Method for Isolating Mitochondrial DNA from Etiolated Tissue of Cabbage
2014-03-07WangShuaiWangChaoandZhangXiaoxuan
Wang Shuai, Wang Chao, and Zhang Xiao-xuan
College of Horticulture, Northeast Agricultural University, Harbin 150030, China
Method for Isolating Mitochondrial DNA from Etiolated Tissue of Cabbage
Wang Shuai, Wang Chao*, and Zhang Xiao-xuan
College of Horticulture, Northeast Agricultural University, Harbin 150030, China
Isolation of high-quality mitochondrial DNA (mtDNA) is an important premise for researching molecular mechanisms in cytoplasmic male sterility of cabbage (Brassica oleracea L.var.capitata). An efficient protocol for separation and purification of mitochondria and extraction of mitochondrial DNA (mtDNA) from etiolated tissues of cabbage was developed. We took a method combined mannitol density gradient with differential centrifugation, selected appropriate rotational speed, extended DNase I treating time and changed mitochondria cracking condition. The results showed that the extracted mitochondria in this protocol had complete structure, appeared to ellipsoid and had not been contaminated with other impurities under the Jannus Green B staining. The isolated mitochondrial DNA had high purity and yield through detecting the optical density, nuclear specific primer PCR and agarose gel electrophoresis. The results indicated that mitochondrial DNA extracted by this protocol had high quality and enabled to be used in futher genetic studies.
cabbage, cytoplasmic male sterility (CMS), mitochondrial DNA (mtDNA), isolation
Introduction
Cytoplasmic male sterility (CMS) is expressed as a maternally inherited trait in more than 150 plant species whereby plants inability to produce functional pollen attribute to mitochondrial dysfunction during meiosis or microsporogenesis, although vegetative development is unaffected (Hanson 1991; Schnable et al., 1998; Zhang and Stewart 2001; Dieterich et al., 2003). As a convenient and economic method, CMS plays an important role in the production of hybrid seeds in cabbage (Fang et al., 2001) since it prevented self-fertilization, while production of others depended upon manual or mechanical emasculation.
So far, there are plenty of evidence to support that mitochondrial DNA rearrangements lead to novel loci which can be responsible for CMS (Janska et al., 1998; Gong et al., 2006; Shedge et al., 2007). This phenomenon of mitochondrial DNA rearrangements occurs in many plant species and is often originating from these altered open reading frames which were translated into unique proteins and then interfered with mitochondrial function and pollen development, as a result, leading to CMS (Schnable et al., 1998; Fujii et al., 2009). Meanwhile, some scholars proved that there were differences between amplification products of sterile line and its maintainer line in quantity and quality (Liu et al., 2004; Zhang et al., 2012; Wang et al., 2012).
Many studies are focused on the difference of mitochondrial genome between sterile line and maintainerline in order to provide theoretical basis for the mechanism of CMS (Li et al., 2011; Jee et al., 2013). Therefore, how to extract high-quality mtDNA quickly and easily becomes the key to research on the molecular mechanism of CMS in cabbage (Brassica oleracea L.var.capitata).
However, because of the interference of cell walls, big vacuole and chloroplast DNA in plant cells, it becomes more difficult for mtDNA separating. Traditional mtDNA extraction procedures mainly included density gradient centrifugation and differential centrifugation (Scotti et al., 2001; Tanaka et al., 2011). It is easy for differential centrifugation to extract a large amount of mtDNA, but has the problem including low purity and contamination of nuclear genes or chloroplast genome. Density gradient centrifugation also has some disadvantages, such as timeconsuming, laborious and high cost. The most popular method currently used in extraction of mtDNA is a combined method of sucrose density gradient and differential centrifugation, and has been reported at many species, such as potato (Scotti et al., 2001), rice (Pei et al., 2002), kenaf (Zhao et al., 2011) and Chinese cabbage (Zhang et al., 2006; Li et al., 2011). Compared with many studies on other species, there are few reports on mtDNA extraction of cabbage.
For mtDNA extraction in different plants, influence and constraint by various complicated factors in cells, the methods reported had some disadvantages including low yield and purity that could not meet the following studies. In this text, we devised a protocol which chose the appropriate buffer, centrifugation and DNase I processing time, based on the former studies and biochemical principles to set up an effective method to extract mtDNA from etiolated tissues of cabbage.
Materials and Methods
Plant materials
Cabbage seeds used in this study were provided by Cabbage Group, College of Horticulture, Northeast Agricultural University, Harbin, China. The seeds were sterilized by immersion in 75% alcohol for 30 s with rinsing in distilled sterile water twice and 2% sodium hypochlorite solution for 20 min, with three rinses in distilled sterile water, then germinated in dark condition (26℃) and the etiolated seedings were harvested after 7-10 days as shown in Fig. 1 (Li et al., 2011).

Fig. 1 Etiolated seedling after 7-10 days cultivation in dark condition
Mitochondria extraction
Mitochondria and mitochondrial DNA were extracted according to the method described by Virupakshi et al. (2007) and Zhao et al. (2011) with some modifications. All steps must be carried out at 4℃unless otherwise stated.
(1) About 60 g etiolated seedlings were soaked in 75% ethanol for 2-3 min, cleaned by sterile water, then ground in a breaker with 300 mL of homogenization buffer (0.4 mol • L-1mannitol, 50 mmol • L-1Tris-acetate pH 7.5, 20 m mol • L-1EDTA, 0.1% cysteine, 0.2% bovine serum albumin, 1 % PVP-40) and incubated at 4℃ for 15-30 min in dark condition.
(2) Homogenized cells were obtained by using high speed homogenizer, 15 s once and repeated 5 times, then filtered through eight layer cheese cloth and centrifuged at 1 000 g for 10 min. The suspension was centrifuged twice at 2 000 g for 10 min. The crude mitochondria were pelleted from the supernatant by centrifuging at 20 000 g for 15 min. The pellet wasgently resuspended in homogenization buffer and centrifuged again.
(3) The obtained pellet was resuspended with 12 mL of digestion buffer comprised 0.4 mol • L-1mannitol, 50 mmol • L-1Tris-acetate pH 7.5, 10 mmol • L-1magnesium chloride and 0.1% bovine serum albumin and mixed up with 30 µL (1 U • µL-1) DNase I. The mixture were incubated at room temperature for 30 min (shaking every 5 min) and 4℃ for 1 h, then added 1.2 mL 0.5 mol • L-1EDTA to stop the enzyme digestion reaction.
(4) The suspension was flatted on 25 mL of washing buffer (0.5 mol • L-1mannitol, 50 mmol • L-1Tris-acetate pH 7.5, 20 mmol • L-1EDTA) and centrifuged at 18 000 g for 20 min. The pellet was resuspended in digestion buffer and repeated centrifuging. After centrifugation, high-purity mitochondria was obtained and stored at–80℃ until required or treated for mtDNA extraction immediately.
Mitochondria integrity detection
A small amount of purified mitochondria were resuspended in digestion buffer, blended with same volume of 1% Jannus Green B and used microscope for detecting integrity after 10 min treatment in dark condition.
Mitochondrial DNA extraction
(1) Isolated mitochondria were treated with 0.9 mL lysis buffer (50 mmol • L-1Tris-acetate pH 8.0, 20 mmol • L-1EDTA, 5% sodium dodecyl sulfonate, 0.1 mol • L-1sodium chloride, 0.01% beta mercaptoethanol) and 5 µL (20 mg • mL-1) proteinase K each pellet. Probes were incubated for 1 h at 65℃ with gentle shaking to avoid mechanical breaking of DNA.
(2) 100 µL of 8 mol • L-1ammonium acetate and an equal volume of TE-saturated phenol/chloroform/ isoamyl alcohol (25 : 24 : 1) were added to the lysate and rest for 10 min at room temperature. The nucleic acids were extracted at 12 000 g for 10 min and repeated this procedure once.
(3) The supernatant was digested by RNase I for 1 h at 37℃ with gentle shaking and RNase I was eliminated by an equal volume of TE-saturated phenol/ chloroform /isoamyl alcohol (25 : 24 : 1) as step 2.
(4) Nucleic acids were precipitated from the aqueous fraction by the addition of two volumes of precooling ethanol and one-tenth volume of 8 mol • L-1ammonium acetate overnight at –80℃.
(5) Mitochondrial DNA was concentrated after centrifugation at 18 000 g for 20 min, washed 3 times with 70% ethanol and resuspended in 30-50 µL of sterile ultrapure water after drying in clean bench. mtDNA was either immediately subjected to detect or stored at –80℃ until required.
Mitochondrial DNA concentration determination
2 µL mtDNA were added ultrapure water to 100 µL. Ultraviolet spectrophotometer was used to determine A260nm/A280nm, A260nm/A230nm and the concentration of mtDNA.
Agarose gel electrophoresis
Electrophoresis was performed in 0.8% agarose gel using TAE buffer (0.04 mol • L-1Tris-acetate, 1 mmol • L-1EDTA). Gels were run at 100 V for 30 min, stained with ethidium bromide and photographed over a UV light sourse.
PCR detection
According to NCBI gene sequences in the database, mitochondrial cob gene specific primers and the specific primers of nuclear actin gene (Table 1) were designed by using PrimerPremier5.0 software, and primers synthesized by Suzhou Genewiz Company. mtDNA extracted from the extraction of the mitochondria with DNase I digested and without DNase I digested as the templates for PCR amplification. 50 µL PCR reaction system contained 2 µL mtDNA, 1 µL upstream and downstream primers (10 mmol • L-1), 25 µL 2×EcoTaq PCR Super Mix and 21 µL ddH2O2. PCR products were detected in 1% agarose gel electrophoresis. Gene actin PCR cycle: 94℃ for 2 min; 94℃for 30 s, 30 s to 61℃, 72℃ for 1 min (30 cycles), andfinally extended at 72 ℃ for 5 min, stored at 4℃; gene cob PCR cycle: 94℃ for 2 min; 94℃ for 30 s, 30 s to 56℃, 72℃ for 1 min (30 cycles), and finally extended at 72℃ for 5 min, stored at 4℃.
Results
Effects of different homogenization buffers on mitochondria extraction
The composition in homogenization buffer and concentration of each component were crucial to maintain the integrity of mitochondria. We detected buffer A as the best homogenization buffer in the four different buffers through a detection for the extracted mitochondria by microscope (Table 2).
Effects of different centrifugation on isolation and purification of mitochondria
In this test, we designed five different combinations of the centrifugal force to isolate mitochondria (Table 3). The results showed that high and pure mitochondria were obtained by the fifth combination.
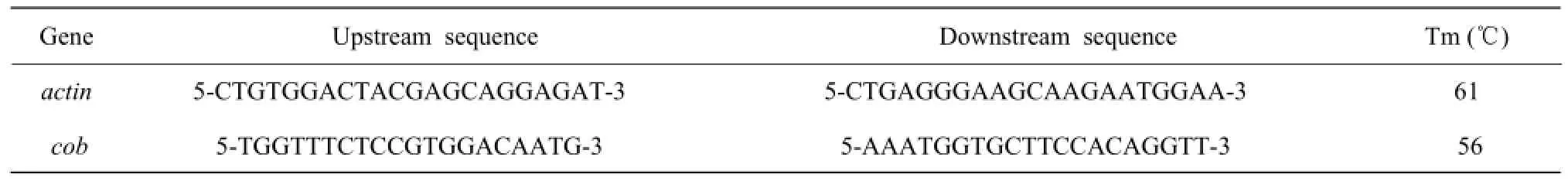
Table 1 Unique primer sequence of mitochondrial gene and nuclear gene

Table 2 Mitochondria-isolating performance at different buffers
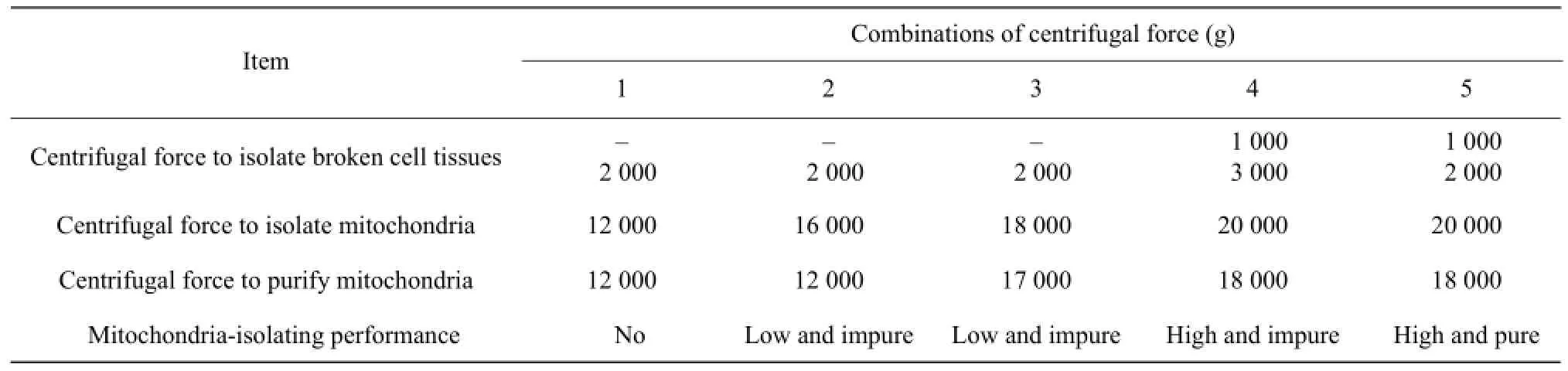
Table 3 Mitochondria-isolating performance at different centrifugal forces
Jannus Green B staining observation
The mitochondria with activity could spread green fluorescence after staining by Jannus Green B. We observed several spots with green fluorescence (Fig. 2A) under the fluorescence microscope and plenty of oval spots (Fig. 2B) under optical microscope. Both of the two figures could illustrate that the extracted mitochondriawere complete and had biological activity.
mtDNA quality analysis
The spectrophotometry of extracted mtDNA showed that A260nm/A280nm was about 1.80-1.90, A260/A230above 2.0, and concentration of mtDNA about 189 ng • µL-1. The results indicated that there were few proteins or phenol pollutions and the quality of mtDNA could fulfill the requirement for molecular marker technology analyses and researches.
mtDNA electrophoresis analysis
The lanes in agarose gel were observed clearly and the size of mtDNA fragment was about 23 kb (Fig. 3). It also declared that large fragment of mtDNA with high purity was successfully obtained.
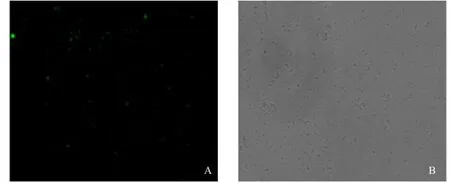
Fig. 2 Observation of mitochondria by Janus Green B reactive dying

Fig. 3 Agarose gel electrophoresis patterns of mtDNA
PCR analysis
Specific product could be obtained after PCR amplification by designing specific primers. Fig. 4 showed that the product of mtDNA of mitochondria with DNase I by specific primer PCR amplificating included cob gene sequence (about 505 bp), but no actin gene sequence (about 340 bp) and the product of mtDNA of mitochondria without DNase I by specific primer PCR amplificating contained both of the two gene sequences. mtDNA extracted by this protocol had high purity without any nuclear genes.
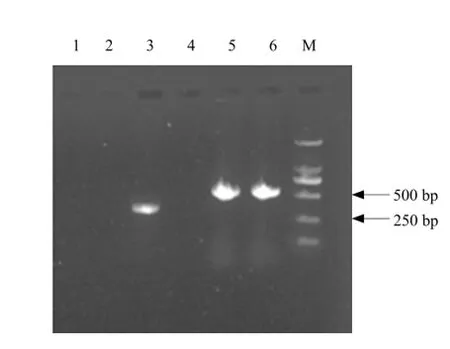
Fig. 4 Agarose gel electrophoresis patterns of mtDNA by specific primers of nucleus gene and mitochondrial gene PCR amplificating
Discussion
CMS plants have a maternally inherited trait and its implementation needs a complex process. People had never stopped to explore its mechanism since it was found. The development of molecular biology supplied an effective way for researching CMS. Many studies had showed that CMS had a close relationship with the mutation of mtDNA (Janska et al., 1998; Gong et al., 2006; Shedge et al., 2007). The research for mtDNA at molecular level could not only find out the relationship between different species, but also cloned the mitochondrial genes which related to some special phenomena, such as CMS.
Separately adopting the method of density gradient or differential centrifugation had a high requirement for experimental conditions and existed many shortcoming such as low purity, time-consuming or high cost so that both of them could not be suitable for general laboratory to extract mitochondrial DNA. Because of the difference in types and contents of inclusions, the experimental procedures of extracting mitochondria were diverse in different species by the method combined sucrose density gradient with differential centrifugation (Pei et al., 2002; Zeng et al., 2005; Zhang et al., 2006). There almost no any one specific efficient method was suitable for all species to extract mtDNA, so that it needs some meliorations.
According to the method described above, few mitochondria appeared at the bottom of the tube and the purity could not conform to the requirements of the subsequent experiment. We used mannitol as the material of the density gradient instead of sucrose and achieved a better result. We changed the centrifugal rotational speed in order to fit for extracting cabbage mitochondria such as removing pieces of large organizations at 1 000 g, disposing small particles like fibers at 2 000 g, precipitating the mitochondria at 20 000 g and enriching the mitochondria precipitation at 18 000 g. As a result, mitochondria formed a large deposit on the surface of the tube. Nuclear DNA still existed after 1 h DNase I treatment with the extracted mitochondria as others, but would be deleted thoroughly after a treatment at room temperature for 30 min (shaking every 5 min) and at 4℃ for 1 h. The concentration of DNA increased after a treatment for mitochondrial under cracking condition at 65℃ for 1 h instead of 37℃ for 4 h.
Conclusions
In this study, we took advantages of the protocol of combining the density gradient with the differential centrifugation to obtain mitochondria with the high quality. The extracted mitochondria was treated with DNase I to eliminate the influence of nuclear gene and dissolved by SDS. Proteins and RNA were digested by protein K and RNase A. We used TE-saturated phenol/ chloroform/isoamyl alcohol (25 : 24 : 1) to separate mtDNA from protein. Finally, mtDNA was concentrated by washing with 75% ethanol. Under the testing of microscope, gel electrophoresis and PCR amplification, we got a result that the purify and quality of mtDNA samples were high enough for the further relative researches. It provided a premise condition for the further researches on the molecular mechanism of CMS in cabbage.
Dieterich J H, Braun H P, Schmitz U K. 2003. Alloplasmic male sterility in Brassica napus (CMS 'Tournefortii-Stiewe') is associated with a special gene arrangement around a novel atp9 gene. Mol Genet Genomics, 269: 723-731.
Fang Z Y, Sun P T, Liu Y M, et al. 2001. Investigation of different types of male sterility and application of dominant male sterility in cabbage. China Veg, 1: 6-10.
Fujii S, Yamada M, Toriyama K. 2009. Cytoplasmic male sterility related protein kinase, OsNek3, is regulated downstream of mitochondrial protein phosphatase 2C, DCW11. Plant Cell Physiol, 50: 828-837.
Jee Y P, Lee Y P, Lee J, et al. 2013. Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containingDCGMS cytoplasm. Theor Appl Genet, 126: 1763-1774.
Janska H, Sarria R, Woloszynska M, et al. 1998. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. The Plant Cell, 10: 1163-1180.
Hanson M R. 1991. Plant mitochondrial mutations and male-sterility. Annu Rev Genet, 25: 461-486.
Gong Y, Cang X, Chao Z. 2006. Research on molecular mechanism of cytoplasmic male sterility in plants. Molecular Plant Breeding, 6(4): 51-56.
Liu J, Cui C R, Cui C S, et al. 2004. RAPD analysis of mitochondrial DNA of CMS line and its maintainer in onion. Journal of Northeast Agricultural University, 35(3): 322-324.
Li S S, Xue L F, Su A G, et al. 2011. Progress on sequencing and alignment analysis of higher plant mitochondrial genomes. Journal of China Agricultural University, 16(2): 22-27.
Li Z X, Zhang D S, Si L T, et al. 2011. A method for mitochondrial isolation in Chinese cabbage. Acta Agriculture Boreali-Sinica, 26(2): 138-142.
Pei D S, Cai P Z, Li M Y, et al. 2002. A simple method for isolation of rice mitochondrial DNA. Journal of Sichuan University (Natural Science Edition), 39(4): 18-20.
Schnable P S, Wise R P. 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci, 3: 175-180.
Scotti N, Cardi T, Marechaldrouard L. 2001. Mitochondrial DNA and RNA isolation from small amounts of potato tissue. Plant Molecular Biology Reporter, 19: 670-671.
Shedge V, Arrieta-Montiel M, Christensen A C, et al. 2007. Plant mitochondrial recombination surveillance requires unusual recA and muts homologs. The Plant Cell, 19: 1251-1264.
Tanaka M N, Fujita H, Uemura H M.2004. Proteomics of the rice cell: systematic identification of the protein populations in subcellularcompartments. Mol Gen Genomics, 271: 566-576.
Virupakshi S, Naik G R. 2007. Purification of DNA from chloroplast and mitochondria of sugarcane. Current Science, 92(11): 1613-1619.
Wang Q B, Zhang Y Y, Fang Z Y, et al. 2012. Chloroplast and mitochondrial SSR help to distinguish allo-cytoplasmic male sterile types in cabbage (Brassica oleracea L. var. capitata). Mol Breeding, 30: 709-716.
Zeng X, Sun W, Meng Y, et al. 2005. Extraction and purification of mtDNA in crucifer. Acta Botanica Boreali-Occidentalia Sinica, 25(6): 1137.
Zhang J F, Stewart J M D. 2001. Inheritance and genetic relationships of the D8 and D2-2 restorer genes for cotton cytoplasmic male sterility. Crop Science, 41(2): 289-294.
Zhang X, Meng Z G, Zhou T, et al. 2012. Mitochondrial SCAR and SSR Markers for distinguishing cytoplasmic male sterile lines from their isogenic maintainer lines in cotton. Plant Breeding, 131(4): 563-570.
Zhao Y, Chen P, Zhou R Y. 2011. Extraction of high purity mitochondrial DNA from kenaf for genome sequencing. Journal of Northeast Agricultural University, 42(4): 98-101.
Zhang Z, Zhang L G, Wang Q, et al. 2006. An effective and quick protocol for mitochondrial DNA extraction from Chinese cabbage. Biotechnology, 16(3): 48-50.
S634; S330
A
1006-8104(2014)-03-0023-07
Received 5 December 2013
Supported by Funding of Utilization of Heterosis and Breeding of New Variety in Brassicaceous Vegetable (2012BAD02B01)
Wang Shuai (1988-), male, Ph. D, engaged in the research of molecular breeding and biotechnique. E-mail: wangshuai10log@126.com
* Corresponding author. Wang Chao, professor, supervisor of Ph. D student, engaged in the research of molecular breeding and biotechnique. E-mail: wangchao504@126.com
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Effect of Seed Soaking with Exogenous Proline on Seed Germination of Rice Under Salt Stress
- Physiological Changes and Cold Tolerance of Three Camphor Species During Natural Winter Temperature Fluctuations
- Effects of Methylated Soybean Oil Adjuvant on Fomesafen Efficacy to Weeds
- Lentivirus Mediated Gene Manipulation in Trophectoderm of Porcine Embryos
- Effects of Allicin on Lipid Metabolism and Antioxidant Activity in Chickens
- Design and Implementation of Stud-farm Daily Management System Based on C/S Structure
