米根霉脂肪酶基因pro-ROL和m-ROL在毕赤酵母中的密码子优化、表达和酶学性质的比较分析
2011-02-26杨江科严翔翔黄日波张搏
杨江科,严翔翔,黄日波,张搏
1 武汉工业学院生物与制药工程学院,武汉 430023
2 广西科学院国家非粮生物质能源工程技术研究中心,南宁 530070
Introduction
Rhizopus orgzae lipases (EC 3.1.1.3) have been widely utilized in hydrolysis of triglycerides, synthesis of aromatic esters and biofuel, and kinetic resolutions of prochiral compounds[1-3]. Typically, Rhizopus orgzae lipase (ROL) consists of three parts that includes a signaling peptide responsible for the translocation and secretion of ROL, pre-sequence, and mature lipase (m-ROL) domain[4]. The pre-sequence is an intramolecular chaperone involved in the correct folding and efficient secretion of m-ROL. Although intramolecular chaperones have been found in several protein families[5-6], to our knowledge, Rhizopus lipase is the only member of the lipase family to possess intramolecular chaperone-like pre-sequences. It remains unknown how the pre-sequence contributes to folding, post-translational processing and maturation of Rhizopus lipase.
Currently, Rhizopus lipase has been successfully expressed in Escherichia coli, Saccharomyces cerevisiae and Pichia pastoris. In E. coli, pro-ROL and m-ROL exists as insoluble inclusion bodies. To obtain their active forms, a complex refolding process was needed[4,7]. m-ROL exhibited no enzymatic activity when expressed in S. cerevisiae, suggesting the pre-sequence is necessary for correct protein folding of m-ROL[8-9]. However, m-ROL displayed enzymatic activity when expressed in Pichia[10-11]. It means that without the help of pre-sequence the mature domain of ROL can form the active conformation in P. pastoris by itself. But questions such as whether pro-ROL would actively express in P. pastoris and whether pro-ROL and m-ROL were enzymatically identical characteristics still need to be clarified.
P. pastoris expression system is an easy and efficient system suitable for high-density fermentation, and now broadly used to produce recombinant heterologous proteins[12-14]. Like most organisms, Pichia displays a non-random pattern of synonymous codon usage and showed general bias towards a subset of codons. This bias can affect the heterogenous expression efficiency of genes in Pichia. Codon optimization by substituting rare codons with frequently used ones has been established as an efficient measure to enhance the expression level[15-17].
In this study, we cloned and expressed R. orzyae lipase genes, pro-ROL and m-ROL, in P. pastoris. To improve its expression levels, we conducted overlap extension PCR to optimize the codons of m-ROL gene. We further compared the enzymatic activity of pro-ROL and m-ROL to assess the function of the pre-sequence on the maturation of ROL, and to acquire parameters for potential biotechnological applications.
1 Materials and methods
1.1 Reagents, media and strains
All molecular biological reagents were purchased from TaKaRa, Dalian. p-Nitrophenyl acetate (pNPA, C2), p-Nitrophenyl butyrate (pNPB, C4), p-Nitrophenyl caprate (pNPC, C10), p-Nitrophenyl laurate (pNPL, C12), and p-Nitrophenyl palmitate (pNPP, C16) were purchased from Sigma.
R. oryzae strains HU3005 was obtained from the China Centre of Industrial Culture Collection (CICC) with deposition number CICC3005. E. coli strain DH10B was cultured at 37 °C in Luria-Bertani (LB) medium unless otherwise noted. Ampicillin (100 mg/L) and kanamycin (50 mg/L) were added when necessary.
1.2 Lipase genes cloning
R. oryzae pre-mature lipase gene pro-ROL was amplified using the primer pairs ROL1 (5¢-CTGAATTCGTTCCTGTTTCTGGTAAATC-3¢, EcoR I site) and ROLA1 (5¢-CTGCGGCCGCTTA CAAACAGCTTCCTCGT-3¢, Not I site), and mature lipase gene m-ROL was amplified with the primer pairs MROL2 (5¢-CTGAATTCTCTGAT GGTGGTAAGGTTG-3¢, EcoR I site) and MROLA2 (5¢-CTGCGGCCGCTTACAAACAGC TTCCTTCGT-3¢, Not I site) (Fig. 1).
1.3 RNA extraction
Total RNA from R. oryzae was extracted by Trizol reagent (Gibcol) according to the manufacturer’s protocol. The first strand cDNA was synthesis by using the RevertAid First Strand cDNA Synthesis Kit (Fermentas). PCR was carried out in a 50 μL reaction containing 200 mmol/L dNTPs, 0.1 mmol/L primers, 1.5 mmol/L MgCl2, and 1 U of pfu DNA polymerase (TaKaRa). The PCR conditions is as followed: denaturation at 94 °C for 4 min, 28 cycles of 94 °C for 30 s, 56 °C for 50 s and 72 °C for 1 min, and final elongation at 72 °C for 6 min. The PCR product was cloned into the pMD18-T simple vector (TaKaRa), and then sequenced by Sangon Ltd., Shanghai. The sequence of R. oryzae lipase gene was deposited into GenBank with the accession number GQ502721.
1.4 Codon optimization
Overlap extension PCR amplifications were conducted to substitute eight less-frequently used codons with the frequently used codons (Table 1). Three fragments which overlap each other were amplified with the primers containing the mutant nucleotides (Table 2), and then assembled into the full-length m-ROL gene by a second PCR using 5¢- and 3¢-end primers (Fig. 2).

Fig. 1 Primary structure of R. oryzae lipase ROL.

Table 1 The optimized codons of R. oryzae lipase gene m-ROL and their usage in Pichia
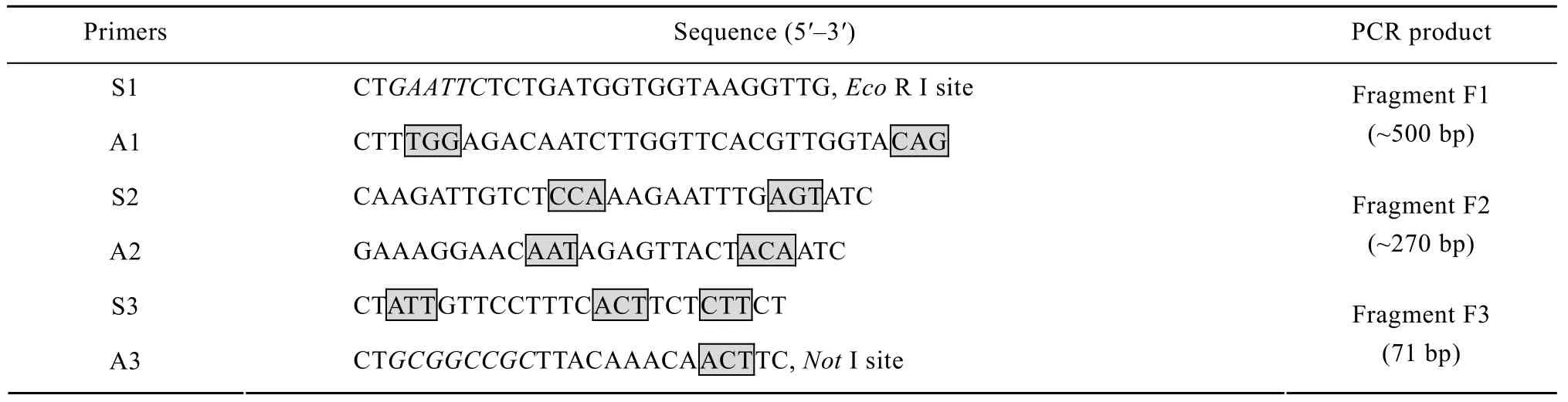
Table 2 The primers used for codon optimization of ROL and the size of PCR products.
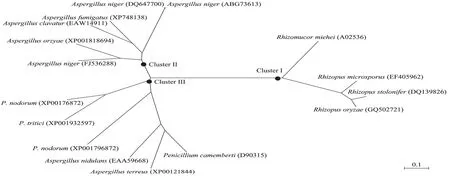
Fig. 2 Phylogeny tree of R. oryzae lipase (ROL) and the related species. Bracketed are the GenBank accession numbers. P. nodorum: Phaeosphaeria nodorum; P. tritici: Pyrenophora tritici.
1.5 Plasmid construction, transformation, and recombinants selection
PCR product of m-ROL and pro-ROL were digested with EcoR I and Not I, and then inserted into vector pPIC9K (Invitrogen) to obtain plasmids pPIC9K-m-ROL and pPIC-pro-ROL, with a fusion expression of α-factor. Sac I was used to linearize the plasmid pPIC9K-m-ROL and pPIC-pro-ROL for the single crossover with P. pastoris genome to generate the methanol-utilized phenotype (Mut+). About 6 μg of linearized DNA was mixed with 80 μL of yeast competent cells, and transformed into P. pastoris GS115 cells by electroporation conducted on Gene Pulser (Bio-rad) according to the manufacturer’s suggestion for S. cerevisiae. Positive clones were initially selected by MD medium (1.34% yeast nitrogen base, 4×10−5% biotin, 2% dextrose) plates and then checked by colony PCR.
The lipase activity of the recombinants was initially checked by BMMY agar plate containing olive oil and rhodamine B. Positive clones displayed orange fluorescent halos visible upon UV irradiation[18].
1.6 Fermentation
A single colony of recombinant bacteria was selected and inoculated into 50 mL BMGY (1% yeast extract, 2% peptone, 100 mmol/L potassium phosphate buffer with pH 6.0, 1.34% yeast nitrogen base, 4×10−5% biotin, 1% glycerol) medium, and grown at 28 °C in a shaking incubator (250 r/min) until the culture reached an OD600of 3.0. The cells were harvested, and a portion was transferred into 50 mL BMMY (1% yeast extract, 2% peptone, 100 mmol/L potassium phosphate buffer pH 6.0, 1.34% yeast nitrogen base, 4×10−5% biotin, and 0.5% methanol) medium to obtain a cell suspension with OD600=1.0. The cells were grown for another 5 d and expression of lipase was induced by methanol at a final concentration of 0.5%. Lipase activity of the supernatant of fermentation broth was checked at intervals. For every type of recombinant, three colonies were randomly selected for fermentation.
1.7 Lipase activity and characterization
Lipase activity was quantified at pH 7.5 by free fatty acid titration with 50 mmol/L NaOH after incubation in a thermostated vessel for 10 min. The assay mixture consisted of 5 mL 50 mmol/L Tris-HCl buffer, 50 mmol/L NaCl, 4 mL emulsified olive oil and 1 mL enzyme solution. One unit (U) of the activity was defined as the amount of enzyme liberating 1 micromole of fatty acid per min at 40 °C. Properties of lipase such as its thermal stability and optimal pH were determined as previously reported[13-14]. All experiments were carried out in triplicate. Protein content of the fermentation broth was determined by the Bradford method[25].
1.8 Phylogenic analysis
Phylogenetic analysis was performed by molecular evolutionary genetics analysis (MEGA3.1) software, which was also used to produce a phylogenetic dendrogram reflecting the evolutionary relationship between the cloned genes and other reference strains by the neighbor-joining method according to the Kimura 2-parameter model[19].
2 Results
2.1 Cloning of R. oryzae lipase genes
R. oryzae premature lipase gene pro-ROL and mature lipase gene m-ROL were amplified by RT-PCR. The ORFs of pro-ROL and m-ROL genes were 1 101 bp and 810 bp, respectively. Phylogenetic analysis based on the similarity of amino acid sequences between R. oryzae and the related reference lipases revealed three distinct clades (I to III). Clade I consisted of lipases from Rhizopus and Rhozomucor. Cloned R. oryzae lipase exhibited a 95% amino acid similarity with R. stolonifer (Fig. 2).
2.2 Codon optimization and expression in P. pastoris
Bias codon usages between different microorganisms are one of the main factors that restrict gene expression in heterogeneous host. To achieve a high level expression of m-ROL in Pichia, the less frequently used codons consisting of eight amino acids were substituted with the frequently used ones by overlap extension PCR (Fig. 3). After transformation, yeast recombinants carrying codon-optimized m-ROL, pro-ROL and original m-ROL were acquired. Both m-ROL and pro-ROL showed lipase activity on plates. However, the enzymatic activity of the recombinants carrying codon-optimized m-ROL (optimized-ROL) is significantly higher than the non-optimized m-ROL and pro-ROL (Fig. 4A). After methanol-inducible expression in Pichia, the molecular weight of the enzymes m-ROL and pro-ROL determined by PAGE were 30 kDa and 35 kDa, respectively (Fig. 4B). The expression level of codon-optimized m-ROL is higher than the original m-ROL and pro-ROL. After 72 h of fermentation, the enzymatic activity (Fig. 5A) and protein content (Fig. 5B) of the codon-optimized m-ROL reached 132.7 U/mL and 50.4 mg/L, while the activity of the parental m-ROL and pro-ROL are 28.7 U/mL and 14.4 mg/L, 29.6 U/mL and 14.1 mg/L, respectively.
2.3 Characterization of pro-ROL and m-ROL
We determined the optimal substrate, pH and temperature of pro-ROL and m-ROL (Fig. 6). pNP esters with different carbon chain lengths were selected as substrates. pro-ROL and m-ROL showed differential preferences to these substrates. pro-ROL preferred short- and middle-chain substrates (C4and C10), while m-ROL preferred middle-chain substrates with optimal activity observed using C10substrates (Fig. 6A).
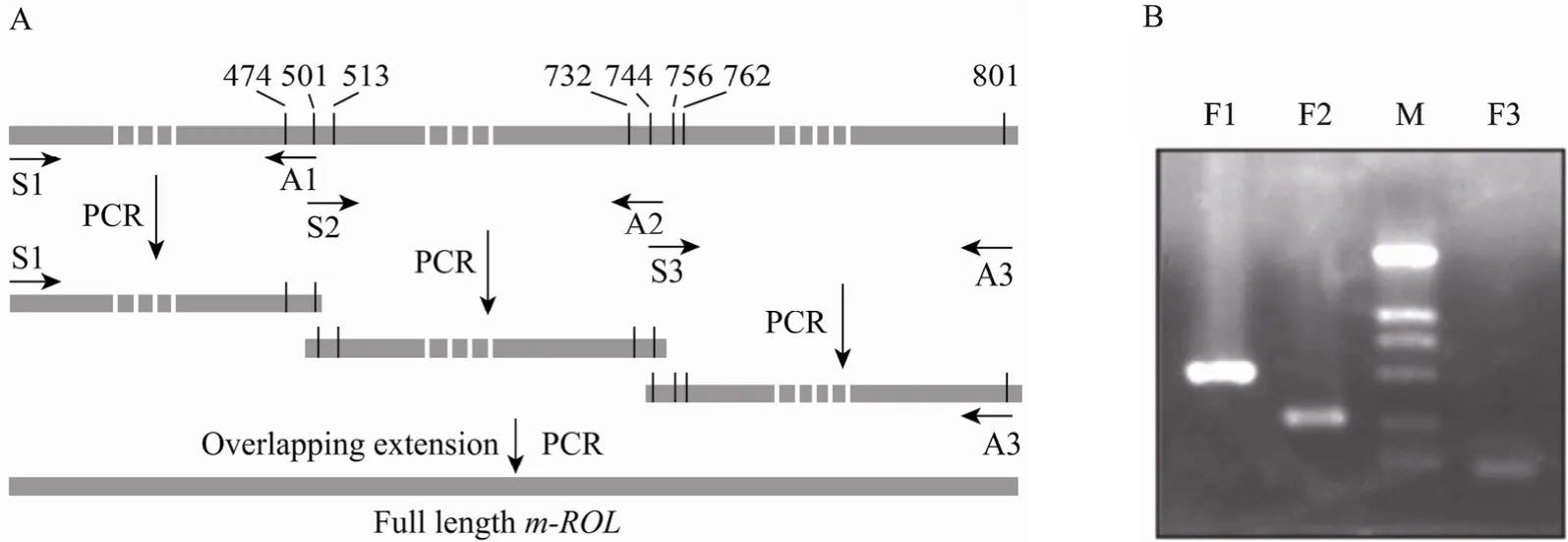
Fig. 3 Flow chart of codon optimization of m-ROL by overlap extension PCR (A), and the gel picture of the three fragments (B), F1 (500 bp), F2 (270 bp ) and F3 (71 bp). M: DNA marker (2 000, 1 000, 750, 500, 250, 100 bp)

Fig. 4 Expression of codon-optimized ROL (optimized ROL), pro-ROL and original m-ROL in P. pastoris. (A) The morphology of different recombinants on the plate under the UV light. (B) SDS-PAGE of supernatant product of recombinant lipase.
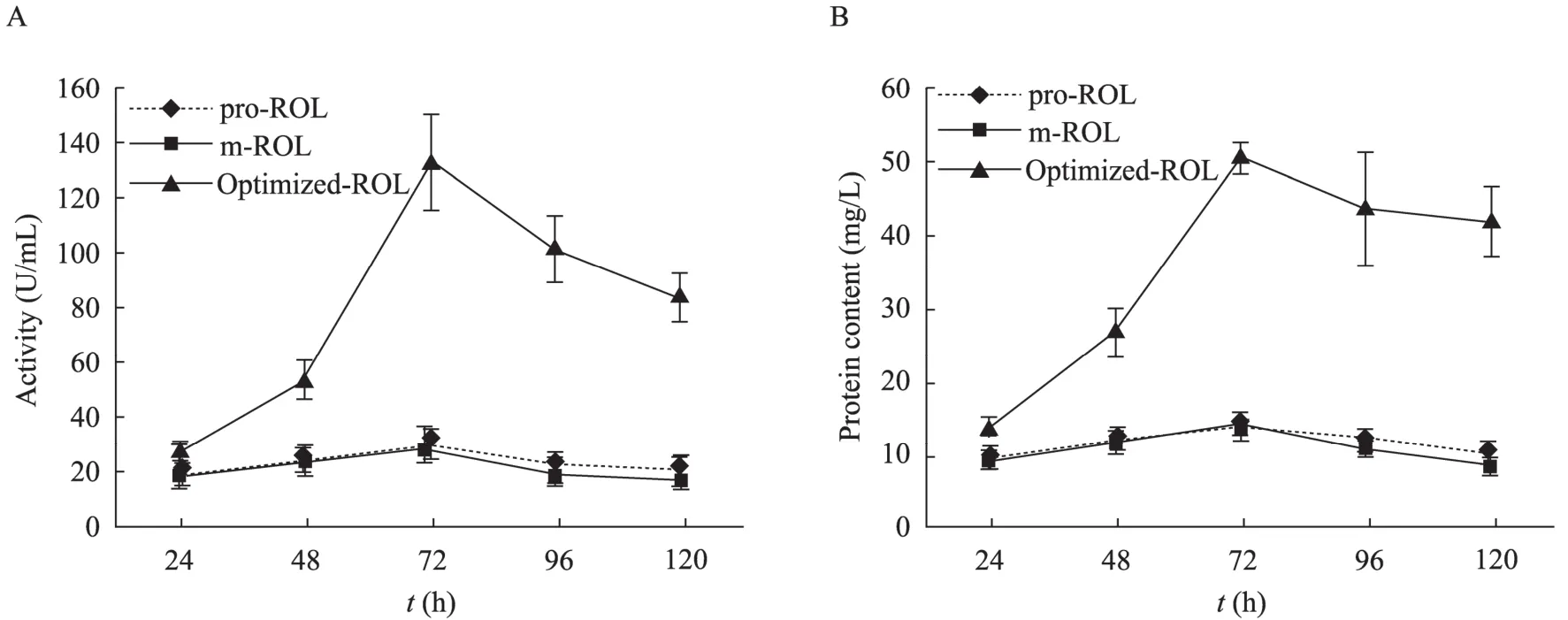
Fig. 5 Time course of the production of lipases by recombinants carrying pro-ROL, m-ROL and the codon-optimized m-ROL (optimized-ROL). (A) and (B) directed out lipase activity and protein content of the fermentation broth, respectively.
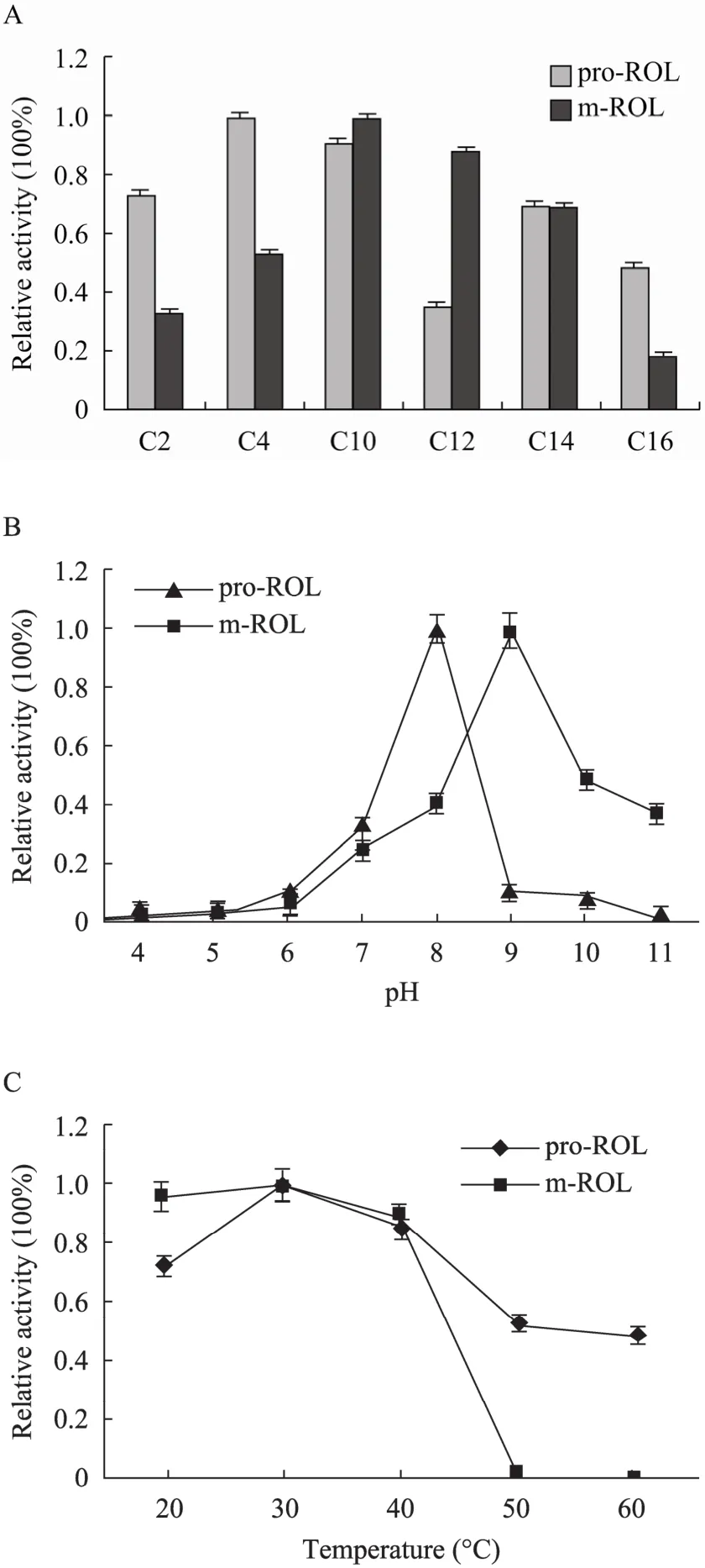
Fig. 6 Enzymatic characterization of pro-ROL and m-ROL. (A) Substrate specificity of pro-ROL and m-ROL towards ester of nitrophenol. (B) and (C) Effects of temperature and pH on lipase activity.
The optimal pH was determined by incubating lipases in reaction buffer with different pH values. As shown in Fig. 6B, both pro-ROL and m-ROL preferred alkaline environments. Unlike pro-ROL, which has an optimal pH value 8.0, the optimal pH value of m-ROL was 9.0. The suitable pH range, determined as having>60% remaining activity, of m-ROL is 8.5−9.5, while the suitable pH range of pro-ROL is from 7.5 to 8.5. Enzymatic activity significantly decreased when the lipases were not maintained at their optimal pH.
To determine the optimal reaction temperature and thermal stability, lipase activities were assessed under different temperatures (20 °C−60 °C) and calculated as percentages of the maximal activity. As shown in Fig. 6C, the optimal temperatures for both pro-ROL and m-ROL were 30 °C. At 50 °C, enzymatic activity was not detected for m-ROL, whereas pro-ROL had 51% activity. Additionally, pro-ROL was more thermally stable than m-ROL.
3 Discussion
Pre-sequence plays an important role on the folding, post-translational processing and secretion of R. oryzae lipase[4,7,9]. Previous reports showed lack of enzymatic activity when m-ROL was expressed in S. cerevisiae[8]. This suggested that the lack of the pre-sequence in m-ROL may have inhibited formation of its active conformation. In our study, we actively expressed pro-ROL and m-ROL in P. pastoris. The enzymatic characteristics of pro-ROL in Pichia are different than those in S. cerevisiae. In Pichia, pro-ROL prefers short and middle-length carbon chains (C4-C10), while in S. cerevisiae, pro-ROL favors middle-chain substrates[8]. It is possible that in P. pastoris, post-translational modification may modulate the function of the pre-sequence and facilitate correct protein folding of m-ROL. Moreover, intramolecular chaperones may also affect enzymatic properties by mediating the folding and conformational changes of the mature protein domain[20]. In this study, pro-ROL was more thermally stable than m-ROL. Similar phenomenon was observed by Beer and his colleague. In their study, they found that in contrast to m-ROL, pro-ROL and prepro-ROL had considerably higher thermostability[7]. This suggests that the pre-sequence domain may also protect the m-ROL region from dramatic conformational change when exposed to high temperatures.
A special characteristic of lipase is the interfacial activation. Generally, the active site (catalytic triad) is completely buried beneath a short amphiphilic helical segment, which prevents substrate access. When the enzyme contact with the hydrophobic substrates, it will go through a series structural rearrangement, and the enzyme changes from a closed to a open conformation that make the substrates entre into the activity site[21]. Although there were no report on the effect of pre-sequence on the substrate selectivity of lipase before, but it is possible that this pre-sequence of pro-ROL can spatially retard the conformational change of mature ROL domain. When the long-chain substrates contacted with enzymes, although the enzymes have conformational change, the open size of enzyme is still not enough to permit the long-chain substrates entre into the active site. On the contrary, this half-open conformation will be more accessible to the shorter chain substrate (C4). As observed in this study, the favorable substrate of pro-ROL and m-ROL are C4, while for m-ROL is C10 (Fig. 6A).
Codon-usage frequency is a major impediment to the efficient expression of foreign gene in heterologous host. In ROL gene, four codons 155Leu (CTC), 171Ser (AGC), 254Leu (CTC) and 267Ser (AGC), which have the lowest usage frequency, are the bottleneck of gene expression in P. pastoris. Our study shows that codon optimization by overlap extension PCR to substitute less frequently used codons with frequently used ones could significantly improve the expression level of ROL. Compared to other methods such as chemical gene synthesis, overlap extension PCR is simple and effective. The initial enzymatic activity achieved based on the flask fermentation method could reach 132.7 U/mL. Although our product yield is not as high as a previous study[22]using a large-scale bioreactor (644.0 U/mL), it is still significantly better than other reported results[23-24]. We believe that lipase production and enzymatic activity from recombinants carrying optimized ROL gene may be improved under a batch-induced mode with further optimization in pH, methanol concentration and aeration during the fermentation process.
In this study, we actively expressed pre-mature (pro-ROL) and mature lipase gene (m-ROL) of R. oryzae in P. pastoris. The differences in enzymatic characteristics between these two forms of lipase may be caused by the intramolecular chaperone-like pre-sequence in pro-ROL, which could affect the conformation of m-ROL domain. Codon optimization by overlap extension PCR has effectively improved the expression level by 4.6-fold. This newly codonoptimized ROL gene may be useful for future large-scale production of R. oryzae lipase. Further characterization of pro-ROL and m-ROL may offer a valuable tool in industrial applications.
[1] Gotor-Fernández V, Brieva R, Gotor V. Lipases: useful biocatalysts for the preparation of pharmaceuticals. J Mol Catal B: Enzym, 2006, 40(3/4): 111−120.
[2] Fukuda H, Hama S, Tamalampudi S, et al. Whole-cell biocatalysts for biodiesel fuel production. Trends Biotechnol, 2008, 26(12): 668−673.
[3] Tamalampudi S, Talukder MR, Hama S, et al. Enzymatic production of biodiesel from Jatropha oil: a comparative study of immobilized-whole cell and commercial lipases as a biocatalyst. Biochem Eng J, 2008, 39(1): 185−189.
[4] Beer HD, Wohlfahrt G, Schmid RD, et al. The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence. Biochem J, 1996, 319(Pt 2): 351−359.
[5] Shinde U, Inouye M. Folding mediated by an intramolecular chaperone: autoprocessing pathway of the precursor resolved via a substrate assisted catalysis mechanism. J Mol Biol, 1995, 247(3): 390−395.
[6] Chen YJ, Inouye M. The intramolecular chaperonemediated protein folding. Curr Opin Stru Biol, 2008, 18(6): 765−770.
[7] Beer HD, McCarthy JEG, Bornscheuer UT, et al. Cloning, expression, characterization and role of the leader sequence of a lipase from Rhizopus oryzae. Gene Struct Expr, 1998, 1399(2/3): 173−180.
[8] Takahashi S, Ueda M, Tanaka A. Function of the prosequence for the in vivo folding and secretion of active Rhizopus oryzae lipase in Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 2001, 55(4): 454−462.
[9] Ueda M, Takahashi S, Washida M, et al. Expression of Rhizopus oryzae lipase gene in Saccharomyces cerevisiae. J Mol Catal B: Enzym, 2002, 17(3/5): 113−124.
[10] Minning S, Schmidt-Dannert C, Schmid RD. Functional expression of Rhizopus oryzae lipase in Pichia pastoris: high-level production and some properties. J Biotechnol, 1998, 66(2/3): 147−156.
[11] Cos O, Resina D, Ferrer P, et al. Heterologous production of Rhizopus oryzae lipase in Pichia pastoris using the alcohol oxidase and formaldehyde dehydrogenase promoters in batch and fed-batch cultures. Biochem Engin J, 2005, 26(2/3): 86−94.
[12] Fernández L, Pérez-Victoria I, Zafra A, et al. High-level expression and characterization of Galactomyces geotrichum (BT107) lipase I in Pichia pastoris. Protein Expr Purif, 2006, 49(2): 256−264.
[13] Shu ZY, Duan MJ, Yang JK, et al. Aspergillus niger lipase: heterologous expression in Pichia pastoris, molecular modeling prediction and the importance of the hinge domains at both sides of the lid domain to interfacial activation. Biotechnol Prog, 2009, 25(2): 409−416.
[14] Yang JK, Zhang B, Yan YJ. Cloning and expression of Pseudomonas fluorescens 26-2 lipase gene in Pichia pastoris and characterizing for transesterification. Appl Biochem Biotechnol, 2009, 159(2): 355−365.
[15] Müller M. Codon optimization of papillomavirus genes. Methods Mol Med, 2005, 119: 433−444.
[16] Tokuoka M, Tanaka M, Ono K, et al. Codon optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl Environ Microbiol, 2008, 74(21): 6538−6546.
[17] Daniell H, Ruiz G, Denes B, et al. Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function. BMC Biotechnol, 2009, 9(1): 33.
[18] Kouker G, Jaeger KE. Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol, 1987, 53(1): 211−213.
[19] Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform, 2004, 5(2): 150−163.
[20] Ma BY, Tsai CJ, Nussinov R. Binding and folding: in search of intramolecular chaperone-like building block fragments. Protein Eng Des Sel, 2000, 13(9): 617−627.
[21] Chapus C, Semeriva M, Bovier-Lapierre C, et al. Mechanism of pancreatic lipase action. 1. Interfacial activation of pancreatic lipase. Biochemistry, 1976, 15(23): 4980−4987.
[22] Surribas A, Stahn R, Montesinos JL, et al. Production of a Rhizopus oryzae lipase from Pichia pastoris using alternative operational strategies. J Biotechnol, 2007, 130(3): 291−299.
[23] Ramchuran SO, Vargas VA, Hatti-Kaul R, et al. Production of a lipolytic enzyme originating from Bacillus halodurans LBB2 in the methylotrophic yeast Pichia pastoris. Appl Microbial Biotechnol, 2006, 71(4): 463−472.
[24] Yao HY, Yu SW, Zhang LD, et al. Isolation of a novel lipase gene from Serratia liquefaciens S33 DB-1, functional expression in Pichia pastoris and its properties. Mol Biotechnol, 2008, 38(2): 99−107.
[25] Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem, 1996, 236: 302−308.
