Genetic diversity and differentiation of masu salmon (Oncorhynchus masou masou) between and within cultured populationsinferred from microsatellite DNA analysis
2012-12-25ZhiyingJIAYuyongZHANGShuqiangCHENLianyuSHI
Zhiying JIA, Yuyong ZHANG, Shuqiang CHEN,, Lianyu SHI,*
1. Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin 150070, China;2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China
Genetic diversity and differentiation of masu salmon (Oncorhynchus masou masou) between and within cultured populations
inferred from microsatellite DNA analysis
Zhiying JIA1, Yuyong ZHANG1, Shuqiang CHEN1,2, Lianyu SHI1,*
1.Heilongjiang River Fisheries Research Institute,Chinese Academy of Fishery Sciences,Harbin150070,China;
2.College of Fisheries and Life Science,Shanghai Ocean University,Shanghai201306,China
Masu salmon,Oncorhynchus masou masou, is one of the most valuable fishery species that has been introduced to China,though to date no studies on the genetic diversity and genetic relationship among hatchery populations has been performed with molecular markers. We undertook such a study and sampled 120 individuals from three hatchery stocks and analyzed 20 microsatellite loci. All loci were polymorphic and a total of 91 alleles were detected. A relatively low level of genetic diversity was revealed with effective number of allele of 3.1094, 3.3299 and 3.1894 and expected heterozygosity of 0.6600, 0.6648 and 0.6638 in the three stocks, respectively. Deviations from Hardy-Weinberg equilibrium were found due to heterozygote deficit. Accordingly,evidence of genetic bottlenecks were found in the three stocks. An individual assignment test demonstrated that 85% of individuals were correctly assigned into their original stocks. PairwiseFstrevealed that significant differentiation occurred between these three stocks. The results of the study indicated that disequilibrium of genetic structure and differentiation has occurred in all three stocks.This information collectively provides a basis for measures to avoid of loss of genetic diversity and introgression in Chinese aquaculture.
Oncorhynchus masou masou; Microsatellite; Genetic diversity; Genetic differentiation
Masu salmon,O. masou masou,were widely distributed in Japanese mountain streams (Kano et al,2010) before being introduced to China in the 1990s(Wang, 1998). The fish is now one of the most valuable fishery species in cold-water aquaculture. At present,commercial catches ofO. masoumasouin Japan have decreased and hatchery programs have been established to release fry to its habitat (Yu et al, 2010). In China,because of its good taste and flesh quality, the fish has been an important aquaculture species (Wang, 1998).However, successive artificial selection and serious stress from inbreeding have led to the remarkable degradation in the fish: early sex maturity, smaller size and silvered body color (Zhang et al, 2011). A second batch ofO. masoumasouwas introduced into China in 2008 to counter these changes.
High genetic diversity is important for the population conservation because it is closely related to population’s fitness and adaptability to environment as well as selection (Filgueira et al, 2010). Loss of genetic variation among hatchery stocks is a common phenomenon caused by genetic drift in many species, including mrigal (Aung et al, 2010), Japanese flounder (Sekino et al, 2002) and common carp (Kohlmann et al, 2005). At present, there are threeO. masoumasouhatchery stocks in China, but their genetic diversity and relationship have not been reported1despite clear indications of genetic abnormalities that prompted the second importation of stock in 2008. To avoid a repeat of the earlier incidence,information on genetic variation of these hatchery stocksis urgently needed to develop a coherent management and conservation plan for this species.
Microsatellites DNA markers have been proven to be useful markers in researching genetic diversity and population structure in fish such as Japanese flounder(Sekino et al, 2002) and common carp (Kohlmann et al,2005). We aim to investigate the genetic diversity and population relationships of the Chinese three stocks ofO.masoumasouusing 20 pairs of microsatellite primers developed for rainbow trout to assess the genetic diversity and their relationship within and among stocks ofO. masoumasou.
MATERIALS AND METHODS
Sample collection and history of stocks studied
Two batches of fertilized eggs ofO. masoumasouwere introduced into China, the first in 1996 and the second 2008. Both were kept in Bohai Experimental Station. In 1998, parts ofO. masou masouindividuals were taken to Beijing Shuntong Rainbow Trout Culture Center and regarded as a separate stock population after several generations of artificial propagation, resulting in three stocks. We named the three stocks we used BO1996, Beijing, and BO2008. In 2010, we collected 40 samples from each of the presumptive stocks. And the samples of the BO2008 stock were collected directly from the introduced population. Fin-clip tissues were preserved in 75%ethanol (Jia et al, 2008) for BO1996 and Beijing stocks.BO2008 stock samples were collected as fry just before beginning to float.
DNA extraction and microsatellite DNA analysis
Genome DNA was extracted using the standard proteinase K, phenol-chloroform-isoamyl procedure(Wei et al, 2001). 20 microsatellite loci (Table1)developed for other salmon were used in the study. The OMM3005 primer was from NCBI (http://www.ncbi.nlm.nih.gov/nuccore/20142355) and others were from the articles of Rexroad et al (2002a,b;2005). Primer conditions, mainly annealing temperatures, were optimized for amplification. PCR was performed using a reaction volume of 15 μL containing 50 ng of genomic DNA, 0.2 μmol/L of each dNTP (TaKaRa, China),0.1 μmol/L of each primer, 1.5 mmol/L MgCl2, 1×PCR buffer (TaKaRa, China) and 1UTaqDNA polymerase. The thermal cycling (Biometra) began with 3 min at 94 ºC, followed by 27 cycles of denaturing at 94ºC for 30 s, annealing temperature from 50 ºC to 62 ºC depending on different primers for 30 s, elongation for 30 s at 72 ºC, followed by a final extension for 5 min at 72 ºC. PCR products were separated in 10%polyacrylamide gels.
Statistical analyses
The effective number of alleles per locus (Ne),number of detected alleles (Na), observed heterozygosity(Ho), expected heterozygosity (He) and allele frequency were calculated using POPGENE 1.32(Yeh et al, 2000).To test the difference between populations ofNeandHe,non-parametrict-tests (Sokal & Rohlf, 1995) were performed using Microsoft Excel.
Deviations from Hardy–Weinberg equilibrium(HWE) were estimated using GENEPOP 4.0 (Raymond& Rousset, 1995). ExactP-values were evaluated with Markov chain method (dememorization=10000;batches=100; iterations per batch=5 000). Additionally,deviations fromHWEin the direction of heterozygote excess or deficit were also carried out.
Analysis of molecular variance (AMOVA),implemented in GenAlEx (Peakall & Smouse, 2006),was used to partition total genetic variation hierarchically among stocks.
Genetic distance between populations was estimated by the PHYLIP software package, version 3.69(Felsenstein, 1993). Bootstrap re-sampling (1 000 replicates) was performed to test the reliability of dendrogram implemented in PHYLIP software.
STRUCTURE 2.2 (Pritchard et al, 2000) was used to determine the most likely number of genetic clusters(K=1−5). An assignment test was also performed in order to determine the extent to which individuals could be correctly assigned to their population of origin. In the analysis, the simulation was used assuming admixture and correlated allele frequencies between populations with a 10 000 replications burn-in period and 100 000 MCMC replicates.
Evidence of population bottlenecks were tested with Bottleneck 1.2.02 (Cornuet & Luikart, 1997) and the Wilcoxon sign-rank test was used to determine if a population exhibits a significant number of loci with heterozygote excess with a two-phased model of mutation (TPM, as recommended in the program). The mode-shift indicator was also employed to discriminate bottlenecked populations from stable populations.
RESULTS
PCR results of 20 pairs of microsattelite primers
According to the clarity and size of banding patterns, 20 pairs of primers showed good images and all were polymorphic in the sampled populations (Figure 1).
Genetic diversity and linkage disequilibrium
Summary statistics showing allelic variation within loci and population are presented in Table 1. A total of 91 alleles were detected in the 20 loci among 120 individuals with an average of 4.55, ranging from threealleles at loci AF352740, AF352758 and OMM5020 to 7 alleles at locus AF352763. All loci were polymorphic in all samples from three stocks.HoandHewithin stocks ranged from 0.2973 (BO2008) to 0.9714 (BO1996)and from 0.3225 (Beijing) to 0.8290 (Beijing),respectively.

Figure 1 Polyacrylamide gel results at the OMM5017 locus for three stocks of masu salmon
Exact tests for genotypic linkage disequilibrium showed significant deviations in the stocks of BO1996(OMM1036 and OMM1035, OMM1036 and AF352744,AF352738 and AF352750 pairs) and Beijing (OMM1045 and AF352763, AF352763 and af352746, AF352750 and AF352744 pairs) at the 5% level. But disequilibria were not consistently observed in the same locus pairs in each population.
Genetic structure with and among populations
The effective number of allele per population was 3.1094, 3.3299 and 3.1894 in BO1996, BO2008 and Beijing respectively. The mean expected heterozygosity per population was 0.6172, 0.6171 and 0.6299 in BO1996, BO2008 and Beijing respectively. No significant difference of genetic diversity was found between stocks.
Deviations fromHWEwere found in all three populations (Table 1). Heterozygote deficit was observed in BO2008 stock while for the other two, however,heterozygote deficits existed at only few loci and did not achieve population level. The BO1996 population exhibited the highest number of loci (11) deviated fromHWEand then BO2008 (10) and Beijing (9) stock populations. Two loci (OMM5017 and OMM5010)deviated fromHWEin all populations but a heterozygote deficiency existed in different population (OMM5017 for BO2008 and Beijing, OMM5010 for BO2008 at 5%level). A significant bottleneck was found for three stocks (P [heterozygote excess] = 0.00000, 0.00001 and 0.00136 for BO1996, BO2008 and Beijing, respectively and the shifted model).
For the real number of clusters, once the cluster (K)is reached,L(K) at largerKsplateaus or continues increasing slightly. In our analysis,L(K) at largerKsplateaus whenK=3, which is consistent with the groups as individual sampled (Figure 2). And theΔKvalue that could give a clear peak at the true value ofKaccording to procedures of Evanno et al (2005) was in Figure 3.The self-assignment test resulted in correct assignment of 102 of the 120 individuals in the three sampled groups,corresponding to 85.0% (Figure 4). The number of misclassi fi ed individuals in the three groups was four,seven, and seven with the ratios of 3.33%, 5.83%, and,5.83% respectively.
The locality of origin for each individual is indicated in the X-axis.
Genetic relationships
The global test for population differentiation was significant withFstvalue of 0.055 (P=0.01). PairwiseFstestimates and level of significance for all population pairs are presented in Table 3. AllFstvalues were significant (allP=0.01) and the highestFstwas foundbetween the BO1996 and BO2008 populations. Global subdivision with the use of AMOVA revealed that the percentage of variation among populations was 5%, 4%among individuals, and 91% within individuals. This indicates that the total variation of microsatellite loci was due to variation among individuals, rather than among stocks.

Figure 3 ΔK value estimated at the K ranged from 1 to 5
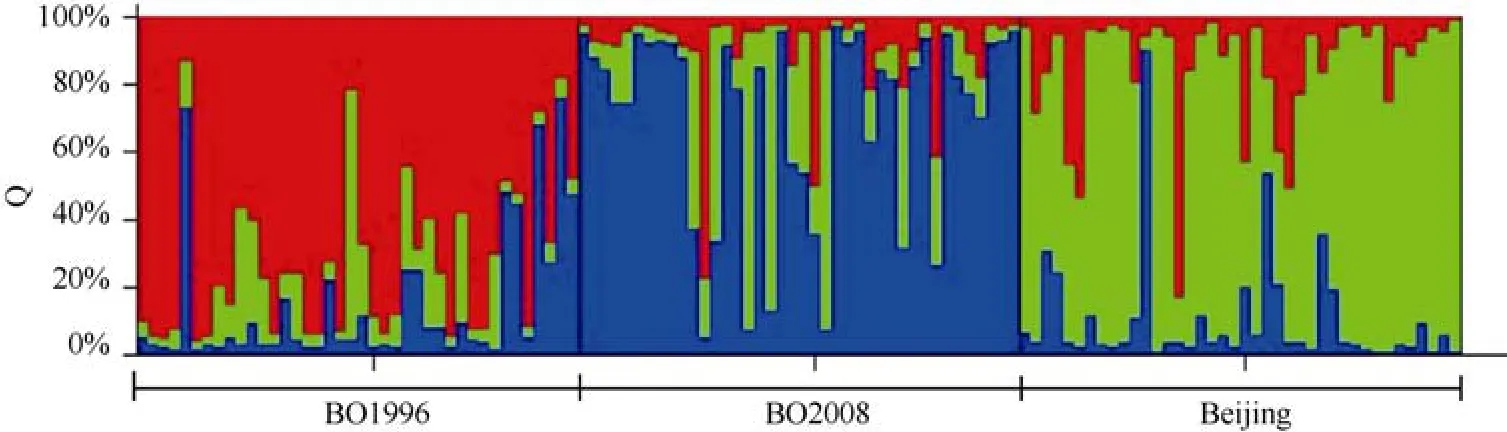
Figure 4 Proportional membership (Q) of each individual in the three clusters
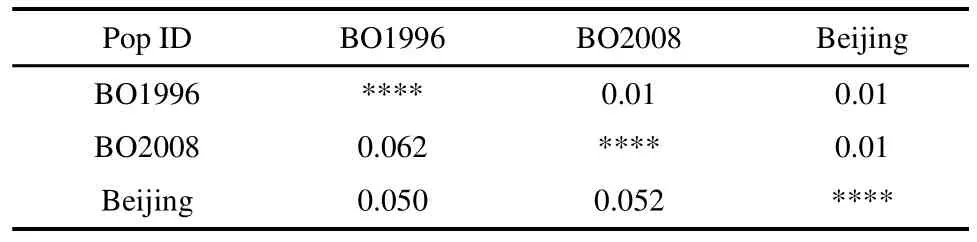
Table 3 Pairwise Population Fst Values at 20microsatellite loci
Pairwise genetic distances among hatchery populations (Table 4) ranged from 0.1282 (BO1996–Beijing) to 0.1349 (BO1996–BO2008), which was consistent withFstresults.
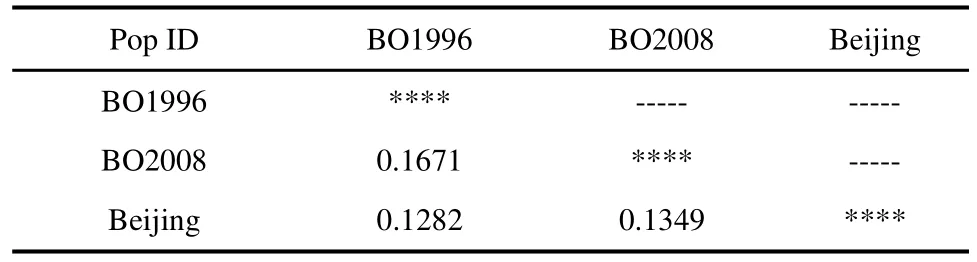
Table 4 Nei's genetic distance (below diagonal)
DISCUSSIONS
Our study provides useful insights into the genetic diversity ofO. masou masouand the data indicates relative low levels of genetic diversity but significant population differentiation the threeO. masou masoustocks.
Within-population diversity and population structure
Low levels of allele diversity amongO. masou masouwere suggested by the 20 microsatellite loci. The number of alleles ranged from 3.1094 to 3.3299 per locus,compared to the average value from 12 species of freshwater fish (A=9.1) (De Woody & Avise, 2000) and fromO. masou masoupopulations in Japanese and Korean populations (A=6.42−9.71) (Yu et al, 2010).However, the observed and expected heterozygosity(Ho=0.6001−0.7149 ,He=0.6600−0.6648) was moderate, slightly higher than the average of freshwater fish (H=0.54 averaged of 13 species) (De Woody &Avise, 2000) and lower thanO. masou masou(Ho=0.698−0.817,He=0.722−0.814) (Yu et al, 2010).For three stocks, samples were introduced as fry and they may have comprised a few full-sib families rather than representatives of their original populations (Na-Nakorn et al, 2004), which resulted in the low level of alleles.We also used primers developed for other salmon. Cross-amplified primers and the loss of low-frequency alleles after several generations of artificial propagation may result in low genetic variation. BO1996 and Beijing population showed even lower genetic diversity compared to BO2008 in terms of number of alleles and heterozygosity. BO1996 and Beijing populations experienced a longer period of being cultured since introduction into China as compared to the BO2008 population. This time difference might lead to a loss of diversity as a result of inbreeding and genetic drift.
In general, a small number of alleles are a signature of population bottleneck (Norris et al, 1999) due to allele loss. The present results indicate that significant genetic bottlenecks were detected with the shifted model in all populations, which implies that a dramatic loss of alleles happened in our samples. In the next breeding or hatcheries, a sufficient number of individuals are necessary to avoid serious bottleneck.
Deviations fromHWEwere found in three populations due to heterozygote deficiencies, likely caused by null alleles, a mixture of independent populations,non-random mating, or artificial or natural selection. In this study, half of loci deviated fromHWEin the sampled populations and heterozygote deficits did not occur in all the loci and populations, which did not reflect features of null allele effect but likely of mixture of independent populations (for BO2008 stock), nonrandom mating, or artificial or natural selection (for BO1996 and Beijing stock).
Genetic differentiation between stock populations
Genetic differentiation was observed between/among different stocks. In all inter-stock tests, deep differentiation was found between BO1996 and BO2008 and then Beijing and BO2008, which was consistent with the different stocks’ origin.
Significant population differentiation was observed between BO2008 and the other two. As a newly introduced stock, BO2008 might be genetically different from original populations. It is interesting that two other stocks, BO1996 and Beijing, are also differentiated significantly from each other, as Beijing samples were recruited from BO1996 stocks. This could be due to genetic drift phenomenon such as non-random mating,inbreeding or selection that occurred in hatcheries.
Identifying individuals to their origin is important for breeding programs to improve desirable quantitative traits and to conserve gene resources to avoid vast gene introgression during broodstock management. In our study, the assignment accuracy of individuals to their stock of origin was more than 80%. Cornuet et al (1999)calculated the accuracy of assignment of an individual to the population of its origin according to the number of loci used and the number of individuals sampled and the degree of population differentiation and pointed out that the best combination was 20-30 loci and 8-12 individuals per population when the value ofFstwas more than 0.05.In our analysis withFst≥0.05, 20 loci and 40 individuals were used, which is sufficient for the assignment. The high percentage of correct assignment (85%) in our assignment test also demonstrated that the individual assignment tests performed well with the level of differentiation.
Implications for the management
Although the results of moderate heterozygosity were revealed, the presence of bottleneck and relatively low levels of allele diversity implied that domestication had greatly reduced the genetic diversity ofO. masou masouin China. Sufficient genetic diversity is crucial in ensuring the long-term adaptability for domesticated stocks ofO. masou masou. Consequently, a large number of individuals to avoid inbreeding will be necessary for the management and conservation of this species.Furthermore, we recommend genetic monitoring be performed regularly on these and any new stocks in order to avoid a loss of genetic diversity.
Genetic differentiation revealed byFstwas observed between/among different stocks, which was supported by the realKestimated from STRUCTURE analysis. Therefore, genetic dissimilarity existed in the different stock ofO. masou masouin China and as such,it is necessary to maintain the genetic integrity of each stock to avoid gene introgression between the genetic groups as revealed in the dendrogram.
Acknowledgement:We thank the Beijing Shuntong Rainbow Trout Culture Center for their help and support in sample collection.
Aung O, Nguyen TTT, Poompuang S, Kamonrat W. 2010.Microsatellite DNA markers revealed genetic population structure among captive stocks and wild populations of mrigal,Cirrhinus cirrhosusin Myanmar [J].Aquaculture,299(1-4): 37-43.
Cornuet JM, Luikart G. 1997. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data [J].Genetics,144(4): 2001-2014.
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. 1999. New methods employing multilocus genotypes to select or exclude populations as origins of individuals [J].Genetics,153(4): 1989-2000.
De Woody JA, Avise JC. 2000. Microsatellite variation in marine,freshwater and anadromous fishes compared with other animals [J].J Fish Biol,56(3): 461-473.
Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study[J].Mol Ecol,14(8), 2611-2620.
Felsenstein J. 1993. PHYLIP (phylogeny inference package), version 3.6 [C] // Department of Genome Sciences and Department of Biology.Seattle, USA: University of Washington.
Filgueira R, Grant J, Strand Ø, Asplinc L, Aurec J. 2010. A simulation model of carrying capacity for mussel culture in a Norwegian fjord:role of induced upwelling [J].Aquaculture,308(1-2): 20-27.
Jia ZY, Zhang YY, Shi LL, Bai QL, Jin SB, Mou ZB. 2008.Amplification of rainbow trout microsatellites inBrachymystax lenok[J].Mol Ecol Resour,8(6): 1520-1521.
Kano Y, Kondou T, Shimizu Y. 2010. Present status and conservation of the markless forms of stream-resident masu salmonOncorhynchus masou(the so-called ‘iwame’) in Japanese mountain streams [J].Ichthyol Res,57(1): 78-84.
Kohlmann K, Kersten P, Flajšhans M. 2005. Microsatellite-based genetic variability and differentiation of domesticated, wild and feral common carp (Cyprinus caprioL.) populations [J].Aquaculture,247(1-4): 253-266.
Na-Nakorn U, Kamonrat W, Ngamsiri T. 2004. Genetic diversity of walking catfish,Clarias macrocephalus, in Thailand and evidence of genetic introgression from introduced farmedC. gariepinus[J].Aquaculture,240(1-4): 145-163.
Norris AT, Bradley DG, Cunningham EP. 1999. Microsatellite genetic variation between and within farmed and wild Atlantic salmon (Salmo salar) populations [J].Aquaculture,180(3-4): 247-264.
Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data [J].Genetics,155(2): 945-959.Rexroad CE III, Coleman RL, Gustafson AL, Hershberger WK,Killefer J. 2002a. Development of rainbow trout microsatellite markers from repeat enriched libraries [J].Mar Biotechnol,4(1): 12-16.
Rexroad CE III, Coleman RL, Hershberger WK, Killefer J. 2002b.Rapid communication: thirty-eight polymorphic microsatellite markers for mapping in rainbow trout [J].J Anim Sci,80(2): 541-542.
Rexroad CE III, Rodriguez MF, Coulibaly I, Gharbi K, Danzmann RG,Dekoning J, Phillips R, Palti Y. 2005. Comparative mapping of expressed sequence tags containing microsatellites in rainbow trout(Oncorhynchus mykiss) [J].BMC Genomics,6: 54.
Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetic software for exact tests and ecumenicism [J].J Hered,86(3):248-249.
Sekino M, Hara M, Taniguchi N. 2002. Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounderParalichthys olivaceus[J].Aquaculture,213(1-4): 101-122.
Sokal RR, Rohlf FJ. 1995. Biometry, 3rd ed. [M]. New York: W.H.Freeman and Company.
Wang ZM. 1998. Masu salmon-A new cold-water cultured fish [J].Chn J Fish,11(2): 96. (in Chinese)
Wei DW, Lou YD, Sun XW, Shen JB. 2001. Isolation of microsatellite markers in the common carp (Cyprinus carpio) [J].Zool Res,22(3):238-241. (in Chinese)
Yeh FC, Yang RC, Boyle TBJ. 1999. POPGENE, version 1.32,Microsoft Window-Based software for Population Genetic Analysis: a quick user’s guide[M]. University of Alberta, Edmonton, Canada.
Yu JN, Azuma N, Yoon M, Brykov V, Urawa S, Nagat M, Jin DH, Abe S. 2010. Population genetic structure and phylogeography of masu salmon (Oncorhynchus masou masou) inferred from mitochondrial and microsatellite DNA analyses [J].Zool Sci,27(5): 375-385.
Zhang YY, Jia ZY, Bai QL, Tang YK. 2011. Properties of silvering salmon inOncorhynchus masou masoupopulation under aquaculture condition [J].Chn J Zool,46(4): 8-15. (in Chinese)
08 December 2011; Accepted: 04 June 2012
s: HRFRI Basic Science Research Special Funds(2009HSYZX-YY-10); Heilongjiang Natural Science Fund (C201044);Heilongjiang Postdoctoral Sustentation Fund (LRB10-081)
*Corresponding author, E-mail: slyfisharmur@163.com; zyjia2010@163.com
