Phenotypic analysis and molecular characterization of an allelic mutant of the D61 gene in rice
2014-02-28XueyongLi
*,Xueyong Li,*
aCollege of Life Sciences,Liaocheng University,Liaocheng,Shandong 252059,China
bNational Key Facility for Crop Gene Resource and Genetic Improvement,Institute of Crop Science,Chinese Academy of Agricultural Sciences, Beijing 100081,China
cShandong Rice Research Institute,Jinan 250100,China
dCollege of Agriculture,Shandong Agricultural University,Tai'an,Shandong 271018,China
Phenotypic analysis and molecular characterization of an allelic mutant of the D61 gene in rice
Yanan Gaoa,b,Guangquan Wanga,Shoujiang Yuanc,Yanling Qinb,Jinfeng Zhaob, Yanpei Zhangb,d,Wenhui Zhanga,*,Xueyong Lib,*
aCollege of Life Sciences,Liaocheng University,Liaocheng,Shandong 252059,China
bNational Key Facility for Crop Gene Resource and Genetic Improvement,Institute of Crop Science,Chinese Academy of Agricultural Sciences, Beijing 100081,China
cShandong Rice Research Institute,Jinan 250100,China
dCollege of Agriculture,Shandong Agricultural University,Tai'an,Shandong 271018,China
A R T I C L E I N F O
Article history:
Received 28 February 2014
Received in revised form
17 April2014
Accepted 21 April2014
Available online 2 June 2014
Rice
Dwarf
Brassinosteroid
D61
OsBRI1
Brassinosteroids(BRs)are a class of plant-specific steroidal hormones that play important roles in multiple biological processes.In this paper,a classic rice mutant gsor300084, showing erect leaves and semi-dwarf stature,was characterized.Morphologicalanalysis in darkness showed that the mesocotyl of the gsor300084 mutant did not elongate when grown in darkness.Coleoptile elongation and root growth were less affected by the exogenous application ofbrassinolide(BL),the mostactive form of BR,in gsor300084 than in the wild-type rice variety Matsumae.Lamina joint bending analysis also showed that gsor300084 was less sensitive to exogenous BL than Matsumae.These results suggested that the gsor300084 mutant is defective in BR sensitivity.Map-based cloning indicated that gsor300084 is a novelallelic mutant of the DWARF61(D61)gene,which encodes the putative BR receptor OsBRI1.A single-base mutation appears in the LRR domain of OsBRI1,changing the 444th amino acid from tryptophan(W)to arginine(R).Subcellular localization analysis suggested that both the wild-type and mutant OsBRI1 protein are localized at the cytoplasmic membrane.Structure modeling revealed that the W444R substitution may affect the perception of BRs by the LRR domain.
©2014 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.All rights reserved.
1.Introduction
Brassinosteroids(BRs)are a class of steroid compounds involved in diverse biological processes during plant growth and development,including seed size and germination,stem elongation,plant height regulation,vascular differentiation, reproductive development,flowering time,male fertility, photomorphogenesis,and stress response[1-9].
A few BR-deficient or-insensitive mutants have been identified in Arabidopsis and rice,exhibiting pleiotropic phenotypes.BR-related mutants in Arabidopsis showed a distinctive dwarf phenotype with dark green leaves and exhibited defects in hypocotyl elongation and cotyledon closing when grown in darkness[10-13].The rice BR-related mutants showed dwarf phenotype,erect leaves,and small and round seeds and exhibited defects in mesocotyl elongation in darkness and leaf angle enlargement in the lamina joint inclination assay[3,4,14].
In plants BRs are perceived at the cell surface by a member of the large family of leucine-rich repeat receptor-like kinases (LRR-RLKs),namely BRASSINOSTEROID INSENSITIVE 1(BRI1) [15-17].The BRI1 gene was first cloned in Arabidopsis[18]and its ortholog D61 was also identified in rice[4].The BRI1 protein contains a hydrophobic signal peptide at the N-terminus,an extracellular leucine-rich repeat(LRRs)domain interrupted by a non-repetitive island domain,a transmembrane domain, and a cytoplasmic serine/threonine kinase domain[18,19].The kinase activity of BRI1 is essential for BR regulation of plant growth and development in rice[20].The N-terminal signal peptide is likely to be required for translocation of the nascent protein across a membrane,while the transmembrane domain is required to anchor the protein in the plasma membrane[21]. The island domain and the adjacent C-terminal LRR repeat of the extracellular domain are responsible for perceiving BRs [22-24].The LRRdomain may be involved in facilitating proteinprotein interactions between individual BRI1 molecules or with other proteins such as BAK1[25].
BR binding can enhance BRI1 heteromerization with BAK1 (BRI1-associated kinase 1),another LRR-RLK that is localized to the plasma membrane[25].In Arabidopsis,BAK1 and BRI1 share similar gene expression and subcellular localization patterns and physically associate with each other.BAK1/BRI1 interaction activates their kinase activities through transphosphorylation [26].Structure analysis reveals that BAK1 acts as a co-receptor to recognize the BRI1-bound brassinolide and the extracellular domains of BRI1 and BAK1 interact with each other in a BL-and pH-dependent manner[27].According to the solved crystal structure of the BRI1LRR-BL-BAK1LRR complex,the C-terminal two LRRs of BRI1LRR make extensive and direct contact with BAK1LRR[27].Thus the structural stability of the BRI1 LRR domain is very important for both BR perception and association with the co-receptor BAK1.
In the present study we characterized a classic semi-dwarf mutant with erect leaves in rice,designated as gsor300084. gsor300084 was insensitive to BRs and shown to be an allelic mutant of D61(OsBRI1).A point mutation in the LRR domain was found in the gsor300084 mutant.The potential effect of this mutation on BRI1 protein structure and function is discussed.
2.Materials and methods
2.1.Plant materials and growth conditions
The gsor300084 mutant and the wild-type variety Matsumae (Oryza sativa ssp.japonica,cv.Matsumae)were kindly provided by the USDA-ARS Dale Bumpers National Rice Research Center.The rice plants were grown in a paddy field at the experimental station of the Shandong Rice Research Institute, Shandong,China.
2.2.Skotomorphologicalanalysis
Rice seeds were soaked in water for 24 h and then sprouted at 37°C.Well-germinated seeds were transferred into 96-well plates supplemented with water and grown in the dark at 28°C for 20 days.
2.3.Response of rice seedling to BL
Seeds of the gsor300084 mutant and Matsumae were grown in half-strength MS solid medium with 0 or 1μmol L-1BL in a dark growth chamber at 28°C for 4 days.Coleoptile and root elongation analysis was performed by measuring the length of coleoptile and root treated with or without BL.
2.4.Lamina joint bending assay
Rice seeds were grown in half-strength MS solid medium with 0 or 1μmol L-1BL at 28°C under continuous light for 15 days. The angle between the second lamina and sheath was measured.
2.5.Map-based cloning
An F2mapping population from a cross between gsor300084 and the indica variety Dular was generated.InDelmarkers(170 pairs)and 20 F2individuals showing mutant phenotypes were used in primary mapping.Seven InDel markers were developed and 358 F2individuals were used for fine mapping. Genomic DNA fragments of the D61 gene from Matsumae and gsor300084 were amplified and sequenced.
2.6.Subcellular localization
The coding sequence of the D61 gene fromthe wild type and the gsor300084 mutant was fused in-frame to the 3′end of the sGFP gene in the transient expression vector pCAMBIA1205-GFP.The 1205-GFP-d61300084and 1205-GFP-D61 fusion constructs were transformed into protoplasts prepared from wild-type rice seedlings by polyethylene glycol treatment.The transformed protoplasts were incubated at 28°Cfor 16 h.Green fluorescence of the GFP fusion protein was observed under a Zeiss LSM 510 META confocalmicroscope.
3.Results
3.1.Pleiotropic phenotypes of the gsor300084 mutant
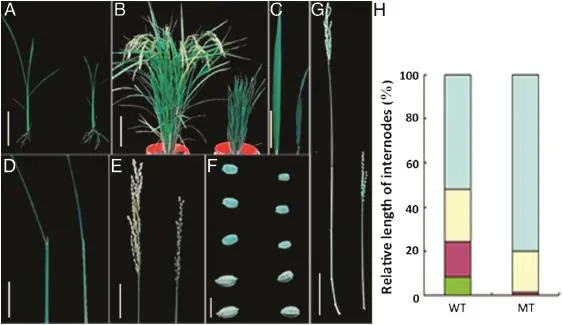
Fig.1-Morphologicalphenotypes of the gsor300084 mutant.(A)Gross morphology ofthe wild type Matsumae(left)and mutant gsor300084(right)at the seedling stage.Bar=5 cm.(B)Gross morphology at the maturity stage.Bar=20 cm.(C)Flag leaf morphology of wild type(left)and mutant(right).Bar=5 cm.(D)Leaf angle of wild type(left)and mutant(right).Bar=5 cm. (E)Panicle structure of wild type(right)and mutant(left).Bar=5 cm.(F)Seed morphology of wild type(left)and mutant(right). The upper three rows:dehusked seeds;the lower two rows:seeds with husk.Bar=5 mm.(G)Phenotypic exhibition of internodes of wild type(left)and mutant(right).Bar=10 cm.(H)Relative length of internode.From the top to the bottomare the first to the fourth internode,respectively.
The phenotype of the gsor300084 mutant was different from that of the wild type variety Matsumae in many respects.The plant height of gsor300084 was less than that of Matsumae from the seedling stage(Fig.1-A).At maturity,the plant height of gsor300084 was only about one half that of the wild type(Fig.1-B and Table 1).In wild-type plants,the leaf blade bent away from the vertical axis of the leaf sheath toward the abaxial side,but in gsor300084 most of the leaves were erect (Fig.1-D).The panicle of gsor300084 was smaller than that of the wild type(Fig.1-E).Moreover,the grains of gsor300084 were smaller and rounder(Fig.1-F and Table 1)and the grain weight was significantly reduced(Table 1).Internode elongation was severely inhibited(Fig.1-G)except for the uppermost internode(Fig.1-H),indicating that gsor300084 is a d6-type dwarf mutant[28].
3.2.Skotomorphogenic phenotype
In rice mutants with defects in BR biosynthesis or sensitivity, elongation of the mesocotyl is inhibited when plants are grown in complete darkness[2].To determine whether gsor300084 is a BR-related mutant,the mesocotyl internode elongation pattern in the darkness was observed.After growth in complete darkness for two weeks,the wild type plants exhibited mesocotyl elongation,whereas no such elongation occurred in gsor300084(Fig.2).The failure ofmesocotyl elongation in gsor300084 is similar to the phenotype ofother rice BR-related mutants grown in darkness[4,29].

Table 1-Comparison of morphological traits between gsor300084 and wild type.
3.3.The gsor300084 seedlings were less sensitive to BL than wild type
Based on the abnormal phenotypes of gsor300084,we suspected that the gsor300084 mutant was defective in BR biosynthesis or sensitivity.To identify the type of BR mutant to which gsor300084 belongs,we evaluated the coleoptile elongation and root length of wild type and 300084 seedlings in response to BL.Rice seeds were germinated in half-strength MS medium with 0 or 1μmol L-1BL in complete darkness. In wild-type plants,coleoptile elongation was promoted,whereas root length was significantly inhibited by the presence of 1μmol L-1BL.However,coleoptile and root length did not vary significantly after BR treatment(Fig.3-A, B and C).

Fig.2-Skotomorphogenic phenotype of a wild type(left)and the gsor300084 mutant(right).Arrowhead indicates elongated mesocotyl.Bar=5 mm.
3.4.Lamina joint inclination assay confirmed the insensitivity of gsor300084 to BL
A more sensitive method,lamina joint inclination assay[30], was used to examine sensitivity of gsor300084 to BL.In the absence of BL,the bending angle of the leaf blade in gsor300084 was smaller than that in wild-type plants (Fig.4-A,B).In the presence of 1μmol L-1BL,the leaf angle increased dramatically in the wild type but remained almost unchanged in the gsor300084 mutant(Fig.4-A,B).This result further confirmed that gsor300084 mutant seedlings were less sensitive to exogenous BL than wild-type seedlings.
3.5.gsor300084 is a novelallelic mutant of the D61 gene
Primary mapping using 20 F2mutant individuals derived from a cross between gsor300084 and the indica variety Dular revealed that the mutation resided on the long arm of chromosome 1 between the InDel markers R1-12 and R1-13 (Fig.5-A and Table 2).Fine mapping using 7 new inDel markers and 358 F2mutant individuals narrowed the location to a 107 kb region.According to the MSU Rice Genome Annotation Project(http://rice.plantbiology.msu. edu/),the D61 gene(LOC_Os01g52050)encoding the putative BR receptor Os BRI1 is located within this region.Accordingly, the genomic DNA fragment of the D61 gene from both gsor300084 and Matsumae was amplified and sequenced. Sequence analysis revealed that only the 1330th base in the D61 coding region was changed from T to A,causing the 444th amino acid tryptophan(W)to be substituted by arginine(R).This mutation was located in the LRR region of OsBRI1 and adjacent to the island domain(Fig.5-A).Sequence alignment among BRI1 orthologs from different plant species revealed that this mutation site is highly conserved (Fig.5-B),indicating that this residue is important for BRI1 protein function.
3.6.Subcellular localization of OsBRI1 was not altered
Although leucine-rich repeats(LRRs)are frequently involved in protein-protein interactions,a previous study had shown that mutations in the LRR domain led to changed protein subcellular localization[31].We accordingly wondered whether the W444R substitution has any effects on the subcellular localization of OsBRI1.The wild-type and gsor300084 mutant allele of the D61 gene fused in-frame with the sGFP gene was transformed into rice protoplasts.Fig.6 presents a confocal microscopy analysis of OsBRI1 expression,showing that OsBRI1::GFP fluorescence is localized to the cell surface. Thus the OsBRI1-directed GFP fluorescence is at the cell wallor plasma membrane but not in the cytoplasm.Since the cell wall has been removed during the preparation of protoplasts from rice seedlings,OsBRI1 should be localized at the plasma membrane.This result is consistent with the subcellular localization analysis of the Arabidopsis BRI1 protein,in which a plasmolysis experiment confirmed that BRI1 was localized at the plasma membrane rather than at the cell wall[24,26].Interestingly,the green fluorescence of both the wild type and mutant sGFP-OsBRI1 was detected at the plasma membrane(Fig.6).This result indicates that the W444R substitution has no effect on the OsBRI1 subcellular localization.
4.Discussion
Brassinosteroids(BRs)are a class of steroid compounds involved in diverse biological processes during plant growth and development.Here we have reported a classic semi-dwarf and erect-leaf rice mutant gsor300084.It belongs to the d6-type dwarf mutants,in which internode elongation was severely inhibited except for the uppermost internode.The gsor300084 mutant was shown to be related to BR and was less sensitive to BRs by assays of coleoptile elongation,root growth inhibition,and lamina joint inclination in the presence of exogenous BL.All these results indicate that gsor300084 is a BR-insensitive mutant.Map-based cloning showed that gsor300084 is a novel allelic mutant of the D61/OsBRI1 gene.The 444th amino acid,tryptophan(W),located in the LRR domain,is substituted by arginine(R)and the mutation site is highly conserved among BRI1 orthologs from different plant species.These results suggest that this mutation site is important for BRI1 protein function and BRI1-mediated plant growth and development.

Fig.3-Response of rice seedlings to BL.(A)Effect of BL on coleoptile and root elongation in wild type(WT)andgsor300084mutant(MT).Arrow indicates the tip of coleoptile.Bar=10 mm.(B)Root length of WT and MT seedling in absence or presence of 1μmol L-1BL.(C)Coleoptile length of WT and MT seedling in the absence or presence of 1μmol L-1BL.**Significantly different atP<0.01 level.Bar on the top of each column represents standard error,n=15.

Fig.4-Lamina joint bending responses to BL in wild type(WT)and gsor300084(MT).(A)Effect of BL on lamina joint bending in WT and MT.(B)Change in leaf angle after BR treatment.**Significantly different at P<0.01 level.n=15.
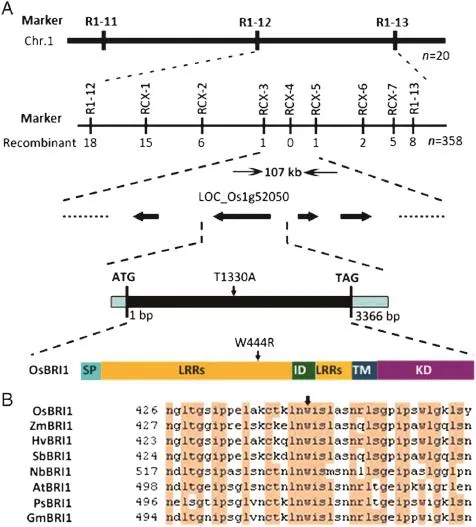
Fig.5-Map-based cloning of the target gene underlying the gsor300084 phenotype and structure of the OsBRI1 protein. (A)Map-based cloning of the causative gene for the gsor300084 mutant.The upper three rows show the map-based cloning process with molecular markers and recombinants indicated above and below the line,respectively.The lower two rows show the gene structure and protein domains of OsBRI1 with a signal peptide(SP)domain,leucine-rich repeat(LRR)domain,island domain(ID),transmembrane(TM)domain and kinase domain(KD).The change of T1330A in gsor300084 results in the conversion of tryptophan(W)to arginine(R)at the 444th residue in the LRR domain.(B)Alignment of partial sequence surrounding the mutation site(indicated by arrow)in BRI1 orthologs from different plant species including Oryza sativa OsBRI1 (BAB68053),Zea mays ZmBRI1(AFW83751),Hordeum vulgare HvBRI1(BAD01654),Sorghum bicolor SbBRI1(XP_002458412), Nicotiana benthamiana NbBRI1(ABO27628),Arabidopsis thaliana AtBRI1(NP_195650),Pisum sativum PsBRI1(BAC99050),and Glycine max GmBRI1(NP_001237411).
Table 2-Primer sequences used for map-based cloning.
More than ten allelic mutants of D61 have been reported in rice[4,20,32,33].Only four(d61-2,d61-3,d61-5,and d61-7)have distinctive mutations in the LRR domain and show various degrees of phenotypic severity.In d61-2 the 491st amino acid, valine,is substituted by methionine,producing an intermediate phenotype[32].d61-3 and d61-5 are two severe mutants that harbor the H420P and N426Y substitution,respectively. The phenotype of d61-7,in which the 467th amino acid is changed from alanine to valine,is milder[33].The gsor300084 mutant described in this study,harboring the W444R substitution,most resembles d61-2,showing an intermediate phenotype.Interestingly,the five mutation sites(H420P, N426Y,W444R,A467V,and V491M)are clustered together in a small portion of the LRR domain,which may be a potential essential motif for BRI1 function.However,the manner in which these mutations affect the OsBRI1 function remains unclear.Our protein localization analysis revealed that defects other than subcellular localization account for the OsBRI1 dysfunction.
The extracellular domain of Arabidopsis BRI1 contains 25 LRR repeats and a 70-amino acid island domain between the 21st and 22nd LRR[18].The crystal structure of the extracellular domain of AtBRI1 has been resolved.The AtBRI1 LRR comprises a helical solenoid structure,while the separate island domain anchors onto the inner surface of the solenoid and spans six LRRs(LRR 17-22)[22,23].The brassinolide molecule binds to a hydrophobic groove between the island domain and the inner surface of the LRRs.Thus both the island domain and the adjacent C-terminal LRR repeat(LRR 17-22)contribute to the formation of the hormone binding site [22,23].Extensive non-covalent interactions occur between the island domain and the solenoid structure.For example, W516,I540,W564,and F658 in LRRs establish close contacts with the island domain[23].Several Arabidopsis mutants in the island domain and adjacent LRRs exhibit a BR-insensitive phenotype.For example,bri1-6,carrying the G644D mutation in the island domain,shows a loss-of-function phenotype[34]; bri1sud1,carrying the G643E mutation in the island domain, stabilizes the island domain and shows a gain-of-function phenotype[35].The loss-of-function allele bri1-9(S662F in the 22nd LRR)has been mapped to the island domain-LRR interface and probably interferes with folding of the island domain[34].
The W444R mutation in the rice gsor300084 mutant is equivalent to the W516 in the 19th LRR of the Arabidopsis BRI1 protein[18],which is involved in the formation of the brassinolide binding site as described above.Thus,although the W444R mutation occurs outside of the island domain (from L508 to F577),it still likely adversely affects the perception of BL.Compared with the Arabidopsis BRI1 (AtBRI1)protein,the rice BRI1(OsBRI1)protein lacks three LRR domains,corresponding to the third to fifth LRR repeats of AtBRI1[4].Thus,the LRRs that contribute to the formation of the hormone binding site are expected to be LRR14-19 in OsBRI1.We performed in silico structure modeling of the extracellular domain of the wild-type and gsor300084 mutantOsBRI1.There was no dramatic change in the BR binding groove formed between the island domain and LRR14-19 (Fig.7).However,the change from the neutral hydrophobic tryptophan to the basic hydrophilic arginine may exert a subtle effect on the hydrophobic environment of the binding groove(Fig.7).So the W444R mutation can perturb local conformations and consequently hinder BRI1 recognition of brassinosteroids.The rice gsor300084 mutant,together with other missense mutations,will play useful roles in assigning functions to specific domains or motifs and allow us to validate the structural model of the BRI1 protein.
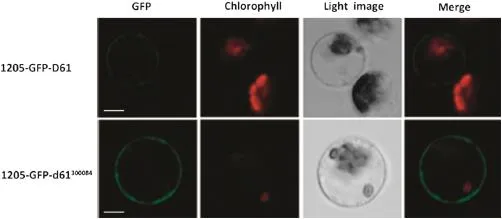
Fig.6-Subcellular localization of the wild type and mutant OsBRI1 protein.The 1205-GFP-D61 and 1205-GFP-d61300084vector was transformed into protoplasts prepared fromrice seedlings.Green fluorescence ofthe GFP fusion protein was viewed under a confocal microscope.Red fluorescence of chlorophyll and light-field images are also shown.Bar=10μm.
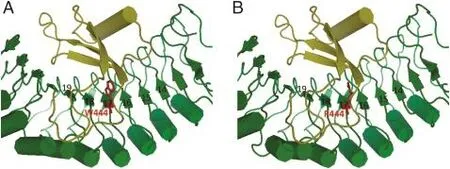
Fig.7-In silico structure modeling ofthe extracellular domains ofthe wild type and gsor300084 mutant OsBRI1.The structure is predicted by CBS Prediction Servers(http://www.cbs.dtu.dk/services/),viewed and compared by PyMOL.(A)In silico structure modeling of the extracellular domain of the wild-type OsBRI1.(B)In silico structure modeling ofthe extracellular domain of the gsor300084 mutant OsBRI1.The LRR and island domain are shown in green and yellow cartoons,respectively.The position of the W444R mutation is shown as a red stick.
Acknowledgments
We thank the USDA-ARS Dale Bumpers National Rice Research Center for providing the rice gsor300084 mutant.This work was supported by grants from the Ministry of Science and Technology of China(Grant No.2013CBA01401),the Ministry of Agriculture of China(Grant No.2011ZX08009-003) and the Agricultural Science and Technology Innovation Program of China.
R E F E R E N C E S
[1]S.D.Clouse,M.Langford,T.C.McMorris,A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development,Plant Physiol.111(1996)671-678.
[2]Z.Hong,M.Ueguchi-Tanaka,K.Umemura,S.Uozu,S. Fujioka,S.Takatsuto,S.Yoshida,M.Ashikari,H.Kitano,M. Matsuoka,A rice brassinosteroid-deficient mutant,ebisu dwarf(d2),is caused by a loss offunction of a new member of cytochrome,Plant Cell15(2003)2900-2910.
[3]Z.Hong,M.Ueguchi-Tanaka,S.Shimizu-Sato,Y.Inukai,S. Fujioka,Y.Shimada,S.Takatsuto,M.Agetsuma,S.Yoshida, Y.Watanabe,S.Uozu,H.Kitano,M.Ashikari,M.Matsuoka, Loss-of-function of a rice brassinosteroid biosynthetic enzyme,C-6 oxidase,prevents the organized arrangement and polar elongation of cells in the leaves and stem,Plant J. 32(2002)495-508.
[4]C.Yamamuro,Y.Ihara,X.Wu,T.Noguchi,S.Fujioka,S. Takatsuto,M.Ashikari,H.Kitano,M.Matsuoka,Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint,Plant Cell 12(2000)1591-1605.
[5]J.Cheon,S.Y.Park,B.Schulz,S.Choe,Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction,BMC Plant Biol.10 (2010)270.
[6]H.Fukuda,Signals that controlplant vascular cell differentiation,Nat.Rev.Mol.Cell Biol.5(2004)379-391.
[7]L.Song,X.Y.Zhou,L.Li,L.Xue,X.Yang,H.W.Xue, Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis, Mol.Plant 2(2009)755-772.
[8]G.M.Steber,P.McCourt,A role for brassinosteroids in germination in Arabidopsis,Plant Physiol.125(2001)763-769.
[9]T.W.Kim,M.M.Michniewicz,D.C.Bergmann,Z.Y.Wang, Brassinosteroid inhibits stomatal development by releasing GSK3-mediated inhibition of a MAP kinase pathway,Nature 482(2012)419-422.
[10]H.S.Yun,Y.H.Bae,Y.J.Lee,S.C.Chang,S.K.Kim,J.Li,K.H. Nam,Analysis of phosphorylation of the BRI1/BAK1 complex in Arabidopsis reveals amino acid residues critical for receptor formation and activation of BR signaling,Mol.Cell.27(2009) 183-190.
[11]Z.Yan,J.Zhao,P.Peng,R.K.Chihara,J.Li,BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling,Plant Physiol.150(2009) 710-721.
[12]R.Karlova,S.Boeren,E.Russinova,J.Aker,J.Vervoort,S.de Vries,The Arabidopsis somatic embryogenesis receptor-like KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1,Plant Cell18(2006)626-638. [13]K.Oki,N.Inaba,K.Kitagawa,S.Fujioka,H.Kitano,Y. Fujisawa,H.Kato,Y.Iwasaki,Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling, Plant Cell Physiol.50(2009)161-172.
[14]L.Y.Zhang,M.Y.Bai,J.Wu,J.Y.Zhu,H.Wang,Z.Zhang,W. Wang,Y.Sun,J.Zhao,X.Sun,H.Yang,Y.Xu,S.H.Kim,S. Fujioka,W.H.Lin,K.Chong,T.Lu,Z.Y.Wang,Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis,Plant Cell 21(2009)3767-3780.
[15]S.D.Clouse,Brassinosteroid signal transduction:from receptor kinase activation to transcriptional networks regulating plant development,Plant Cell 23(2011)1219-1230.
[16]Z.Y.Wang,J.X.He,Brassinosteroid signal transduction-choices of signals and receptors,Trends Plant Sci.9(2004)91-96.
[17]J.Gao,Y.Y.Ma,Y.N.Sun,H.D.Zhao,D.P.Hong,L.M.Yan,Z.Y. Lou,Crystallization and preliminary crystallographic analysis of Arabidopsis thaliana BRI1-associated kinase 1 (BAK1)cytoplasmic domain,Acta Crystallogr.Sect.F:Struct. Biol.Cryst.Commun.68(2012)340-342.
[18]J.Li,J.Chory,A putative leucine-rich receptor kinase involved in brassinosteroid signal transduction,Cell 90 (1997)929-938.
[19]T.Kinoshita,A.Caño-Delgado,H.Seto,S.Hiranuma,S. Fujioka,S.Yoshida,J.Chory,Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1, Nature 433(2005)167-171.
[20]J.F.Zhao,C.X.Wu,S.J.Yuan,L.Yin,W.Sun,Q.L.Zhao,X.Y.Li, Kinase activity of OsBRI1 is essential for brassinosteroids to regulate rice growth and development,Plant Sci.199(2013) 113-120.
[21]N.Geldner,D.L.Hyman,X.Wang,K.Schumacher,J.Chory, Endosomalsignaling of plant steroid receptor kinase BRI1, Genes Dev.21(2007)1598-1602.
[22]M.Hothorn,Y.Belkhadir,M.Dreux,T.Dabi,J.P.Noel,I.A. Wilson,J.Chory,Structural basis of steroid hormone perception by the receptor kinase BRI1,Nature 474(2011) 467-471.
[23]J.She,Z.Han,T.W.Kim,J.Wang,W.Cheng,J.Chang,J.Chai, Structural insight into brassinosteroid perception by BRI1, Nature 474(2011)472-476.
[24]D.M.Friedrichsen,C.A.Joazeiro,J.Li,T.Hunter,J.Chory, Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase,Plant Physiol.123(2000)1247-1256.
[25]J.Li,J.Wen,K.A.Lease,J.T.Doke,F.E.Tax,J.C.Walker,BAK1, an Arabidopsis LRR receptor-like protein kinase,interacts with BRI1 and modulates brassinosteroid signaling,Cell2002(110) (2002)213-222.
[26]K.H.Nam,J.Li,BRI1/BAK1,a receptor kinase pair mediating brassinosteroid signaling,Cell 110(2002)203-212.
[27]Y.D.Sun,Z.F.Han,J.Tang,Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide,Cell Res. 23(2013)1326-1329.
[28]K.Takeda,Internode elongation and dwarfism in some gramineous plants,Gamma Field Symp.16(1977)1-18.
[29]A.Tanaka,H.Nakagawa,C.Tomita,Z.Shimatani,M.Ohtake, T.Nomura,C.J.Jiang,J.G.Dubouzet,S.Kikuchi,H.Sekimoto, T.Yokota,T.Asami,T.Kamakura,M.Mori, BRASSINOSTEROID UPREGULATED1 encoding a helix-loop-helix protein,is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice,Plant Physiol.151(2009)669-680.
[30]S.Tanabe,M.Ashikari,S.Fujioka,S.Takatsuto,S.Yoshida,M. Yano,A.Yoshimura,H.Kitano,M.Matsuoka,Y.Fujisawa,H. Kato,Y.Iwasaki,A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant,dwarf11,with reduced seed length,Plant Cell 17(2005)776-790.
[31]H.Zhi,H.Jin,T.Tzfira,J.Li,Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis,Plant Cell 20(2008) 3418-3429.
[32]Y.Morinaka,T.Sakamoto,Y.Inukai,M.Agetsuma,H.Kitano, M.Ashikari,M.Matsuoka,Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice,Plant Physiol.141(2006)924-931.
[33]A.Nakamura,The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3,in rice,Plant Physiol.140(2006)580-590.
[34]T.Noguchi,S.Fujioka,S.Choe,S.Takatsuto,S.Yoshida,H. Yuan,F.E.Tax,Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids,Plant Physiol.121 (1999)743-752.
[35]J.Santiago,C.Henzler,M.Hothorn,Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases,Science 341(2013)889-892.
*Corresponding authors.
E-mail addresses:whzhang@lcu.edu.cn(W.Zhang),lixueyong@caas.cn(X.Li).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
杂志排行
The Crop Journal的其它文章
- Development and mapping of SSR markers linked to resistance-gene homologue clusters in common bean
- Variation and trends in dough rheologicalproperties and flour quality in 330 Chinese wheat varieties
- Effect of stripe rust on the yield response of wheat to nitrogen
- Effects of free-air CO2enrichment on adventitious root development of rice under low and normal soil nitrogen levels
- Evaluation of maize inbred lines currently used in Chinese breeding programs for resistance to six foliar diseases
- Effect of different levels of nitrogen deficiency on switchgrass seedling growth
