle to tip; relatively short, extending to the eighth subcaudal; unforked, sulcus single and prominent, extending to the tips of the organ; the base to middle of the organ covered with larger hard spines, but changing to tiny spines after middle to the tip; no nick at the tip.
The hemipenis characteristics of O. multizonatum (KIZ01623) are similar to the description of the type specimen of O. multizonatum (CIB9964) provided by Zhao and Jiang (1981). Based on hemipenis morphology, O. multizonatum can be seperated from the species of the genus Oligodon that lack a hard spine, such as O. joynsoni (Smith, 1917), but is similar to most species of the genera Oligodon, Lycodon and Dinodon, which have a hard spine on the hemipenis (Zhao et al., 1998; Zhao, 2008; Green et al., 2010). Green et al. (2010) reported the hind teeth of snakes of the genus Oligodon are broad and strongly recurved. However, our analysis showed that the hind teeth of both new and type specimens of O. multizonatum are not strongly recurved. In addition, a striking characteristic of Oligodon is the large rostral scale that is clearly visible when viewed from above (Zhao et al., 1998; Zhao, 2006). Nevertheless, such observations are to some extent subjective and might even be dependent on the viewing angle (Figures 1–2). Actually the rostral scale of O. multizonatum is not aslarge as that in members of the genus Oligodon, and is only clearly visible from above. The placement of O. multizonatum in the genus Oligodon was based on it having a large rostal scale (ref to original description), but our analysis suggests that the rostal scale of O. multizonatum is not particularly prominent.
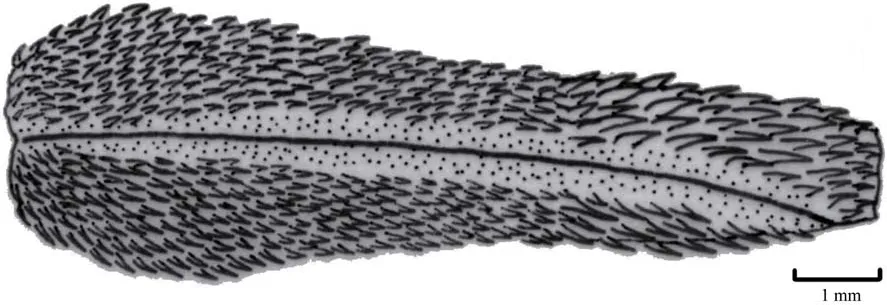
Figure 3 The left hemipenis of Oligodon multizonatum (specimen number KIZ01623). Drawing by Ke JIANG..
3.2 PhylogenyThe initial aligned data set contained 1085 bp of cyt b, 648 bp of ND4 and 564 bp of c-mos for 41 ingroup and two outgroup taxa, whereas the f i nal aligned data set contained 722 bp of cyt b, 432 bp of ND4 and 564 bp of c-mos after deleting the 3rdcodon positions of mitochondrial genes.
The topologies of trees derived from each dataset and analytical method were nearly identical (see Figures 4–5). BI and ML trees showed strong support (100% PP and 99% BS respectively) for the monophyly of Oligodon, adding some novel molecular sequence data of O. chinensis, O. formosanus, but excluding O. multizonatum. Unexpectedly, all analyses demonstrated that O. multizonatum is not part of the genus Oligodon but is instead nested within the genus Lycodon, where it is most closely related to L. liuchengchaoi in our sampling. These two species together formed a highly supported clade (100% PP and 99% BS), which are themselves sister to L. ruhstrati in the BI tree but to other clades including two species of L. butleri and L. fasciatus in ML tree. All these five species formed a monophyletic group with support values of 100% PP and 99% BS. However, most of the main nodes were not well solved within Oligodon and Lycodon in both BI and ML trees. In addition, the genetic distance (uncorrected P-distance) between O. multizonatum and L. liuchengchaoi is 0.066 (cyt b gene) and the minimum genetic distance of valid species between L. aulicus and L. capucinus is 0.047 (cyt b gene).
4. Discussion
4.1 Phylogenetic position and morphology of O. mulizonatum Although our sampling was incomplete relative to the sampling of Colubridae, multilocus phylogenetic reconstruction has indicated that all representatives from Oligodon except O. multizonatum formed a strongly supported clade, and those from Lycodon with O. multizonatum clustered into another highly supported group (Figure 1), in which the species within Lycodon and Dinodon were shown to be paraphyletic or polyphyletic. Previous studies support the conclusion that Lycodon is paraphyletic with respect to Dinodon (Siler et al., 2013). In addition, Guo et al. (2013) concluded that Lycodon and Dinodon are paraphyletic based on molecular and morphological data, and suggested synonymizing Dinodon with Lycodon. Moreover, Pyron et al. (2013) indicated the genus of Dryocalamus which had previously been identified as Lycodon nested within the group Dinodon and Lycodon, suggesting that Dinodon and Lycodon are not monophyletic. In agreement with our molecular phylogenetic results, previous morphological studies have noted the difficulty of separating Dinodon and Lycodon (Pope, 1935; Smith, 1943; Vogel and Brachtel, 2008), and thus the validity of these two genera has triggered debate. This is the reason why we sampled widely within these taxa and used different methods of analysis to make our conculsions. Our results support synonymizing the genus Dinodon and Lycodon.

Figure 4 The 50% majority-rule consensus tree from Bayesian analysis based on c-mos, cyt b and ND4 combined sequences. Values at nodes are posterior probability support values. Black bar: Lycodon; Open bar: Oligodon; Gray bar: old Dinodon.

Figure 5 Maximum likelihood inferred phylogeny of the c-mos, cyt b and ND4 combined data. Bootstrap values are shown at the corresponding nodes. Support values below 50% were not shown in this figure. Black bar: Lycodon; Open bar: Oligodon; Gray bar: old Dinodon.
Unexpectedly, our analysis indicates O. multizonatum as the sister species of L. liuchengchaoi which is clustered within Lycodon based on both mitochondrial and nuclear genes. Based on morphology, Zhao and Jiang (1981) suggested that O. multizonatum is closely related to the Indochinese O. joynsoni, and then assigned this species tothe genus Oligodon. However, there are many differences between these two species. The former species differs from the latter by having: 1) more subcaudal scales, 68–75 pairs vs 40–50 pairs; 2) a divided vs entire anal plate; 3) eight upper labial scales, the third, fourth and fifth vs the fourth and fifth touching the eye; 4) a hemipenis with spines vs without spines. The colour pattern and markings of these two species are also quite different (Zhao and Jiang, 1981). The hind teeth are also different being broad and strongly recurved, much like the shape of the kukri knife in Oligodon (Green et al., 2010), but not recurved in O. multizonatum. The recurved shaped teeth of Oligodon are used to open reptile eggs (Green et al., 2010), upon which they mainly feed. The combined morphological data also indicate that O. multizonatum is neither a close relative to O. joynsoni nor a member of the genus Oligodon.
In terms of pattern, O. multizonatum is most similar to L. liuchengzhaoi except for the fact that the number and color of bands of the former are greater and deeper than those of the latter (5–73 orange rings spaced along the black body, and 16–19 orange rings spaced along the black tail vs 40-45 well-def i ned yellow rings evenly spaced along the entire length of the black body, and more than 10–15 yellow rings evenly spaced along the black tail), but differs by the following traits: more maxillary teeth (10–11 vs 8–9), fewer ventrals (190–195 vs 202–206) (Zhao and Jiang, 1981; Zhang et al., 2011).
Therefore, we suggest that the species previously assigned to O. multizonatum needs to be transferred to Lycodon. Zhao (2006) reported that O. multizonatum feed on reptile eggs, but no analysis of the stomach content of this species has been reported. Further studies on prey types consumed by species within Oligodon and Lycodon are needed to conf i rm or provide some new evidence to support the view that most of Oligodon feed on retile eggs whereas snakes and lizards are the major food of Lycodon. Considering that the genus Dinodon has been merged into Lycodon, we suggested that the scientific name of O. multizonatum should be renamed as Lycodon multizonatum. Consistent with this we propose a new common English name, the Luding wolf snake, referring to the type locality, Luding County, China.
4.2 The validity of O. multizonatum and L.liuchengzhaoi For many species, selective or developmental constraints either prevent morphological divergence (Colborn et al., 2001) or promote convergence (Wake, 1991), complicating our understanding of group composition based on evolutionary relationships inferred from morphology (Guo et al., 2013). On the other hand, within species individual variations in morphology can make species identification difficult. A good example is Lycodon, one of the most diverse genera of Asiatic colubrids (sensustricto, see Pyron et al., 2011). Recently, L. futsingensis (Pope, 1928), which was subsequently synonymized with L. ruhstrati by Pope himself (Pope, 1935) was revalidated by Vogel et al. (2009). In 2010 and 2011, two new endemic species were described from China: L. synaptor (Vogel and David, 2010) and L. gongshan (Vogel and Luo, 2011). Based on the specimens collected from northern Sichuan Province, China, Zhang et al. (2011) described L. liuchengzhaoi. They are similar to L. fasciatus in shape and were identified as L. fasciatus previously. By careful examination of the specimens it was noticed that they could be distinguished from L. fasciatus and other species of the L. fasciatus group by several morphological characters (Vogel et al., 2009). However, O. multizonatum was not compared with these specimens. Thus it should be cautioned that O. multizonatum is the closest related specie to L. liuchengchaoi from our molecular phylogenetic analysis (Figures 4–5). Although they shared very similar morphlogical characters, such as same dorsal scale rows, loreal enters eye, same infralabial, similar temporals and similar subcaudals (Table 3), the genetic distance between these two species reached the level of interspecific differentiation. Our molecular data showed that the genetic distance (uncorrected P-distance) between O. multizonatum and L. liuchengchaoi is 0.066 (cyt b gene), which is greater than the minimum genetic distance of the valid species difference between L. aulicus and L. capucinus which is 0.047 of cyt b gene. Therefore, we strongly suggest that O. multizonatum and L. liuchengchaoi are valid as distinct species.
Currently, these two species of Lycodon are known in Sichuan Province. The three sites where L. liuchengchaoi was found are from the east of the Hengduan mountains, at the eastern edge of Qinghai-tibet Plateau. O. multizonatum is known only from Luding county, Sichuan Province, Tianshui and Kang counties, Gansu Province. These specimen records and published literature suggest that O. multizonatum might be distributed in the middle eastern and northern edge of Hengduan mountains which is sympatric with the L. liuchengchaoi. However, it is interesting to notice that the specimen of O. multizonatum sourced from Gansu province had fewer rings in its body on the photograph (Zhao, 2006) is rather similar to L. liuchengchaoi. Therefore, we suggest the distribution of O. multizonatum in Gansu province might be questionable.
Unfortunately, the molecular phylogeny presented here did not resolve the relationships among Lycodon and Dinodon. Considering the morphological and phylogenetic results in this study, we suggest future studies need to add more markers to resolve the relationships among Lycodon and Dinodon.
Acknowledgements This work was supported by the National Natural Science Foundation of China (NSFC 31071913).
Arévalo E., Davis S. K., Sites J. W. 1994. Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central Mexico. Syst Biol, 43: 387–418
Branch W. R. 1986. Hemipenial morphology of African snakes: a taxonomic review, Part I Scolecophidia and Boidae. J Herpetol, 20 (3): 285–299
Burbrink F. T., Castoe, T. A. 2009. Molecular Snake phylogeography. In Mullin S., and Seigel R. (Eds.), Snakes: Applied ecology and conservation. Ithaca: Cornell University Press, 33–77
Burbrink F. T., Lawson, R., Slowinski J. B. 2000. Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): A critique of the subspecies concept. Evolution, 54: 2107–2118
Campden S. M. 1969. The status of Oligodon taeniatus (Guenther), 1861, and Oligodon mouhoti (Boulenger), 1914 (Serpentes, Colubridae). Herpetologica, 25: 295–299
David P., Vogel G., Rooijen J. 2008. A revision of the Oligodon taeniatus (Gunther, 1861) group (Squamata: Colubridae), with the description of three new species from the Indochinese region. Zootaxa, 1965: 1–49
Dowling H. G. 1951. A proposed standard system of counting ventrals in snakes. Brit J Herpetol, 1: 97–99
Dowing H. G., Savage J. M. 1960. A guide to snake hemipenis: A survey of basic structure and systematic characteristics. Zoologica, 45: 17–28
Erixon P. B., Svennblad T. B., Oxelman B. 2003. The reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol, 52: 665–673
Green M. D., Orlov N. L., Murphy R. W. 2010. Toward a Phylogeny of the Kukri Snakes, Genus Oligodon. Asian Herpetol Res, 1: 1–21
Guo P., Zhang L., Liu Q., Li C., Pyron R. A., Ke J., Burbrink F. T. 2013. Lycodon and Dinodon: One genus or two? Evidence from molecular phylogenetics and morphological comparisons. Mol Phylogenet Evol, 68: 144–149
Huelsenbeck J. P., Ronquist F. 2001. MrBayes 2.01 (Bayesian analysis of phylogeny)
Jiang K. 2010. A method for evaginating the hemipenis of preserved snakes. Sichuan J Zool, 29 (1): 122–123 (In Chinese)
Kishino H., Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. Mol Phylogenet Evol, 29: 170–179
Kishino H., Hasegawa M. 1999. Multiple comparisons of loglikelihoods with applications to phylogenetic inference. Mol Phylogenet Evol, 16: 1114–1116
Lawson R., Slowinski J. B., Crother B.I., Burbrink F. T. 2005. Phylogeny of Colubroidea (Serpentes): New evidence from mitochondrial and nuclear genes. Mol Phylogenet Evol, 37: 581–601
Leviton A. E. 1963. Contribution to a review of Philippine snakes, I. The snakes of the genus Oligodon. Philipp J Sci, 91: 459–484
Huang S., Liu S. Y., Guo P., Zhang Y. P., Zhao E. M. 2009. What are the closest relatives of the hot-spring snakes (Colubridae, Thermophis), the relict species endemic to the Tibetan Plateau? Mol Phylogenet Evol, 51: 438–446
Pesantes O. S. 1994. A method for preparing the hemipenis of preserved snakes. J Herpetol, 28: 93–95
Pope C. H. 1935. The reptiles of China. Turtles, crocodilians, snakes, lizards. Natural History of central Asia, Vol. X. American Museum of Natural History, New York
Posada D., Crandall K. A. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics, 14: 817–818
Pyron R. A., Kandambi H. K. D., Hendry C. R., Pushpamal V., Burbrink F. T. Somaweera R. 2013. Genus-level molecular phylogeny of snakes reveals the origins of species richness in Sri Lanka. Mol Phylogenet Evol, 66: 969–975
Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: A laboratory manual, 2ndEd. New York: Cold Spring Harbor Laboratory Press
Siler C. D., Oliveros C. H., Santanen A., Brown R. M. 2013. Multilocus phylogeny reveals unexpected diversif i cation patterns in Asian wolf snakes (genus Lycodon). Zool Scr, 42(3): 262–277
Slowinski J. B., Lawson R. 2002. Snake phylogeny: Evidence from nuclear and mitochondrial genes. Mol Phylogenet Evol, 23: 194–202
Smith M. A. 1943. The fauna of British India including Ceylon and Burma. Reptilia and Amphibia, Vol. III, Serpentes. London: Taylor and Francis, 583
Stamatakis A. 2005. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688–2690
Swofford D. L. 2003. PAUP*bv10: Phylogenetic analysis using parsimony (*and other methods) v. 4. Sinauer Assoc, Sunderland, MA
Tamura K., Dudley J. Nei. M., Kuma, S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol, 24, 1596–1599
Tillack F., Günther R. 2009. Revision of the species of Oligodon from Sumatra and adjacent islands with comments on the taxonomic status of Oligodon subcarinatus (Günther, 1872) and Oligodon annulifer (Boulenger, 1893) from Borneo (Reptilia, Squamata, Colubridae). Russ J Herpetol, 16: 265–294
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res, 25: 4876–4882
Vogel G., Brachtel N. 2008. Contribution to the knowledge of Lycodon ruhstrati (Fischer, 1886) in Vietnam – taxonomy and biology of a little-known species. Salamandra, 44: 207–224
Wall F. 1923. A review of the Indian species of the genus Oligodon suppressing the genus Simotes (Ophidia). Calcutta: Records of the Indian Museum, 25: 305–334
Wallach V., Bauer A. M. 1996. On the identity and status of Simotes semicinctus Peters, 1862 (Serpentes: Colubridae). Hamadryad, 21: 13–18
Zhang F. J., Hu S. Q., Zhao E. M. 1984. Comparative studies and phylogenetic discussion on hemipenial morphology of the Chinese Colubrinae (Colubridae). Acta Herpetol Sinica, 3(3): 23–44
Zhang J., Jiang K., Vogel G., Rao D. Q. 2011. A new species of the genus Lycodon (Squamata, Colubridae) from Sichuan Province, China. Zootaxa, 2982: 59–68
Zhao E. M., Huang M. H., Zong Y. 1998. Fauna Sinica: Reptilia, Vol. 3: Squamata: Serpentes). Beijing: Science Press (In Chinese)
Zhao E. M., Jiang Y. M. 1981. Studies on amphibians and reptiles of Mt. Gongga Shan, Sichuan, China, I. A new species and a new subspecies of snakes from Sichuan. Acta Herpetol Sinica, 5 (7): 53–58 (In Chinese)
Zhao E. M. 2006. Snakes of China. Hefei: Science Press, 232 (In Chinese)
10.3724/SP.J.1245.2014.00026
*Corresponding author: Dr. Li DING, from Chengdu Institute of Biology, Chinese Academy of Sciences, Sichuan, China, with his research focusing on taxonomy and systematics of snakes, molecular phylogeny and phylogeography of reptiles, faunal survey and biodiversity, animal behavior of reptile, and conservation and public awareness of snakes.
E-mail: dingli@cib.ac.cn
31 December 2013 Accepted: 11 March 2014
Asian Herpetological Research 2014, 5(1): 26–37
杂志排行
Asian Herpetological Research的其它文章
- Molecular Assessment and Taxonomic Status of the Rapid Racerunner (Eremias velox complex) with Particular Attention to the Populations in Northwestern China
- Molecular Characterization and Expression Analysis of Matrix Metalloproteinase 3 in the Asian Yellow Pond Turtle Mauremys mutica
- Structure Organization of Urinary System in the Yellow Spotted Mountain Newts (Salamandridae: Neurergus microspilotus)
- A New Species of the Genus Tylototriton (Urodela: Salamandridae) from Northeastern Hunan Province, China
- Body Surface Area Prediction in Odorrana grahami
- Preliminary Insights into the Habitat Preferences of the Centralian Bandy Bandy (Vermicella vermiformis) (Squamata: Elapidae) in Central Australia
