Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase(CKX)gene family in foxtail millet(Setaria italica)
2014-02-28*
*
aInstitute of Genetics and Developmental Biology,Chinese Academy of Sciences,Beijing 100101,China
bPostdoctoral Research Center of Shandong Shengfeng Seeds Co.,Ltd.,Jiaxiang 272400,China
cYantai Academy of Agricultural Sciences,Yantai 265500,China
Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase(CKX)gene family in foxtail millet(Setaria italica)
Yuange Wanga,Huaihua Liub,Qingguo Xinc,*
aInstitute of Genetics and Developmental Biology,Chinese Academy of Sciences,Beijing 100101,China
bPostdoctoral Research Center of Shandong Shengfeng Seeds Co.,Ltd.,Jiaxiang 272400,China
cYantai Academy of Agricultural Sciences,Yantai 265500,China
A R T I C L E I N F O
Article history:
Received 9 February 2014
Received in revised form29 April2014
Accepted 12 May 2014
Available online 22 May 2014
Foxtail millet
Cytokinin oxidase/dehydrogenase(CKX;EC.1.5.99.12)regulates cytokinin(CK)level in plants and plays an essentialrole in CK regulatory processes.CKXproteins are encoded by a smallgene family with a varying number of members in different plants.In spite of their physiological importance,systematic analyses of SiCKX genes in foxtailmillet have not yet beenexamined.In this paper,we report the genome wide isolation and characterization of SiCKXs using bioinformatic methods.Atotalof11 members ofthe family were identified in the foxtailmillet genome.SiCKX genes were distributed in seven chromosomes(chromosome 1,3,4,5,6,7,and 11).The coding sequences of all the SiCKX genes were disrupted by introns,with numbers varying from one to four.These genes expanded in the genome mainly due to segmental duplication events.Multiple alignment and motifdisplay results showed thatallSiCKXproteins share FAD-and CK-binding domains.Putative cis-elements involved in Ca2+-response,abiotic stress response,light and circadian rhythm regulation,disease resistance and seed development were present in the promoters of SiCKX genes.Expression data mining suggested that SiCKX genes have diverse expression patterns.Real-time PCR analysis indicated that all 11 SiCKX genes were up-regulated in embryos under 6-BAtreatment,and some were NaClor PEG inducible.Collectively,these results provide molecular insights into CKX research in plants.
©2014 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.All rights reserved.
1.Introduction
The plant hormone group known as cytokinins(CKs)play a significant role not only in the regulation of proliferation and differentiation of plant cells,but also controlvarious aspects of plant growth and development,such as leaf senescence,lateral bud growth,shoot or root branching,photosynthesis,seed germination,transduction of nutritional signals,chloroplastformation and crop productivity[1-6].Natural CKs are mainly N6-substituted adenine derivatives that generally contain an isoprenoid or aromatic side-chain.The fine-tuning of hormone levels in individual cells must be under proper control by biosynthetic and metabolic enzymes[7].It was reported that homeostasis of CK concentration in cells is regulated by the rates ofbiosynthesis and degradation[2].CK synthesis in plants is catalyzed by the enzyme isopentenyltransferase via the methylerythritol phosphate and mevalonate pathways[8-10]. Irreversible degradation of CKs and theirderivatives is catalyzed by CKXs,which are encoded in plants by a small gene family [11].
The CKXenzyme degrades CKs by cleaving the N6-substituted side chain to produce adenine and unsaturated aldehyde 3-methyl-2-butenal[12,13].CKX enzyme is a flavoenzyme, containing flavin adenosine dinucleotide(FAD)bound domain, and catalyzes degradation of CKs with molecular oxygen as the oxidant or with other electron acceptors in a dehydrogenase reaction[14,15].The CKX enzyme was reported to be an important regulatory factor regulating local CK contents and to contribute to the controlof CK-dependent processes[16].
CKX activity was first discovered in crude extracts from tobacco plants[17].Subsequently,a number of CKX genes were cloned and identified in different plants,including maize[18,19],Arabidopsis thaliana[5,20],Dendrobium orchid [21],Hordeum vulgare[22],Oryza sativa[23],Populus trichocarpa [24],Pisum sativum[25],Brassica rapa[26],Triticum aestivum [27-31]and Glycine max[32].Although the similarity of the entire protein sequences is not very high among different plant species,these CKX proteins have FAD-and CK-binding domains,located at the N and C termini,respectively[31]. These conserved motifs are thought to be related by CKX enzymatic activity[33].
In order to understand their biological functions in plant growth and development,CKX genes have been studied extensively.In developing maize kernels,CKX activity is much higher after pollination,and correlated with the increased CK levels[34].Transgenic CKX tobacco plants displayed stunted shoots and enlarged root meristems with more branched roots compared to the wild-type[5].In Arabidopsis,ckx3/ckx5 double mutants have higher endogenous CK levels,and the mutants have larger flowers,more siliques,more flower primordia,and more seeds;the total seed yield of these mutants was increased by 55%,compared to the wild-type[35].Furthermore,different Arabidopsis CKX genes showed different expression patterns,which suggests that differential expression of CKX gene family members may play an important role in the control of CK levels[20].In barley,reduction in HvCKX1 gene expression led to higher plant productivity and a greater root mass[36].Most importantly,Ashikari et al.characterized a rice yield quantitative trait locus(QTL),Gn1a or OsCKX2,encoding a CKX protein. Further analysis of Gn1a showed that naturalgenetic variants of OsCKX2 conferred increased grain numbers per panicle. Reduced expression of OsCKX2 increased the number of flowers,resulting in enhanced grain yield[23].
Genome-wide analyses of the CKX gene family have been conducted following the release of full genome sequences in many plants,There are at least 13 CKX family members in maize[37],11 in rice[23],7 in Arabidopsis[38],12 in Chinese cabbage(Brassica rapa ssp.pekinensis)[39],5 in potato[40],13 in Brachypodium distachyon[41],12 in Sorghum bicolor[41],and at least 4 in Hordeum vulgare[41].Genome duplication events [37,39],phylogenetic and comparative genomic analysis[41], enzymatic properties[40],and biochemical characterization [38,40]have been studied in this gene family.
Foxtail millet(Setaria italica)was an important foodstuff in China in the past and continues to be grown in semi-arid areas [42].The release of foxtail millet genome information[42,43] made it possible to identify allthe CKX family members in this species.Mameaux et al.previously performed a genome-wide search for the members of this family in foxtail millet[41],but their results lacked a detailed bioinformatic analysis.In this paper,we also conducted a genome-wide search and identified 11 CKX genes.Their distribution in the genome,evolutionary expansion pattern,motif characteristics and distribution,evolutionary relationships,presence of cis-elements in the promoter regions and expression profiles were also analyzed in detail. Our bioinformatic analysis will provide useful information for further functionaldissection of CKX genes in plants.
2.Materials and methods
2.1.Isolation of CKX genes in foxtail millet
The sequences of 11 rice and 7 Arabidopsis CKX proteins were downloaded from the TIGR(http://rice.plantbiology.msu.edu/) and TAIR(http://www.arabidopsis.org/)databases.To obtain all the CKX genes in foxtailmillet,BLASTP searches were conducted in the Phytozome(http://www.phytozome.net/),and NCBI (http://www.ncbi.nlm.nih.gov/)databases with the rice and Arabidopsis CKX proteins as queries.First,we selected the sequence as a candidate SiCKX protein if it satisfied the query with E-value<10-10.Redundant sequences were removed.Then, the Pfam(http://www.sanger.ac.uk/Software/Pfam/)and SMART (http://smart.embl-heidelberg.de/smart/batch.pl)databases were used to identify the FAD-and CK-binding domains of all the candidate proteins.Genes that did not contain the FAD-and CK-binding domains were excluded from further analysis.
2.2.Gene structure analysis of foxtailmillet CKX genes
The information for SiCKX genes,including chromosomal location,open reading frame(ORF)length,and full length cDNAsequence,were obtained from the foxtailmillet sequencing database(http://www.phytozome.net/).Structures of SiCKX genes were determined by the GSDS tool(http://gsds.cbi.pku. edu.cn/)[44].
2.3.Motif display and phylogenetic analysis of CKX proteins
The multiple expectation maximization for the motif elicitation(MEME)utility program[45]was used to display motifs in SiCKX proteins.A phylogenetic tree was constructed in ClustalX[46]based on the full sequence of the proteins with default parameters from Arabidopsis,rice,and foxtail millet and the tree was constructed by the neighbor-joining(NJ) method using MEGA4 software[47].

Table 1-CKXgenes identified inSetaria italicaand their sequence characteristics.
2.4.Analysis of the promoter regions of SiCKX genes
To identify cis-elements in the promoter sequences of SiCKX genes,2 kb offoxtailmillet genomic DNAsequence upstreamof the initiation codon(ATG)was downloaded from Phytozome and PLACE(http://www.dna.affrc.go.jp/PLACE/)was used to analyze the cis-elements[48].
2.5.Chromosomal localization and gene duplication
Eleven SiCKX genes were mapped on chromosomes by identifying their chromosomal positions in Phytozome.Tandem and segmental duplications have impacts on gene family amplifications[49].Tandem duplications and large-scale block duplications(segmentalduplication)were identified according to the methods of Wang et al.[49]and Zhang et al.[50]
2.6.Expression profiles of SiCKX genes
The coding sequences of SiCKX genes were used to query the NCBIESTdatabase(http://www.ncbi.nlm.nih.gov/dbEST/)using the megablast tool.Parameters were as follows:maximum identity>95%,length>200 bp and E-value<10-10.
2.7.Expression of SiCKX genes under stress treatments
To understand the expression of SiCKX genes in germinating embryos under stress seeds of foxtail millet cultivar Yugu 1 imbibed for 12 h were transferred to Petridishes fitted with filter papers that were soaked in 10μmol L-16-BA,200 mmol L-1NaCl,and 20%PEG-6000 and then cultured for 10 h in a growth chamber at 23°C.Embryos were separated from endosperms after the treatment.Water treatment served as the control(CK).
3.Results
3.1.The CKX gene family in foxtail millet
After carefully searching the foxtail millet genome,11 members were defined as SiCKX genes(Table 1).The length of CKX ORF in foxtailmillet ranged from 720 to 1620 bp.BLAST analysis against the Pfam and SMART database indicated that all of them belonged to the SiCKX gene family.The predicted SiCKX proteins had a typical FAD-and CK-binding domains, which were specific to CKX family members.The 11 SiCKX genes were distributed on seven foxtail millet chromosomes: chromosomes 1,3,4,6,7,and 11 each contains one gene, while chromosome 5 contains five genes.
The tool of NetOGlyc(http://www.cbs.dtu.dk/services/ NetOGlyc/)was used to predict the number of glycosylation sites.SiCKX1,SiCKX3,SiCKX5,and SiCKX10 each contains two glycosylation sites,SiCKX7,SiCKX8 and SiCKX11 each contains five glycosylation sites,SiCKX4 contains three sites,SiCKX6 contains one site and SiCKX2 and SiCKX9 contain no glycosylation sites.Five of the 11 SiCKX proteins showed localization on the chloroplast thylakoid membrane by a PSORT analysis (http://psort.nibb.ac.jp/).Of the remaining proteins,SiCKX2, SiCKX5 and SiCKX9 showed localization in the cytoplasm, SiCKX4 located in the nuclear,and SiCKX10 and SiCKX11 showed localization in the extracellular and vacuole,respectively(Table 1).The cDNA sequences were compared with the corresponding genomic DNA sequences to detect the numbers and positions of exons and introns within each SiCKX gene by using the GSDS program(http://gsds.cbi.pku.edu.cn/) (Fig.1).The coding sequences of all the SiCKX genes were disrupted by introns,with numbers varying from one to four.
3.2.Motif analysis and protein architecture
The motif distribution in SiCKX proteins was analyzed based on the MEME program.Three putative conserved motifs were identified,each with 50 amino acids.Allthree were present in each SiCKX member except SiCKX9;motif 2 appeared twice in SiCKX8,and SiCKX9 contained only motif 1 and motif 2(Fig.2).
3.3.Phylogenetic analysis
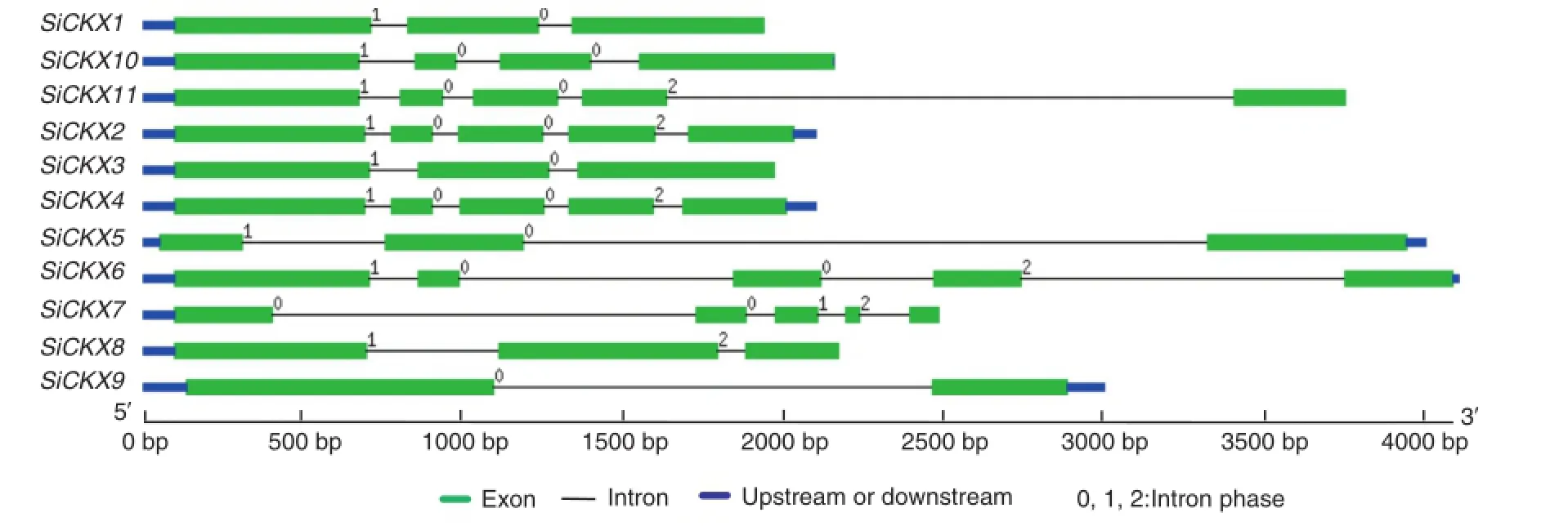
Fig.1-Gene structure ofthe foxtailmillet CKX family.Exons and introns are shown by filled boxes and single lines,respectively. UTRs are displayed by thick blue lines at both ends.Intron phases 0,1,and 2 are indicated by numbers 0,1,and 2.
In order to uncover the evolutionary relationships among foxtail millet,rice and Arabidopsis CKXs,the amino acid sequences of CKX genes were compared by ClustalX.In the phylogenetic tree constructed by the NJ method(Fig.3)the proteins clustered into three major groups(I,II,and III).Group I contained 22 members(with 8,9,and 5 members of foxtail millet,rice,Arabidopsis,respectively)and further divided into subclusters IA and IB.Group IIincluded 6 members(with 3,3, and 1 members of foxtail millet,rice,Arabidopsis,respectively).Group III contained the SiCKX7 gene only.
3.4.Genome duplication of SiCKX genes
Based on phylogenetic results(Fig.4),four paralogs(SiCKX1/ SiCKX3,SiCKX2/SiCKX4,SiCKX5/SiCKX8,and SiCKX10/SiCKX11), were identified in SiCKX genes.According to the foxtail millet genome annotation results,we found one tandemly duplicated pair,namely SiCKX5/SiCKX8,on chromosome 5.Segmental duplications might have contributed to the other three paralogous genes(Fig.5).
3.5.cis-element analysis
To identify the putative cis-acting regulatory elements,about 2000 bp sequences upstreamfromthe start codon were isolated. The cis-elements included Ca2+-responsive,abiotic stress responsive,light and circadian rhythm regulation,disease resistance and seed development,such as:ABRERATCAL,ABRELATERD1, ACGTATERD1,BOXLCOREDCPAL,CARGCW8GAT,CIACADI ANLELHC,CTRMCAMV35S,DPBFCOREDCDC3,EECCRCAH1, DOFCOREZM,GT1CONSENSUS,INRNTPSADB,POLASIG2, RYREPEATGMGY2,RYREPEATLEGUMINBOX,SEBFCONSST PR10A,SEF4MOTIFGM7S,SP70A,and TATABOX2(Table 2).
3.6.Expression patterns of the SiCKX gene family
EST data could provide useful information for gene expression research.There are about 66,051 foxtail millet ESTs in the NCBI EST database.Thus,EST mining and thr whole-genome shotgun(WGS)database were employed to analyze transcript levels of SiCKX genes.The coding sequences of SiCKX genes were used to query the EST database using the megablast tool.Based on tissue and organ types, the EST data were classified into eight groups(Table 3).The results indicated that all SiCKX genes had expression data support.Expression data for all the SiCKX genes were obtained for seedlings and leaves.The EST number for SiCKX3 was highest,up to 25.Expression data for SiCKX genes SiCKX1 and SiCKX10 were checked in eight tissue types.

Fig.2-Putative motif distribution in 11 SiCKX proteins.Motifs of SiCKX proteins were searched by the MEME program (http://meme.nbcr.net/meme/).
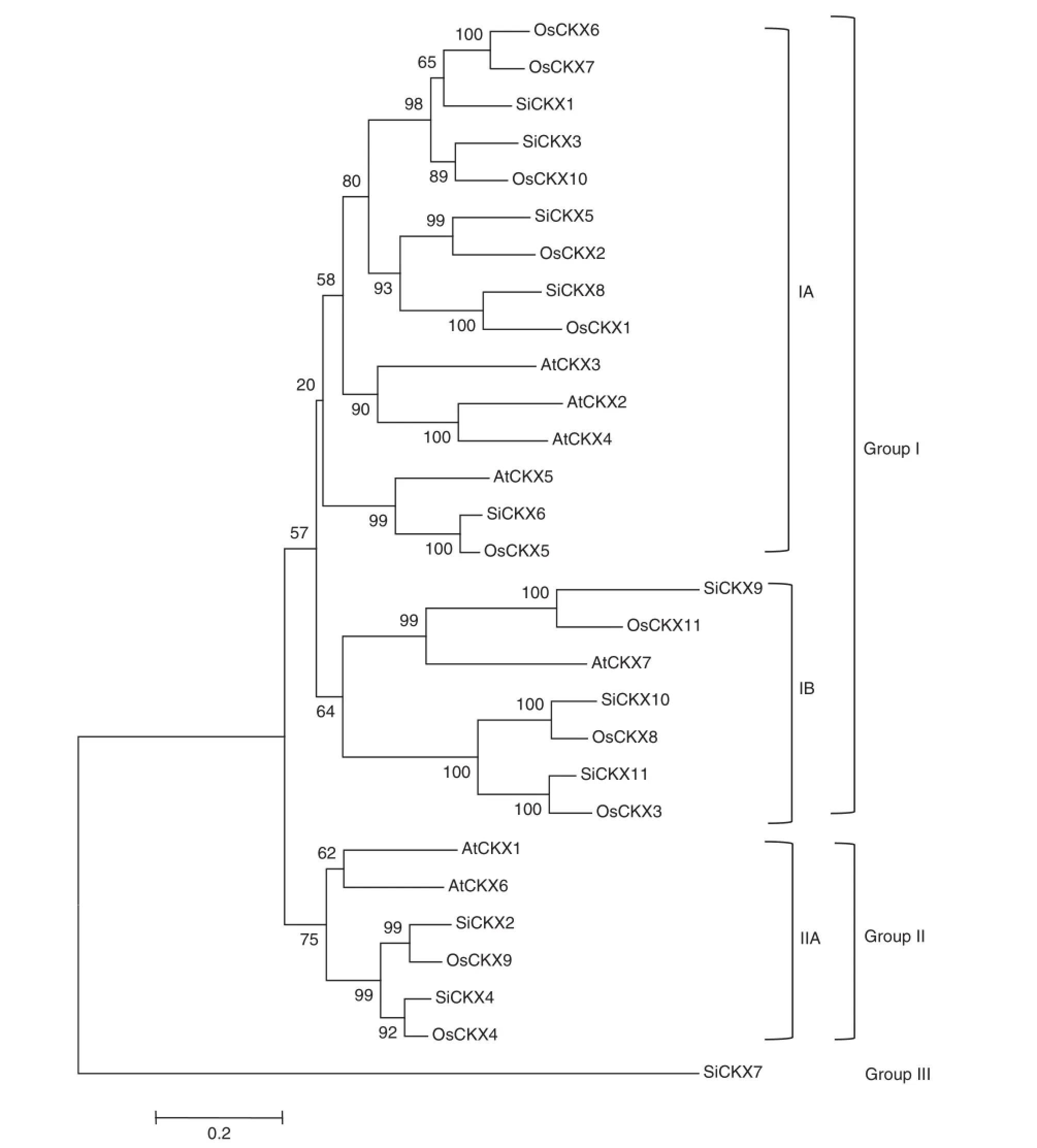
Fig.3-Phylogenetic tree for foxtailmillet,Arabidopsisand rice CKXproteins.The jointunrooted tree was generated using MEGA4 by the neighbor joining method.Bootstrap values from 1000 replicates were indicated ateach branch.The protein sequences ofrice andArabidopsiswere from TIGR(http://rice.plantbiology.msu.edu/)and TAIR(http://www.arabidopsis.org/),respectively.Abbreviations: Os,Oryza sativa;At,Arabidopsis thaliana;and Si,Setaria italica.
3.7.Expression levels of SiCKX genes in germinating embryos
The relative transcript levels of the SiCKX genes in germinating embryos under 6-BA,NaCl,and PEG treatments are shown in Fig.6.Transcripts of SiCKX1,SiCKX3,SiCKX4,SiCKX5, SiCKX8,SiCKX9,and SiCKX10 were induced by all three treatments.SiCKX6 and SiCKX7 were up-regulated in embryos treated with 6-BA and PEG,whereas the expression levels were unchanged in embryos under NaCl treatment,and SiCKX2 and SiCKX11 expressions were induced only by 6-BA.
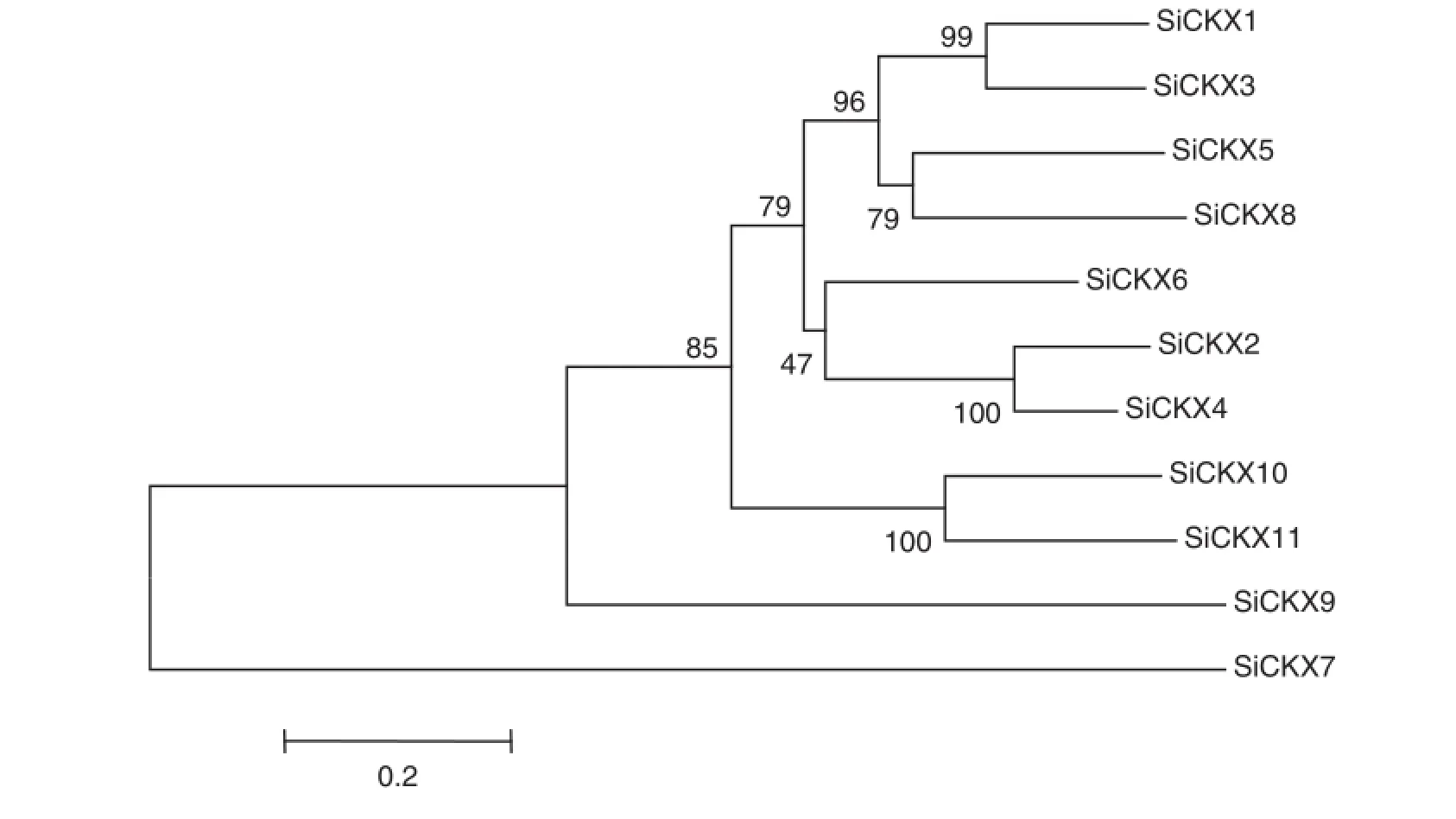
Fig.4-Phylogenetic tree for 11SiCKXgenes in foxtailmillet.The joint unrooted tree was generated using MEGA4 by the neighbor joining method.Bootstrap values from 1000 replicates were indicated at each branch.Si:Setaria italica.
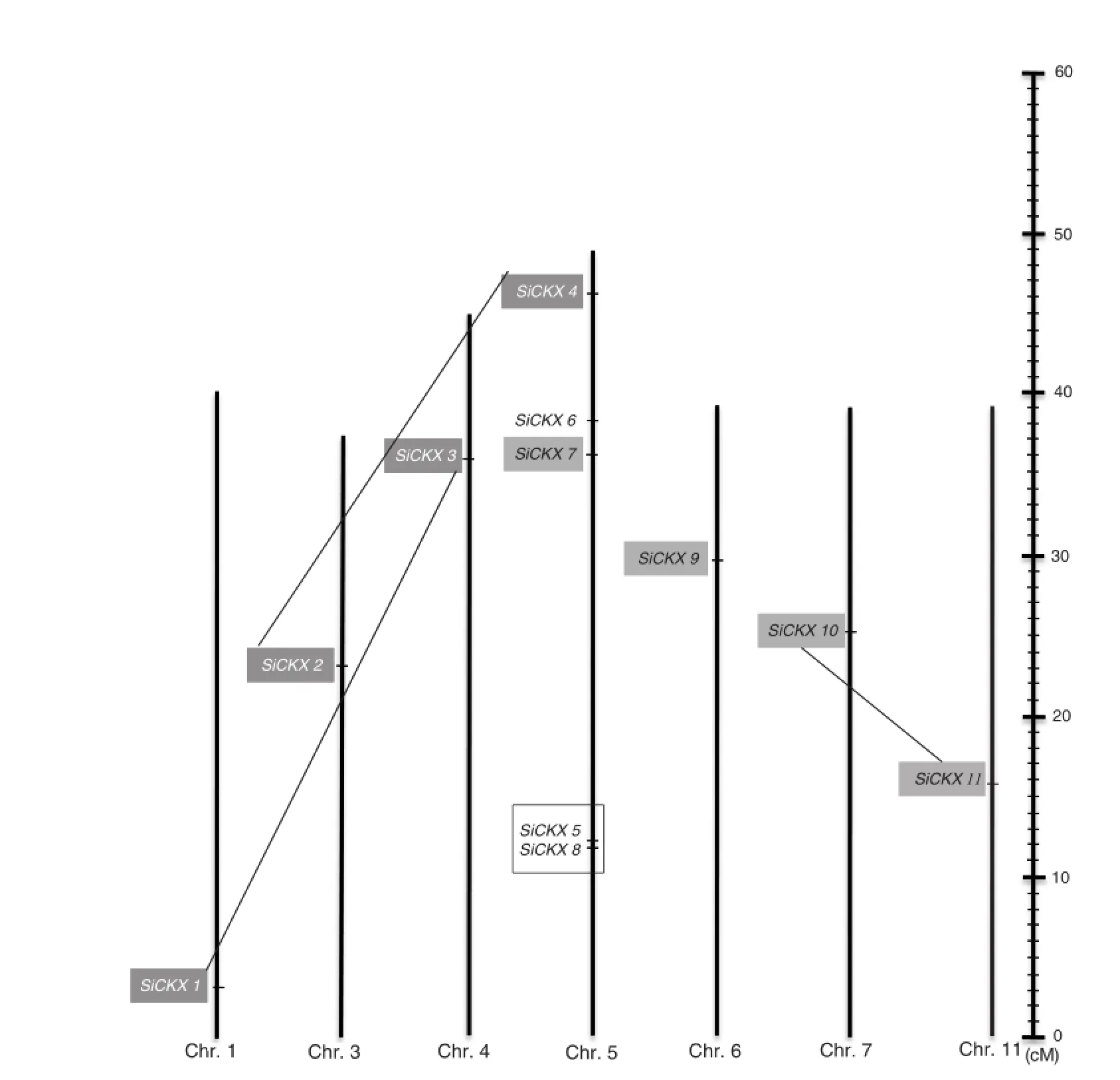
Fig.5-Chromosomallocalization ofSiCKXgenes.Segmentalduplicates,includingSiCKX1/SiCKX3,SiCKX2/SiCKX4,andSiCKX10/SiCKX11,are connected by lines and the tandemly duplicated genes(SiCKX5/SiCKX8)are marked by hollow boxes.
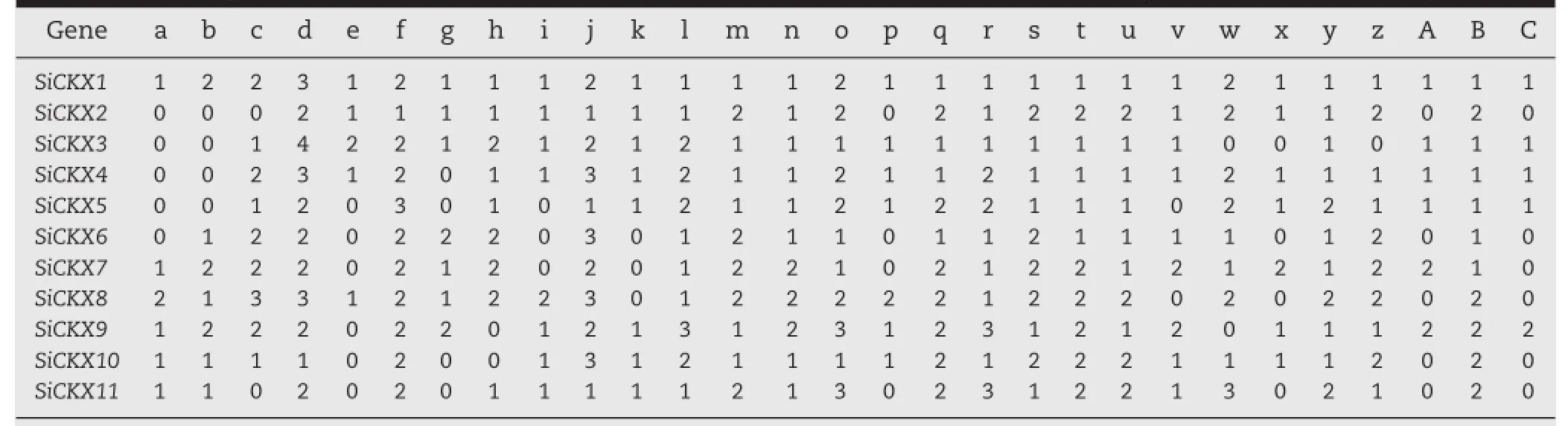
Table 2-Twenty-nine putativecis-elements,more than 6 bp,were identified in 11SiCKXgenes in foxtail millet by PLACE.
4.Discussion
In 1971,Pačes et al.described CKX activity in crude tobacco cell culture extracts[17].Later,similar activity was found in maize kernels[51].Since then many papers have reported CKX activity from a variety of tissues and species.In recent years,with whole genome sequencing,the CKX gene family was thoroughly investigated in many plant species using comparative genomic approaches.CKX enzymes are encoded by a multigene family with varying numbers of members in different plant species[37].In the present work,we isolated 11 CKX genes from foxtail millet in silico.
cis-elements play essential roles in the regulation of gene expression by controlling the efficiency of promoters. Thus research on cis-elements could lay a foundation for further functional analysis of the SiCKX genes.Among putative cis-elements,the water-stress responsive element (ACGTATERD1),light-responsive motifs(-10PEHVPSBD and BOXIINTPATPB),mesophyll-specific expression element(CACTF TPPCA1),copper-stress responsive element(CURECORECR), stem specific expression element(DOFCOREZM),ABA-responsive element(EBOXBNNAPA),CO2-responsive element (EECCRCAH1),light-responsive element(GT1CONSENSUS and INRNTPSADB),salt-responsive element(GT1GMSCAM4),pollenspecific cis-element(GTGANTG10),cold-responsive element (MYCCONSENSUSAT),nodule-specific element(NODCON2GM), androotspecific expression element(ROOTMOTIFTAPOX1)were found in all11 SiCKX genes(Table 2).Diverse cis-elements in the promoters of SiCKX genes suggest that CKX proteins are expressed in different plant tissues.For example,SiCKX1, SiCKX3,SiCKX4,SiCKX5,SiCKX8,SiCKX9,and SiCKX10 all have salt-responsive element(GT1GMSCAM4)(Table 2)and their gene expressions were obviously up-regulated under salt condition (Fig.6).Conversely,other genes(SiCKX2,SiCKX6,SiCKX7,and SiCKX11)were not responsive to salt stress compared to the control(Fig.6)probably due to the lack of the salt-responsive element(Table 2).
Paralogs are genes derived from duplication events within a genome.Segmental(chromosomal segments)duplication, tandem duplication(duplications in a tandem pattern),and transposition events,can result in duplication of gene families[52].Duplicate genes provide raw materials for evolution of new gene functions.Phylogenetic analysis has been commonly used to identify gene families and predict their functional orthologs[37,53,54].However,there is far less evolutionary information about the CKX gene family in foxtail millet.To detect the expansion of this family in S.italica in ourstudy a phylogenetic tree was reconstructed using full-length SiCKX protein sequences(Fig.4).The phylogenetic tree divided the SiCKX genes into several distinct groups.Among the 11 proteins,three pairs of paralogous proteins(SiCKX1/SiCKX3, SiCKX2/SiCKX4,and SiCKX10/SiCKX11)and one tandemly duplicated protein(SiCKX5/SiCKX8)were found,suggesting that divergence in each protein pair occurred relatively late.Each of other three SiCKX proteins,including SiCKX 6,SiCKX7,and SiCKX9,occupied a distinct branch.Furthermore,SiCKX6 was basal to SiCKX2/SiCKX4.These results suggested that SiCKX6, SiCKX7,and SiCKX9 may have diverged earlier from the other SiCKX proteins.Further investigation suggests that both segmental duplication and tandem duplication led to expansion of CKX gene family in the foxtail millet genome(Fig.5).
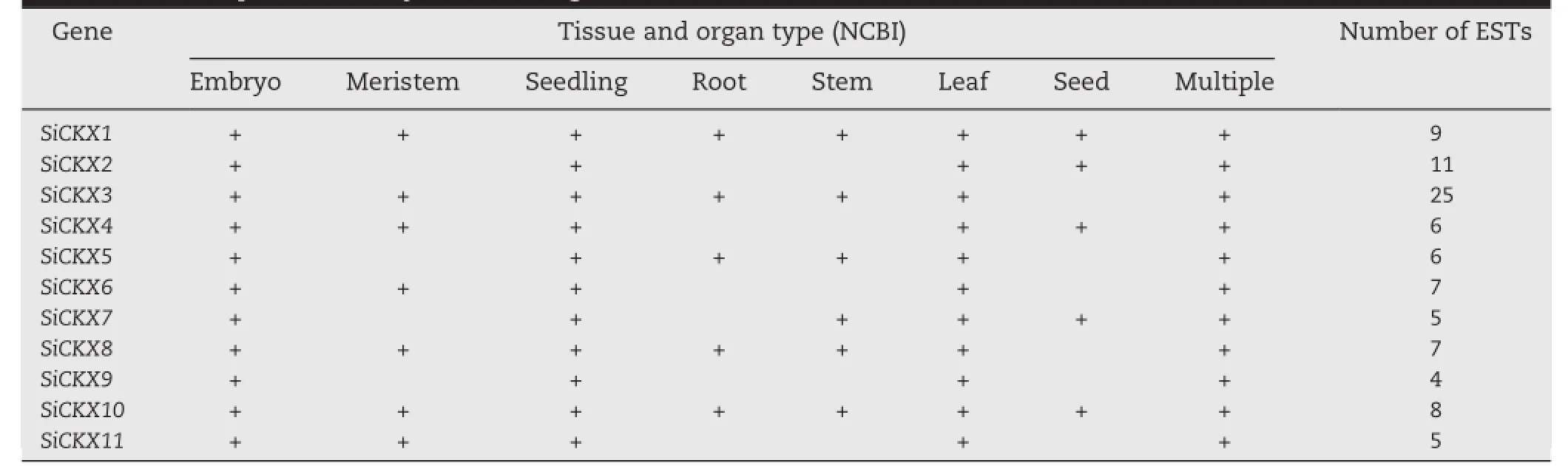
Table 3-EST expression analysis ofSiCKXgenes in different tissues.
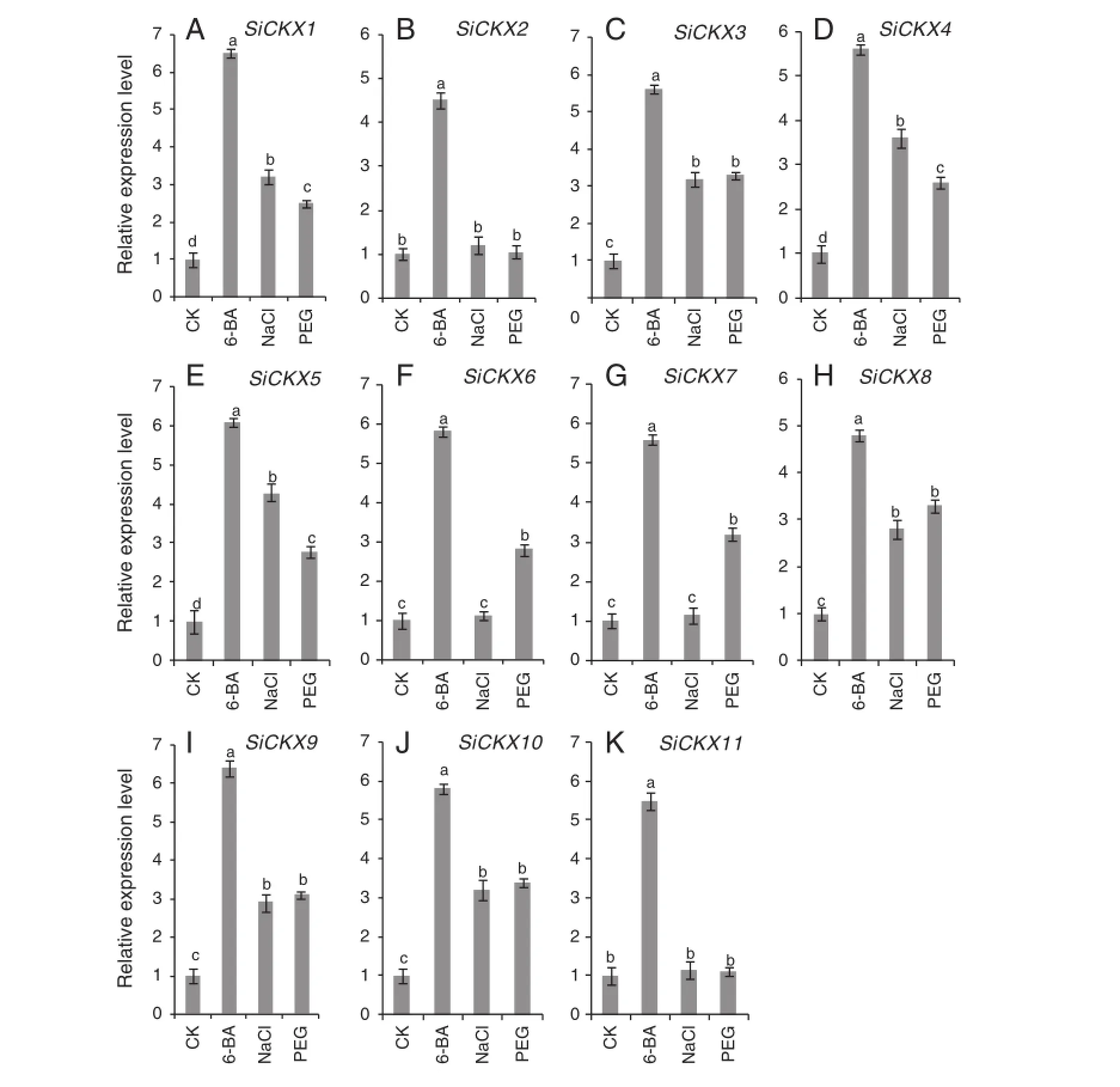
Fig.6-Real-time PCR analysis ofSiCKXstranscripts in germinating embryos after treatment with H2O,10μmol L-16-BA, 200 mmol L-1NaCl,and 20%PEG-6000 for 10 h.The data are mean±SE of three biological replicates.Different letters above the column indicate statistical significance at the 0.05 probability level.
Members of the CKX family in wheat,soybean,cotton, Arabidopsis,and Zea mays showed tissue-specific expression patterns.Wheat TaCKX3 was expressed in embryos,and was strongly up-regulated by 6-BA[31].In soybean,GmCKX12 and GmCKX16 were abundant in leaves,while GmCKX13 and GmCKX14 were highly expressed in young shoots[32].GhCKX transcripts were found in cotton roots,hypocotyls,stems, leaves,and ovules.The highest expression level was found at -1 DPA(day post anthesis)ovule[55].Arabidopsis AtCKX1 had slightly higher expression in roots while AtCKX2 was better expressed in shoots[56].In maize,ZmCKX6,ZmCKX10,and ZmCKX12 were abundant and constitutively expressed in roots,shoots,mature leaves,immature ears,and tassels, whereas ZmCKX2 and ZmCKX3 were preferentially expressed in young leaves and mature leaves,respectively[37].In this study,expression data of allthe SiCKX genes were checked in embryos,seedlings and leaves.Two SiCKX genes,including SiCKX1 and SiCKX10,were examined in all eight tissues.ESTnumbers of SiCKX9 were the lowest,up to 4(Table 3).The above results suggest that some as yet unidentified tissuespecific factors may affect the expression of CKX genes.
Real-time PCR analysis in this work showed that all 11 SiCKX genes were significantly induced by exogenous 6-BAin germinating embryos(Fig.6).This is consistent with other reports of applying exogenous CKs or 6-BA resulting in enhancement of CKX expression levels[31,57].In Fig.4,four protein pairs(SiCKX1 and SiCKX3,SiCKX5 and SiCKX8, SiCKX2 and SiCKX4,and SiCKX10 and SiCKX11)formed distinct subgroups in the phylogenetic tree,suggesting that each protein pair may share the same biological function. However,only four genes(SiCKX1,SiCKX3,SiCKX5,and SiCKX8)were obviously induced under salt and 20% PEG-6000 stresses.This finding indicates that SiCKX genes may have distinct and partially overlapping expression patterns related to their diverse roles.Further work is required in order to illuminate the detailed functions of each CKX gene in abiotic stress.
In summary,11 foxtail millet CKX genes were identified in whole genome analysis.The results of SiCKX gene chromosomal location,expansion pattern,motif distribution,evolutionary relationship,cis-element analysis in promoter regions, and expression profiles under various abiotic treatments provided useful information for CKX research in foxtail millet and other plants.
Acknowledgments
This study was supported by the project of the Modern Seed Industry Enterprise Science and Technology Development of Shandong Province,China(SDKJ2012QF003).
R E F E R E N C E S
[1]P.J.Davies,Plant Hormones:Physiology,Biochemistry,and Molecular Biology,Kluwer Academic Press,Dordrecht,1995.
[2]D.W.S.Mok,M.C.Mok,Cytokinin metabolism and action, Annu.Rev.Plant Physiol.Plant Mol.Biol.52(2001)89-118.
[3]J.P.C.To,J.J.Kieber,Cytokinin signaling:two-components and more,Trends Plant Sci.13(2008)285-292.
[4]H.J.Kim,H.Ryu,S.H.Hong,H.R.Woo,P.O.Lim,I.C.Lee,J. Sheen,H.G.Nam,I.Hwang,Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis,Proc.Natl.Acad.Sci.U.S.A.103(2006)814-819.
[5]T.Werner,V.Motyka,M.Strnad,T.Schmuelling,Regulation of plant growth by cytokinin,Proc.Natl.Acad.Sci.U.S.A.98 (2001)10487-10492.
[6]K.Takei,H.Sakakibara,M.Taniguchi,T.Sugiyama,Nitrogen dependent accumulation of cytokinins in root and the translocation to leaf:implication of cytokinin species that induces gene expression of maize response regulator,Plant Cell Physiol.42(2001)85-93.
[7]I.Frebort,M.Kowalska,T.Hluska,J.Frebortova,P.Galuszka, Evolution of cytokinin biosynthesis and degradation,J.Exp. Bot.62(2011)2431-2452.
[8]H.Sakakibara,Activity,biosynthesis,and translocation, Annu.Rev.Plant Biol.57(2006)431-449.
[9]K.Miyawaki,M.Matsumoto-Kitano,T.Kakimoto,Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis:tissue specificity and regulation by auxin,cytokinin, and nitrate,Plant J.37(2004)128-138.
[10]I.Hwang,H.Sakakibara,Cytokinin biosynthesis and perception, Physiol.Plant.126(2006)528-538.
[11]T.Schmülling,T.Werner,M.Riefler,E.Krupková,I. Bartrina y Manns,Structure and function of cytokinin oxidase/dehydrogenase genes of maize,rice,Arabidopsis and other species,J.Plant Res.116(2003)241-252.
[12]B.A.McGaw,R.Horgan,Cytokinin catabolism and cytokinin oxidase,Phytochemistry 22(1983)1103-1105.
[13]P.Galuszka,I.Frebort,M.Sebela,P.Pe,Degradation of cytokinins by cytokinin oxidases in plants,Plant Growth Regul.32(2000)315-327.
[14]M.Laloue,J.E.Fox,Cytokinin oxidase from wheat:partial purification and general properties,Plant Physiol.90(1989) 899-906.
[15]J.Frebortova,M.W.Fraaije,P.Galuszka,M.Sebela,P.Pec,J. Hrbac,O.Novak,K.D.Bilyeu,J.T.English,I.Frebort,Catalytic reaction of cytokinin dehydrogenase:preference for quinones as electron acceptors,Biochem.J.380(2004)121-130.
[16]D.J.Armstrong,Cytokinin oxidase and the regulation of cytokinin degradation,in:D.W.S.Mok,M.C.Mok(Eds.), Cytokinins:Chemistry,Activity,and Function,CRC Press, Boca Raton,1994.
[17]V.Pačes,E.Werstiuk,R.H.Hall,Conversionof N6-(Δ2-isopentenyl) adenosine to adenosine by enzyme activity in tobacco tissue, Plant Physiol.48(1971)775-778.
[18]N.Houba-Hérin,C.Pethe,J.d’Alayer,M.Laloue,Cytokinin oxidase from Zea mays:purification,cDNA cloning and expression in moss protoplasts,Plant J.17(1999)615-626.
[19]R.O.Morris,K.D.Bilyeu,J.G.Laskey,N.Cheikh,Isolation of a gene encoding a glycosylated cytokinin oxidase from maize, Biochem.Biophys.Res.Commun.255(1999)328-333.
[20]T.Werner,V.Motyka,V.Laucou,R.Smets,H.Van,T. Schmulling,Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity,Plant Cell 15(2003)2532-2550.
[21]S.H.Yang,H.Yu,C.J.Goh,Characterization of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid,Plant Mol.Biol.51 (2003)237-248.
[22]P.Galuszka,J.Frébortová,T.Werner,M.Yamada,M.Strnad, T.Schmülling,I.Frébort,Cytokinin oxidase/dehydrogenase genes in barley and wheat:cloning and heterologous expression,Eur.J.Biochem.271(2004)3990-4002.
[23]M.Ashikari,H.Sakakibara,S.Lin,T.Yamamoto,T.Takashi, A.Nishimura,E.R.Angeles,Q.Qian,H.Kitano,M.Matsuoka, Cytokinin oxidase regulates rice grain production,Science 309(2005)741-745.
[24]G.A.Tuskan,S.Difazio,S.Jansson,J.Bohlmann,I.Grigoriev, U.Hellsten,N.Putnam,S.Ralph,S.Rombauts,A.Salamov,J. Schein,L.Sterck,A.Aerts,R.R.Bhalerao,R.P.Bhalerao,D. Blaudez,W.Boerjan,A.Brun,A.Brunner,V.Busov,M. Campbell,J.Carlson,M.Chalot,J.Chapman,G.L.Chen,D. Cooper,P.M.Coutinho,J.Couturier,S.Covert,Q.Cronk,R. Cunningham,J.Davis,S.Degroeve,A.Déjardin,C. Depamphilis,J.Detter,B.Dirks,I.Dubchak,S.Duplessis,J. Ehlting,B.Ellis,K.Gendler,D.Goodstein,M.Gribskov,J. Grimwood,A.Groover,L.Gunter,B.Hamberger,B.Heinze,Y. Helariutta,B.Henrissat,D.Holligan,R.Holt,W.Huang,N. Islam-Faridi,S.Jones,M.Jones-Rhoades,R.Jorgensen,C. Joshi,J.Kangasjärvi,J.Karlsson,C.Kelleher,R.Kirkpatrick,M. Kirst,A.Kohler,U.Kalluri,F.Larimer,J.Leebens-Mack,J.C. Leplé,P.Locascio,Y.Lou,S.Lucas,F.Martin,B.Montanini,C. Napoli,D.R.Nelson,C.Nelson,K.Nieminen,O.Nilsson,V. Pereda,G.Peter,R.Philippe,G.Pilate,A.Poliakov,J. Razumovskaya,P.Richardson,C.Rinaldi,K.Ritland,P.Rouzé, D.Ryaboy,J.Schmutz,J.Schrader,B.Segerman,H.Shin,A. Siddiqui,F.Sterky,A.Terry,C.J.Tsai,E.Uberbacher,P.Unneberg,J.Vahala,K.Wall,S.Wessler,G.Yang,T.Yin,C. Douglas,M.Marra,G.Sandberg,Y.Van de Peer,D.Rokhsar, The genome of black cottonwood,Populus trichocarpa(Torr.& Gray),Science 313(2006)1596-1604.
[25]A.Pepper,F.Guinel,Abnormalroot and nodule vasculature in R50(sym16),a pea nodulation mutant which accumulates cytokinins,Ann.Bot.99(2007)765-774.
[26]K.Konagaya,S.Ando,S.Kamachi,M.Tsuda,Y.Tabei, Efficient production of genetically engineered,male-sterile Arabidopsis thaliana using anther-specific promoters and genes derived from Brassica oleracea and B.rapa,Plant Cell Rep.27(2008)1741-1754.
[27]L.Zhang,B.S.Zhang,R.H.Zhou,L.F.Gao,G.Y.Zhao,Y.X.Song, J.Z.Jia,Cloning and genetic mapping of cytokinin oxidase/ dehydrogenase gene(TaCKX2)in wheat,Acta Agron.Sin.33 (2007)1419-1425(in Chinese with English abstract).
[28]L.Zhang,B.S.Zhang,R.H.Zhou,X.Y.Kong,L.F.Gao,J.Z.Jia, Isolation and chromosomal localization of cytokinin oxidase/ dehydrogenase gene(TaCKX5)in wheat,Sci.Agric.Sin.41 (2008)636-642(in Chinese with English abstract).
[29]L.Zhang,Y.L.Zhao,L.F.Gao,G.Y.Zhao,R.H.Zhou,B.S.Zhang, J.Z.Jia,TaCKX6-D1,the ortholog of rice OsCKX2,is associated with grain weight in hexaploid wheat,New Phytol.195(2012) 574-584.
[30]D.S.Feng,H.G.Wang,X.S.Zhang,L.R.Kong,J.C.Tian,X.F.Li, Using an inverse PCR method to clone the wheat cytokinin oxidase/dehydrogenase gene TaCKX1,Plant Mol.Biol.Report. 26(2008)143-155.
[31]X.Ma,D.S.Feng,H.G.Wang,X.F.Li,L.R.Kong,Cloning and expression analysis of wheat cytokinin oxidase/dehydrogenase gene TaCKX3,Plant Mol.Biol.Report.29(2011)98-105.
[32]D.T.Le,R.Nishiyama,Y.Watanabe,R.Vankova,M.Tanaka, M.Seki,L.H.Ham,K.Yamaguchi-Shinozaki,K.Shinozaki,L.S.P. Tran,Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels,PLoS One 7(2012) e42411.
[33]E.Malito,A.Coda,K.D.Bilyeu,M.W.Fraaije,A.Mattevi, Structure of michaelis and product complexes of plant cytokinin dehydrogenase:implications for flavoenzyme catalysis,J.Mol.Biol.341(2004)1237-1249.
[34]J.T.Dietrich,M.Kaminek,D.G.Blevins,T.M.Reinbott,R.O. Morris,Changes in cytokinins and cytokinin oxidase activity in developing maize kernels and the effects of exogenous cytokinin on kernel development,Plant Physiol.Biochem.33 (1995)327-336.
[35]I.Bartrina,E.Otto,M.Strnad,T.Werner,T.Schmulling, Cytokinin regulates the activity of reproductive meristems, flower organ size,ovule formation,and thus seed yield in Arabidopsis thaliana,Plant Cell 23(2011)69-80.
[36]W.Zalewski,P.Galuszka,S.Gasparis,W.Orczyk,A. Nadolska-Orczyk,Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity,J.Exp.Bot.61(2010) 1839-1851.
[37]R.L.Gu,J.J.Fu,S.Guo,F.Y.Duan,Z.K.Wang,G.H.Mi,L.X. Yuan,Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase(CKX)gene family,J. Plant Growth Regul.29(2010)428-440.
[38]P.Galuszka,H.Popelková,T.Werner,J.Frébortová,H. Pospíšilová,V.Mik,I.Köllmer,T.Schmülling,I.Frébort, Biochemical characterization of cytokinin oxidases/ dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L,J.Plant Growth Regul.26(2007) 255-267.
[39]Z.N.Liu,Y.X.Lv,M.Zhang,Y.P.Liu,L.J.Kong,M.H.Zou,G.Lu, J.S.Cao,X.L.Yu,Identification,expression,and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage(Brassica rapa ssp.pekinensis),BMC Genomics 14 (2013)594.
[40]J.C.Suttle,L.L.Huckle,S.W.Lu,D.C.Knauber,Potato tuber cytokinin oxidase/dehydrogenase genes:biochemicalproperties, activity,and expression during tuber dormancy progression,J. Plant Physiol.171(2014)448-457.
[41]S.Mameaux,J.Cockram,T.Thiel,B.Steuernagel,N.Stein,S. Taudien,P.Jack,P.Werner,J.C.Gray,A.J.Greenland,W. Powell,Molecular,phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae,Plant Biotechnol.J.10(2012)67-82.
[42]G.Y.Zhang,X.Liu,Z.W.Quan,S.F.Cheng,X.Xu,S.K.Pan,M.Xie, P.Zeng,Z.Yue,W.L.Wang,Y.Tao,C.Bian,C.L.Han,Q.J.Xia,X. H.Peng,R.Cao,X.H.Yang,D.L.Zhan,J.C.Hu,Y.X.Zhang,H.N. Li,H.Li,N.Li,J.Y.Wang,C.C.Wang,R.Y.Wang,T.Guo,Y.J.Cai, C.Z.Liu,H.T.Xiang,Q.X.Shi,P.Huang,Q.C.Chen,Y.R.Li,J. Wang,Z.H.Zhao,J.Wang,Genome sequence of foxtailmillet (Setaria italica)provides insights into grass evolution and biofuel potential,Nat.Biotechnol.30(2012)549-554.
[43]J.L.Bennetzen,J.Schmutz,H.Wang,R.Percifield,J.Hawkins, A.C.Pontaroli,M.Estep,L.Feng,J.N.Vaughn,J.Grimwood,J. Jenkins,K.Barry,E.Lindquist,U.Hellsten,S.Deshpande,X. W.Wang,X.M.Wu,T.Mitros,J.Triplett,X.H.Yang,C.Y.Ye,M. Mauro-Herrera,L.Wang,P.Li,M.Sharma,R.Sharma,P.C. Ronald,O.Panaud,E.A.Kellogg,T.P.Brutnell,A.N.Doust,G.A. Tuskan,D.Rokhsar,K.M.Devos,Reference genome sequence of the model plant Setaria,Nat.Biotechnol.30(2012)555-561.
[44]A.Y.Guo,Q.H.Zhu,X.Chen,J.C.Luo,GSDS:a gene structure display server,Hereditas 29(2007)1023-1026.
[45]T.L.Bailey,M.Boden,F.A.Buske,M.Frith,C.E.Grant,L. Clementi,J.Ren,W.S.Noble,MEME SUITE:tools for motif discovery and searching,Nucleic Acids Res.37(2009)202-208. [46]M.A.Larkin,G.Blackshields,N.P.Brown,R.Brown,R.Chenna, P.A.McGettigan,H.McWilliam,F.Valentin,I.M.Wallace,A. Wilm,R.Lopez,J.D.Thompson,T.J.Gibson,D.G.Higgins, Clustal Wand ClustalX version 2.0,Bioinformatics 23(2007) 2947-2948.
[47]K.Tamura,J.Dudley,M.Nei,S.Kumar,MEGA4:molecular evolutionary genetics analysis(MEGA)software version 4.0, Mol.Biol.Evol.24(2007)1596-1599.
[48]K.Higo,Y.Ugawa,M.Iwamoto,T.Korenaga,Plant cis-acting regulatory DNA elements(PLACE)database,Nucleic Acids Res.27(1999)297-300.
[49]Y.J.Wang,D.X.Deng,Y.L.Bian,Y.P.Lv,Q.Xie,Genomewide analysis of primary auxin-responsiveAux/IAAgene family in maize(Zea mays.L.),Mol.Biol.Rep.37(2010)3991-4001.
[50]Z.B.Zhang,J.W.Zhang,Y.J.Chen,R.F.Li,H.Z.Wang,J.H.Wei, Genome-wide analysis and identification of HAK potassium transporter gene family in maize(Zea mays L.),Mol.Biol.Rep. 39(2012)8465-8473.
[51]C.D.Whitty,R.H.Hall,A cytokinin oxidase in Zea mays,Can.J. Biochem.52(1974)787-799.
[52]S.B.Cannon,A.Mitra,A.Baumgarten,N.D.Young,G.May, The roles of segmentaland tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana,BMC Plant Biol.4(2004)10.
[53]X.Yang,G.A.Tuskan,M.Z.Cheng,Divergence of the Dof gene families in poplar,Arabidopsis,and rice suggests multiple modes of gene evolution after duplication,Plant Physiol.142 (2006)820-830.
[54]Z.Yang,Y.Zhou,X.Wang,S.Gu,J.Yu,G.Liang,C.Yan,C.Xu, Genome wide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis,rice,and poplar,Genomics 92(2008)246-253.
[55]Q.W.Zeng,S.Qin,S.Q.Song,M.Zhang,Y.H.Xiao,M.Luo,L. Hou,Y.Pei,Molecular cloning and characterization of a cytokinin dehydrogenase gene from upland cotton (Gossypium hirsutum L.),Plant Mol.Biol.Report.30(2012)1-9.
[56]M.Trifunović,A.Cingel,A.Simonović,S.Jevremović,M. Petrić,I.Č.Dragićević,V.Motyka,P.I.Dobrev,L.Zahajská,A. Subotić,Overexpression of Arabidopsis cytokinin oxidase/ dehydrogenase genes AtCKX1 and AtCKX2 in transgenic Centaurium erythraea Rafn,Plant Cell Tissue Organ Cult.115 (2013)139-150.
[57]Y.Wang,J.P.Luo,Z.J.Wei,J.C.Zhang,Molecular cloning and expression analysis of a cytokinin oxidase(DhCKX) gene in Dendrobium huoshanense,Mol.Biol.Rep.36(2009) 1331-1338.
*Corresponding author.
E-mail address:20040092@163.com(Q.Xin).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
CKX gene family
Phylogenetic analysis
Real-time PCR analysis
杂志排行
The Crop Journal的其它文章
- Phenotypic analysis and molecular characterization of an allelic mutant of the D61 gene in rice
- Development and mapping of SSR markers linked to resistance-gene homologue clusters in common bean
- Variation and trends in dough rheologicalproperties and flour quality in 330 Chinese wheat varieties
- Effect of stripe rust on the yield response of wheat to nitrogen
- Effects of free-air CO2enrichment on adventitious root development of rice under low and normal soil nitrogen levels
- Evaluation of maize inbred lines currently used in Chinese breeding programs for resistance to six foliar diseases
