16S rRNA基因测序技术在肝脓疡细菌鉴定中的作用
2014-06-09宋伦圭
宋伦圭
·临床研究Clinical research·
16S rRNA基因测序技术在肝脓疡细菌鉴定中的作用
宋伦圭
目的评价16S核糖体RNA(rRNA)基因测序在肝脓疡中细菌鉴定中的应用价值。方法2012年1月—2013年12月间共20例肝脓疡行经皮置管引流的患者,分别行脓液培养,血培养和16S rRNA基因测序。利用454 GS Junior System对脓液基因组DNA行PCR和16S rRNA基因测序。脓液培养,血液培养和16S rRNA基因测序结果进行分别评价。结果脓液和血液培养阳性的患者分别是9例(45%)和4例(20%)。16S rRNA基因测序细菌鉴定率为90%,明显高于传统的培养方法。结论16S rRNA基因测序方法较传统的培养方法能更准确和有效对肝脓疡进行细菌鉴定。
肝脓疡;脓肿培养;宏基因组学;16S rRNA基因测序
INTRODUCTION
A liver abscess is still a severe disease with considerable mortality[1-3].It is often polymicro-bial(PLA).Advanced imaging tools such as high resolution computed tomography(CT)and ultrasound device have improved a possible early diagnosis for PLA[4].Actually,the treatment regimen is changing.Recently,the trend regarding initial treatment choice for PLA has changed into antibiotics and radiological intervention[3-4].Therefore,an antibiotic treatmentwith an accurate identification of bacteria is the basis for PLA treatment.
Traditionally,the identification of bacteria was performed using phenotypic tests,including gram smear and biochemical tests as well as blood or abscess cultures.However,there are some limitations such as rare bacteria,slow-growing bacteria,uncultivable bacteria and culture-negative infection in those traditional identification methods.So the clinicians may have problems in the choice and duration of antibiotic treatments.
In the last decades,molecular-based tools with invention of PCR and DNA sequencing have been employed in the diagnostic fields.Nowadays,the most commonly used and commercially available method for the bacterial identification in clinical laboratories is 16S ribosomal RNA(rRNA)gene sequencing[5-8].It has provided immense information regarding to microbial communities in human and in variety of animals with healthy or pathological conditions.However,the gene sequencing method is not routinely used in hospitals.
Therefore,the purpose of our study was to prospectively evaluate the usefulness of 16S rRNA gene sequencing for more accurate and better identification of the causative aetiology of PLA.
METHODOLOGY
Patients
This prospective study was reviewed and approved by our Institutional Review Board and written informed consent was obtained from all participants.Twenty two patients(10 men and 12 women)at the Samsung ChangWon hospital were initially considered for the study between January 2012 and December 2013.Two patients with hepatic abscess were excluded because the abscess was not large enough to be considered for percutaneous drainage.Finally,20 patients were enrolled in this study.There were 9 male(45%)and 11 female(55%)patients.Themean age(±standard deviation)at the time of examination was 61.84±13.21 years(age range 33-83 years).
The diagnosis of PLA wasmade on the basis of clinical and imaging findings with ultrasound or CT.We performed percutaneous drainage and antibiotic treatment in patients with pyogenic liver abscess in our hospital.All patients started with antibiotics prior to percutaneous abscess drainage.A blood sample was taken and sent to culture before the administration of antibiotics.In all cases,the time interval between antibiotic administration and drainage was less than 8 hours.
Intervention
All percutaneous procedures were performed under ultrasound(Acuson×300,Sequoia 512,Siemens,USA)guidance.18 G P.T.C Chiba needle(Unimed,Lausanne,Switzerland)with varying lengths was used for puncturing the abscess.The aspirated isolates including cultures were sent for microbiological analysis and 16S rRNA gene sequencing.Under fluoroscopic guidance,2-4m l of undiluted contrast media was instilled into the abscess cavity through the 18 G needle and then a 0.035-inch wire(Terumo,Tokyo,Japan)was inserted into the abscess cavity.After serial dilatation,an 8.5 Frenchpigtail catheter(CooK,Bloomington,In,USA)was inserted into the abscess(Fig.1).An ultrasonography of the liver performed 1-2 weeks after the insertion of a percutaneous drainage catheter.The drain catheter was removed when the abscess cavity had collapsed on follow-up ultrasonography and catheter output had decreased<10 ml daily.

Fig.1 43-year-old male with pyogenic liver abscess treated by insertion of percutaneous drainage catheter
Preparation of Genom ic DNAs from Abscess effusions
Genomic DNAs from abscesseswere prepared by the modification method described previously[9].The abscess effusions of 500 ul were mixed by the same volume of TE buffer(pH 7.0).The abscess diluents were added with 15 U mutalysin(Sigma,St.Louis,MO)and 600 ug lysozyme(Genery,Shanghai,China)and then incubated for 1 hour at 37℃.The mixtures were treated with 10 ul 10%SDS solution and 20 ug RNase(RBC,Taipei,Taiwan)for 1 hour at 37℃and then with 120 ug protease K(GeNet Bio,Daejeon,Korea)for 1 hour at 37℃.Thereafter,the mixtures were added with 1/10 volume of 5% cetyltrimethylammonium bromide(BDH Chemicals Ltd.,Poole,England)-0.5 M NaCl solution and allowed to stand for 1 hour at 37℃.The mixtures were added with the same volume of phenolchloroform-isoamyl alcohol(25∶24∶1)solution and vigorously vortexed.
The mixtures were centrifuged for 10 minutes at 12 000 rpm and aqueous layers were subjected to choloroform extraction.After centrifugation the aqueous phase was collected and added with 1/10 volume of 3 M sodium acetate(pH 5.2)and 2 volumes ethanol.The sample tubes were stored at -20℃for 20 minutes and centrifuged for 10 minutes at12 000 rpm.The pelletswere washed with 1m l of 70%ethanol,completely dried and dissolved with 30 ul of TE buffer.
Polym erase chain reaction
Extracted DNA of 60 ng was subjected to PCR amplification of the V1-V3 region of the 16S rRNA gene using AccuPower PCR PreMix(Cat.No K-2016,BiONEER,Daejon,Korea)containing 1 unit of Top DNA polymerase,1 mM dNTPs,10 mM Tris-HCl[pH9.0],30 mM KCl,and 1.5mM MgCl2with 27 F primer(5′-GAGTTTGATCCTGGCTCAG-3′)and 518R primer(5′-ATTACCGCGGCTGCTGG-3′).The 5’ends of forward primers were subsequently attached with an adaptor 1 sequence(5′-CCATCTCTCCCTGCGTGTCTCCGAC-3′),a key sequence(TCAG),and sample-specific multiplex identifiers sequences.Reverse primer was added with an adaptor 2 sequence(5′-CCTATCCCCTGTGTGCCTTGGCAGTC-3′)and a key sequence(TCAG).The PCR reactions were carried out by a pre-denaturation for 4 seconds at 94℃and 20 cycles of 30 seconds at 94℃,30 seconds at 55℃and 30 seconds at 72℃and then followed by a final extension of 7 minutes at 72℃.Amplified DNAs were separated with 1%agarose gel and purified with GeneALL®ExpinTM PCR SV(Geneall Biotechnology Co.Ltd.,Seoul,Korea).
Pyrosequencing and Data Analysis
The barcoded amplified DNAs of 20 samples weremixed with the same concentrations and applied to 454 pyrosequencing.Pyrosequencing was unidirectionally performed from the 27 F primer end on a 454 GS Junior System platform in a single fullplate run.These SFF-files of primary sequencing reads were then de-multiplexed based on the MIDs.Subsequently,MIDs,U-linkers and primers were trimmed away and the sequence quality was filtered.For 16S rRNA sequences,de-multiplexing,trimming and quality filtering was done using AmpliconNoise[10].16sRNA sequences were identified using the Basic Local Alignment Search Tool(BLASTBN)algorithm and the National Center for Biotechnology Information(NCBI)non-redundant(NT)sequence database.The alignment results were further processed by the Metagenome Analyzer(MEGAN)program to statistically analyze the abundance ofmicroorganismsin each sample.
Statistical analysis
Results are presented as arithmetic means(ranges)and its associated standard deviations.Categorical data were expressed as the number of subjects with clinical variables and its corresponding percentages.
RESULTS
Blood and abscess cultures were processed for all 20 patients.Abscess cultures were positive in 9(45%)patients;in which 5(55.6%)were monomicrobial and 4(44.4%)were polymicrobial(≥2 organisms isolated).Klebsiellapneumoniawasmost commonly isolated from the abscess cultures.No culture was obtained in 11 patients.Blood cultures were positive in 4(20%)patients and 50%of these presented asKlebsiellapneumonia.Two patients had concordant blood and abscess culture results and two had positive blood cultures with negative cultures in the abscess(Table 1).

Table 1 Culture Results
Genomic DNA was extracted from purulent fluids of abscesses.Of 63 207 reads remained after the processing,46 261 reads(73.2%)were assigned in the existing 16S rRNA database.Other 16 937 reads(26.8%)were not assigned and couldn’t be classified into known bacteria.Therefore they might represent novel lineages.Of a total of 46,261 assigned reads was Klebsiella themostabundant genus(74.7%)and the remained reads were classified to the genusFusobacterium(9.3%),Streptococcus(5.0%),Bacteroides(1.9%),Prevotella(1.5%),Peptostreptococcus(0.1%),unassignedEnterobacteriaceae(0.1%)andDialister(0.1%),respectively.
Of 20 abscess sampand better identificaabscess,12 samples(01 F,02 F,06 F,07 F,08 F,10 F,13 F,14 F,15 F,17 F,18 F,and 20 F)were exclusively occupied withKlebsiallaspecies(more than 96%of reads)and 1 sample(09 F)was exclusively contained withFusobacteriumwhen excepted the unassigned and no hit reads.The sample 03F was composed ofPrevotella(40.3%),Streptococcus(23.2%),Peptostreptococcus(4.0%),Dialister(3.4%)andEnterobacteriaceae(3.5%).The sample 04 F was composed ofKlebsiella(7.6%),Bacteroides(8.5%),Fusobacterium(44.2%)andStreptococcus(23.2%).The sample 05 F was composed ofBacteroides(37.8%)andStreptococcus(60.7%).The sample 11 F was composed ofFusobacterium(77.7%)andPrevotella(10.6%)and sample 19 F was composed ofFusobacterium(5.1%)andStreptococcus(14.0%).Two samples(12 F and 16 F)did not contain any assigned bacteria(Table 2).
DISCUSSION
Recently there have been advances in interventional radiology,ICU care,antibiotics,culture technique and imaging devices in the diagnosis and treatment of PLA.However,it is still a life-threatening disease[1-4].
The most frequently isolated microorganisms were the polymicrobialE.coliandStreptococcusspp.and were polymicrobial[11-12].Although the cause is mostly a cryptogenic abscess,a highly virulentK. pneumoniaehad emerged as a predominant cause of PLA in Asian countries and areas recently[13-16].Also,K.pneumoniaewas the predominant cause of PLA in our study.
In our study,blood cultures were positive in 20%and pus cultures were positive in 45%of patients.Several studies revealed before positive blood cultures10.2%-55%and pus cultures in 48%-85%(4,17,18).Usually,the positive rate of abscess cultures is higher than that of blood cultures[3].Thecausative bacterial identification was not completely obtained in the blood and pus cultures in the present study.We believe such resultsmay occur due to the previous use of antibiotics and pure pus(neutrophils)collected abscesses.PLA remains as a serious illness with a considerable morbidity and mortality until today[4,19].In our study,the hospitalmortality rate was zero.However,in previous studies,the PLA mortality rate remained high at 9%-25%[11,17].Therefore,an accurate and rapid identification of bacterial isolates and targeted antibiotics are needed to control PLA.
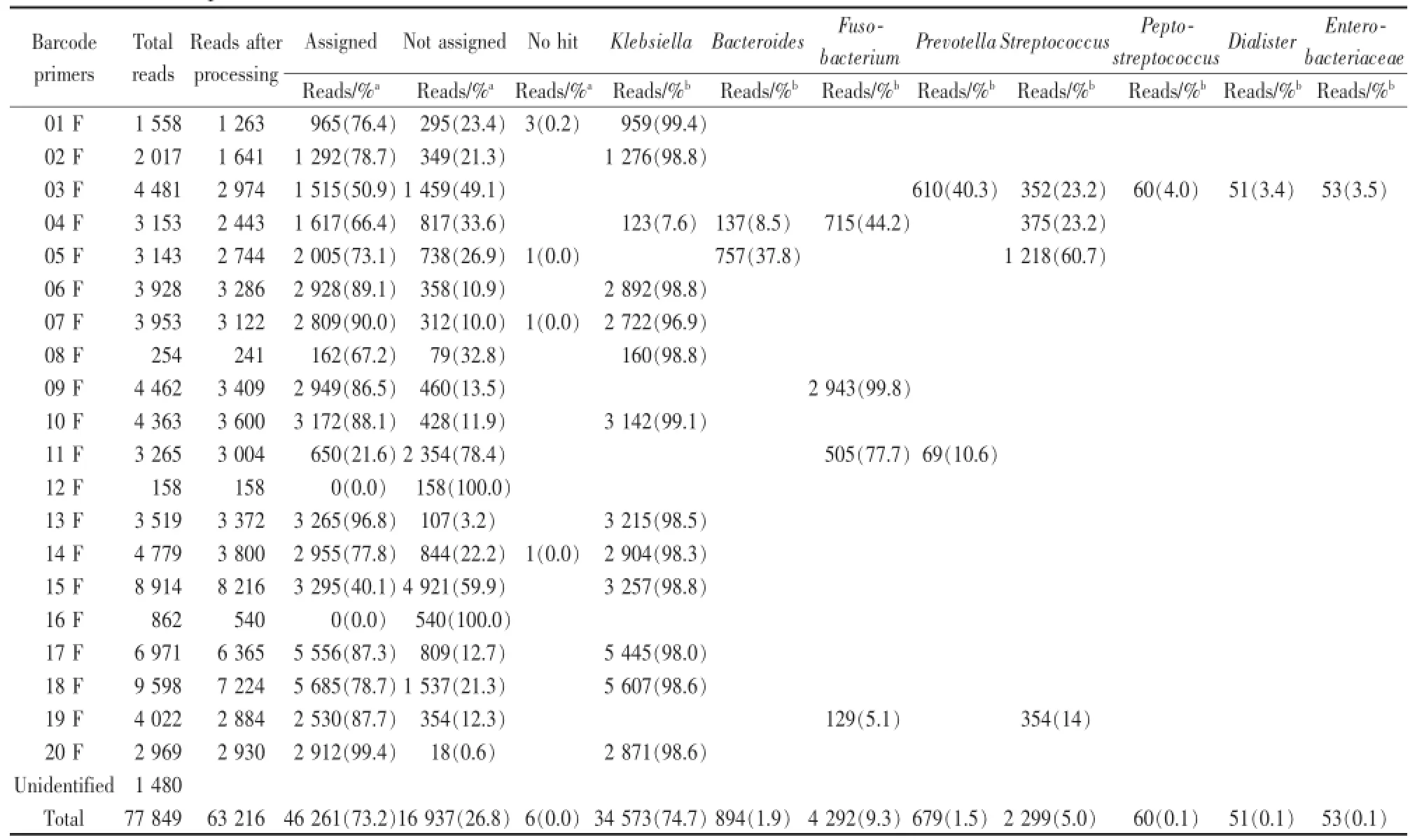
Table 2 Pyrosequencing of the 16S rRNA genes in genomic DNAs extracted from liver abscesses on the 454GSJunior System and classification and species richness of the bacteria in liver abscesses
Nowadays,conventional culture-based methods are used for the identification of bacteria in most hospitals.Themethods are relatively inexpensive,but they have some limitations regarding rare,slowgrowing and uncultivable bacterial identifications.Also,an identification of some particular bacteria,such as anaerobes and mycobacteria,would require additional equipment and expertise that are not available in most of hospitals.Therefore the clinicians have problems in the choice and duration of antibiotic treatments and the appropriate infection control procedures.Non-culture-based techniques(metagenomics)are required.There already exist identification methods by hybridization and sequencing.The hybridization method will be a valuable tool for bacterial identification,but the default of themethod is to identify what an unknown isolate is.Sequencing is a more powerful molecular identification method with the increasing availability of DNA sequencing and PCR[20-24].
Today,16S rRNA gene sequencing is used in clinical laboratories for routine identifications,especially for slow-growing,unusual or fastidious bacteria and also poorly differentiated bacteria by conventional methods.In our study,the 16S rRNA gene sequencing showed 90%identification of bacteria with unusual bacteria such as Dialister.However,the conventional methods showed positive in 20%and 45%,respectively.So this method can provide more definite taxonomic classification than culture-based approaches formany organisms[25-28].
The 16S rRNA gene exits universally among bacteria and includes regions with species specific variability.Also the 16S rRNA function hasconstantly remained over a long period.Therefore,16S rRNA gene sequencing makes it possible to identify bacteria to its genus or species level by comparison with databases in the public domain[20,25,29,30].Also,previous authors have reported its use as a tool for bacterial identification[27,31-37].This approach has been demonstrated to improve a more rapid recognition of novel isolates and the accuracy of organism identification compared to non-molecular testing[20,33,38].
In our study,there was a number of unassigned reads with an average of 26.8%per sample.This unassigned reads were not identified because these sequences were missing from the database.It suggests that novel lineages of pathogens might be one of the pathogenic members responsible for provoking and maintaining the polymicrobial environment contributing to an arising liver abscess.Staphylococcus caprae and staphylococcus epidermidis were cultured in blood and abscess isolates.We do not think that they were causative agents,but rather were a result of a contamination during aspiration procedure.A great number of bacterial species were inhabited in the superior and inferior surfaces of the human body[39].However,limited bacterial species are found in a liver abscess only.It demonstrated that rare species of bacteria hold pathogenic mechanisms for evading or breaking host defense barriers to form an abscess in deep tissues like in the liver.So,the profiling of pathogenic species responsible for forming various tissue abscesses is prerequisite to understand etiologies and to provide new therapeutic approaches against abscesses.
There aremany requirements for this 16S rRNA gene sequence method;i.e.reagent,instrumentation for amplification and sequencing,a database of known sequences and software for sequence editing and database comparison.However,commercial reagents are available,laboratory-developed assays for amplification and sequencing have been reported and there is an increasing number of commercial and public databases[20].
Our study had several limitations.First,16S rRNA gene sequencing provides no information about antibiotic resistance.Second,gene sequencing is a relatively expensive method of identification. However,an introduction ofmore automated methods will decrease the costs of the sequencing method. Third,this method requires accurate and complete genetic databases.
In conclusion,this study showed the greater usefulness of 16S rRNA sequencing than of conventional cultured methods formore accurate and better identification of bacteria in patients with pyogenic liver abscess.
[REFERENCES]
[1]Lee KT,Wong SR,Sheen PC.Pyogenic liver abscess:an audit of 10 years’experience and analysis of risk factors.Dig Surg 2001;18:459-465;discussion 465-456.
[2]Yu SC,Ho SS,Lau WY,et al.Treatment of pyogenic liver abscess:prospective randomized comparison of catheter drainage and needle aspiration.Hepatology 2004;39:932-938.
[3]Cerwenka H.Pyogenic liver abscess:differences in etiology and treatment in Southeast Asia and Central Europe.World J Gastroenterol 2010;16:2458-2462.
[4]Malik AA,Bari SU,Rouf KA,et al.Pyogenic liver abscess:Changing patterns in approach.World JGastrointest Surg 2010;2:395-401.
[5]Kolbert CP,Persing DH.Ribosomal DNA sequencing as a tool for identification of bacterial pathogens.Curr Opin Microbiol 1999;2:299-305.
[6]Fredericks DN,Relman DA.Sequence-based identification of microbial pathogens:a reconsideration of Koch’s postulates.Clin Microbiol Rev 1996;9:18-33.
[7]Kiratisin P,Li L,Murray PR,et al.Identification of bacteria recovered from clinical specimens by 16S rRNA gene sequencing.Eur J Clin Microbiol Infect Dis 2003;22:628-631.
[8]Woo PC,Lau SK,Teng JL,et al.Then and now:use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.Clin Microbiol Infect2008;14:908-934.
[9]Holder RC,Kirse DJ,Evans AK,etal.One third ofmiddle ear effusions from children undergoing tympanostomy tube placement had multiple bacterial pathogens.BMC Pediatr 2012;12:87.
[10]Quince C,Lanzen A,Davenport RJ,etal.Removing noise from pyrosequenced amplicons.BMC Bioinformatics2011;12:38.
[11]Lopez-Cano Gomez M,Laguna Del Estal P,Garcia Montero P,et al.[Pyogenic liver abscess:clinical presentation and predictors of unfavorable outcome].Gastroenterol Hepatol 2012;35:229-235.
[12]Corbella X,VadilloM,Torras J,et al.[Presentation,diagnosis and treatment of pyogenic liver abscess:analysis of a series of 63 cases].Enferm Infecc Microbiol Clin 1995;13:80-84.
[13]Liu Y,Wang JY,Jiang W.An Increasing Prominent Disease of Liver Abscess:Etiology,Diagnosis,and Treatment. Gastroenterol Res Pract 2013;2013:258514.
[14]Li J,Fu Y,Wang JY,et al.Early diagnosis and therapeutic choice of Klebsiella pneumoniae liver abscess.Front Med China 2010;4:308-316.
[15]Siu LK,Fung CP,Chang FY,et al.Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samp les from noninfectious subjects in Hong Kong,Singapore,and Taiwan.J Clin Microbiol 2011;49:3761-3765.
[16]Chung DR,Lee H,Park MH,et al.Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea.Eur JClin Microbiol Infect Dis 2012;31:481-486.
[17]Zibari GB,Maguire S,Aultman DF,et al.Pyogenic liver abscess.Surg Infect(Larchm t)2000;1:15-21.
[18]Ali AH,Smalligan RD,Ahmed M,etal.Pyogenic liver abscess and the emergence of Klebsiella as an etiology:a retrospective study.Int JGen Med 2013;7:37-42.
[19]Ochsner A,DeBakey M,Murray S.Pyogenic abscess of the liver:II.An analysis of forty-seven cases with review of the literature.The American Journal of Surgery 1938;40:292-319.
[20]Patel JB.16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory.MolDiagn 2001;6:313-321.
[21]Lebrun L,Espinasse F,Poveda JD,et al.Evaluation of nonradioactive DNA probes for identification of mycobacteria.J Clin Microbiol 1992;30:2476-2478.
[22]Bourbeau PP,Heiter BJ,Figdore M.Use of Gen-Probe AccuProbe Group B streptococcus test to detect group B streptococci in broth cultures of vaginal-anorectal specimens from pregnant women:comparison with traditional culture method.JClin Microbiol 1997;35:144-147.
[23]Denys GA,Carey RB.Identification of Streptococcus pneumoniae with a DNA probe.J Clin Microbiol 1992;30:2725-2727.
[24]Padhye AA,Smith G,McLaughlin D,et al.Comparative evaluation of a chemiluminescent DNA probe and an exoantigen test for rapid identification of Histoplasma capsulatum.J Clin Microbiol 1992;30:3108-3111.
[25]Mignard S,Flandrois JP.16S rRNA sequencing in routine bacterial identification:a 30-month experiment.J Microbiol Methods 2006;67:574-581.
[26]Salipante SJ,Sengupta DJ,Rosenthal C,et al.Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections.PLoSOne 2013;8:e65226.
[27]Petti CA,Polage CR,Schreckenberger P.The role of 16S rRNA gene sequencing in identification ofmicroorganismsmisidentified by conventional methods.J Clin Microbiol 2005;43:6123-6125.
[28]Clarridge JE,3rd.Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases.Clin Microbiol Rev 2004;17:840-862,table of contents.
[29]Vandamme P,Pot B,Gillis M,et al.Polyphasic taxonomy,a consensus approach to bacterial systematics.Microbiol Rev 1996;60:407-438.
[30]Woese CR.Bacterial evolution.Microbiol Rev 1987;51:221-271.
[31]Bosshard PP,Abels S,Zbinden R,et al.Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory(an 18-month evaluation).JClin Microbiol 2003;41:4134-4140.
[32]Bosshard PP,Abels S,Altwegg M,et al.Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory.J Clin Microbiol 2004;42:2065-2073.
[33]Tang YW,Ellis NM,Hopkins MK,et al.Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli.JClin Microbiol 1998;36:3674-3679.
[34]Tang YW,Von Graevenitz A,Waddington MG,et al. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis.JClin Microbiol 2000;38:1676-1678.
[35]Ferroni A,Sermet-Gaudelus I,Abachin E,et al.Use of 16S rRNA gene sequencing for identification of nonfermenting gramnegative bacilli recovered from patients attending a single cystic fibrosis center.JClin Microbiol 2002;40:3793-3797.
[36]Drancourt M,Berger P,Raoult D.Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans.JClin Microbiol 2004;42:2197-2202.
[37]Coenye T,Goris J,Spilker T,et al.Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen.nov.,sp.nov. JClin Microbiol 2002;40:2062-2069.
[38]Patel JB,Leonard DG,Pan X,et al.Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system.JClin Microbiol 2000;38:246-251.
[39]Huse SM,Ye Y,Zhou Y,et al.A core human microbiome as viewed through 16S rRNA sequence clusters.PLoSOne 2012;7:e34242.
(Recived:2014-07-15)
(Editor:yuanzheng)
Usefulness of 16S rRNA Gene Sequencing for Identification of Bacteria from Pyogenic Liver Abscess
Yun Gyu Song,M.D.,Sang Gun Shim,M.D.,Ph.D.,Kwang Min Kim,M.D.,Dae-Soo Kim,Ph.D.,Sang-Haeng Choi,Ph.D.,Jae-Young Song,Ph.D.,Kon-Ho Lee,Ph.D.,Hyung-Lyun Kang,Ph.D.,Seung-Chul Baik,M.D.,Ph.D.,Woo-Kon Lee,V.M.D.,Ph.D.,Myung-Je Cho,V.M.D.,Ph.D.,Kwang-Ho Rhee,M.D.,Ph.D.,Dong-Hae Lee. Department of Radiology;Department of Internal Medicine,Samsung Changwon Hospital,Sungkyunkwan University School of Medicine,Changwon,Korea;Human Derived Material Center,Korea Research Institute of Bioscience and Biotechnology,Daejeon,Korea;Unitech Science Co.,Ltd.,Daejeon,Korea;Department of Microbiology,Gyeongsang National University College of Medicine,Jinju,Korea
Myung-JeCho,V.M.D.,Ph.D.,E-mail:mjecho@gnu.ac.kr
Background/Aim s To evaluate the usefulness of 16S ribosomal RNA(rRNA)gene sequencing for an accurate and better identification of bacteria from pyogenic liver abscess(PLA). M ethodology 20 patients with PLA were included who underwent percutaneous catheter drainage,abscess culture,blood culture and 16S rRNA gene sequencing for isolates from January 2012 to December 2013. Genomic DNAs of abscess fluidswere subjected to PCR and sequencing of 16S rRNA gene by on a 454 GS Junior System.The resultswere evaluated between abscess cultures,blood and 16S rRNA gene sequencing for isolates.ResultsAbscess and blood cultureswere positive in 9(45%)and 4(20%)patients,respectively. The 16S rRNA gene sequencing showed with 90%identification of bacteria a significantly greater identification than conventional cultured methods.ConclusionThis study showed a greater usefulness of 16S rRNA gene sequencing than conventional cultured methods for accurate and better identification of bacteria from PLA.(J Intervent Radiol,2014,23:906-912)
Pyogenic liver abscess;Abscess culture;Metagenomics;16S rRNA gene sequencing
R575.4
B
1008-794X(2014)-10-0906-07
10.3969/j.issn.1008-794X.2014.10.017
韩国庆尚南道昌原市三星昌原医院,成均馆大学医院
Myung-Je Cho,V.M.D.,Ph.D.E-mail:m jecho@gnu.ac.kr
