Salmonella typhi and gallbladder cancer: report from an endemic region
2010-06-29MallikaTewariRaghvendraMishraandHariShukla
Mallika Tewari, Raghvendra R Mishra and Hari S Shukla
Varanasi, India
Salmonella typhi and gallbladder cancer: report from an endemic region
Mallika Tewari, Raghvendra R Mishra and Hari S Shukla
Varanasi, India
BACKGROUND:Evidence exists of a link between chronic infection bySalmonella typhi(S. typhi) and the development of gallbladder cancer (GBC), but several studies from endemic regions contradict its role in the etiopathogenesis of GBC. This study used various tools to assess the prevalence ofS. typhiin patients with GBC and gallstone disease (GSD) in this region with a high incidence of GBC.
METHODS:S. typhiwas detected in tissue and bile by PCR and culture and in serum by the Widal test and indirect hemagglutination assay (IHA). PCR with two pairs ofS. typhispecific primers (flagellin gene H1d and SOP E gene) could detect 0.6 ng ofS. typhiDNA. Fifty-four patients with GBC (cases) were matched with 54 patients with GSD(controls).
RESULTS:Of the 54 cases, 24 (44.44%) were positive on the Widal test and 12 (22.22%) on IHA, compared to 13 (24.07%) and 5 (9.26%) respectively in the controls. Eighteen (33.33%) cases showed a positive result on PCR (tissue) and 2 on PCR (bile) vs. none in the controls. Bile culture revealed no Salmonella colonies in either cases or controls. Only 3 cases were positive for Salmonella on tissue culture compared to none in the controls. The sensitivity of PCR (tissue) relative to the Widal test, IHA, culture (bile and tissue) and PCR (bile) was 100% vs. 66.67%, 11.11%, and 11.11%, and the specificity was 83.33% vs. 100%, 100%, and 100%, respectively.
CONCLUSIONS:S. typhiis significantly associated with GBC compared to GSD (33% vs. 0%). PCR appears to be the most specific diagnostic tool, the gold standard forS. typhiin tissue samples.
(Hepatobiliary Pancreat Dis Int 2010; 9: 524-530)
Salmonella typhi; gallbladder cancer; polymerase chain reaction; Widal; indirect hemagglutination assay; culture
Introduction
Gallbladder cancer (GBC) is the third most common malignancy in North India[1]but there is no known causative factor. A strong association between chronic bacterial infection of the biliary tract bySalmonella typhi(S. typhi) and GBC has been reported in several studies and they incriminateS. typhias the causative factor.[2]The infected gallbladder harbors microbes on the mucosa which induce immunological responses that are measured by methods such as the Widal test and the indirect hemagglutination assay (IHA) to detect the presence ofS. typhiindirectly. Typhoid carriers have high titers of Vi agglutinins in their sera and Vi antigen is often used to screen forS. typhicarriers. However, all these provide indirect evidence for the presence ofS. typhiand are often associated with false positive and negative results based on the strength of the response.
In comparison, the polymerase chain reaction (PCR) can detect very small amounts of DNA by enzymatic amplification and is a very specific test. The PCR technique using highly specific primers forS. typhiis superior to culture and serology in detecting its presence in blood samples from patients with typhoid fever.[3]It is envisaged that past infection/carrier status can also be detected by PCR.
Therefore, we used PCR in combination with other established detecting tools viz., serology and culture, to assess the prevalence ofS. typhiin bile, serum, and gallbladder tissue in patients with GBC and those with gallstone disease (GSD). The results of PCR were compared with the Widal test and IHA for assessing the comparative efficacy in diagnosing the presence ofS. typhi.
Methods
This prospective case-control study was conducted in the Department of Surgical Oncology, Sir Sunderlal Hospital and Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India, from January 2007 to December 2009.
Patients
A total of 54 patients with GBC (cases) and 54 patients with GSD (controls) matched with respect to age, sex and place of residence were included in the study. The study was approved by the Institute's Ethics Committee and informed consent was obtained from each patient.
Most participants were in the 4th to 6th decade of life (Table 1). The male/female ratio was 1∶1.57 (39% and 61%) in the GBC group and 1∶1.86 (35% and 65%) in the GSD group, showing a female predominance in both goups. Nearly 83% of the GBC patients hailed from rural areas (Table 2).
Thirty patients with GBC and 12 with GSD had a history suggestive of typhoid.
Methods
Five milliliters of blood was collected from each patient before surgery for the Widal and IHA tests. At operation, 5 ml of bile and 5 mg of fresh tissue were obtained from both GBC and GSD patients and the tissue was immediately frozen and stored at -80 ℃ for DNA isolation.
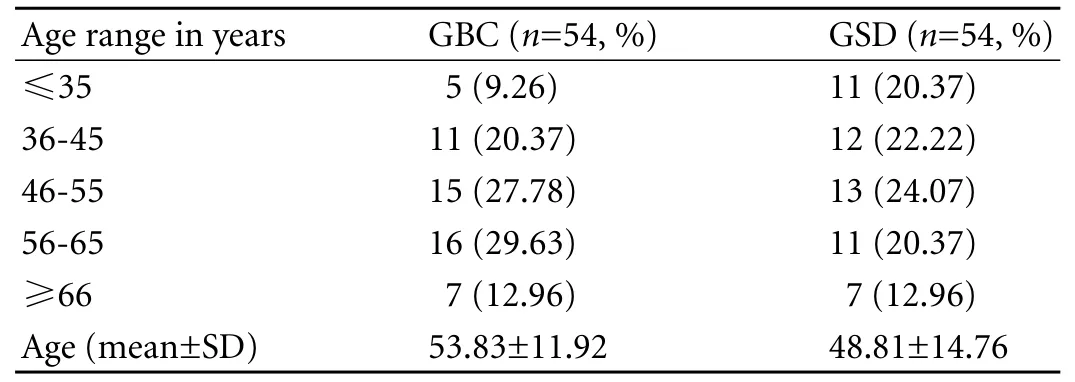
Table 1. Age distribution of cases and controls (GBC/GSD)
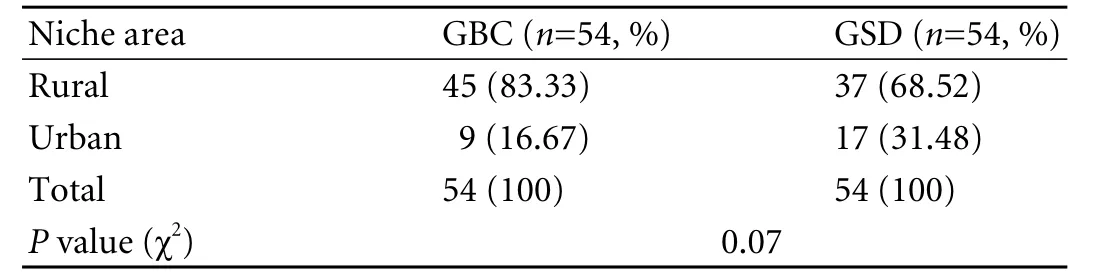
Table 2. Niche area distribution of cases and controls (GBC/GSD)
Serological tests
Widal test
The test was performed to estimate the titer of antibody against somatic (O) and flagellar (H) antigens ofS. typhi. The Sleigh and Duguid (1989) method with some modifications was used.[4]Blood samples were tested in a series of dilution against each of the different Salmonella suspensions, in 5 wells of 300 μl U bottom microtitre plates. The fifth well was used as a non-serum control. Ninety microliters of 0.85% saline was placed in each well of 2 rows. Serum (10 μl) was added to the first well of each row. One hundred microliters of diluent was taken from the first well of each row and added to the second well and mixed by pipetting so that a 1 in 20 dilution was made. Again, 100 μl of diluent was taken from the second well of each row and added to the third well and mixed by pipetting to make a 1 in 40 dilution. Similarly, 10 μl of 5× diluted somatic antigens (TO) and 10× diluted flagellar antigens (TH) were added to 2 of the microtitre plates, so that the final dilutions became 1∶40, 1∶80, 1∶160, 1∶320, and 1∶640. The antigen kit was from Tulip Diagnostics (P). Ltd., India. H-agglutination microtitre plates were incubated in a water bath maintained at 50 ℃ for 2 hours and read after being left for 3 hours at room temperature. O-agglutination microtitre plates were incubated in a water bath, maintained a 37 ℃for 5 hours, and read after being left in a refrigerator overnight. H-agglutination was seen as floccules at the bottom of the tubes and O-agglutination was detected as fine granules. A titer of 1∶80 was regarded as significant.
IHA
The Vi antibody was measured by IHA following the method of Barrett (1985).[5]Pure Vi polysaccharide antigen (BioVac) was used at 10 μl/ml. First, the red blood cells (RBCs) were sensitized with Vi antigen. Fresh sheep blood RBCs were washed three times in PBS (pH 7.2) and the suspension was diluted to 1% (v/v) in PBS. An equal volume of PBS containing Vi antigen (10 μg/ml) was added, mixed, and incubated at 37 ℃ in a water bath for 2 hours. After washing 3 times in PBS, the sensitized cells were finally suspended at a concentration of 0.5% in PBS containing 0.06% BSA. Serial two-fold dilutions of serum were made from 1∶40 to 1∶160 in 100 μl. An equal volume (100 μl) of sensitized sheep RBCs was added and incubated at room temperature for 2 hours. The agglutination patterns were read after incubating overnight at 4 ℃. Clear floccules formation indicated no agglutination, while irregular spreading at the bottom of the well, which was evident from the convex side, showed positive agglutination. Known positive and negative control sera were assayed with eachbatch.[5]All samples with a Vi antibody titer >1∶160 on IHA were taken as indicating typhoid carriers.
Polymerase chain reaction (PCR)Cultivation of reference strain
Pure strains ofS. typhi(ATCC 19430) from the Department of Biochemistry, Institute of Medical Sciences, Banaras Hindu University, Varanasi (India) were cultured as the reference strain for the standardization of PCR. The strain was sub-cultured on nutrient agar and verified biochemically and serologically.
Bacterial suspensions having turbidity matching a standard McFarland concentration of 0.5 were made, i.e. each ml of suspension contained 150 millionS. typhiper ml. Two serial 10-fold dilutions were made to obtain 1.5 million CFU per ml. One milliliter of this standard suspension was mixed with 1 ml of bile from a febrile individual collected in a sterile screw-capped bottle. DNA was isolated from these 2 ml mixtures of bile and bacteria by the phenol/chloroform method described below.
Bacterial DNA isolation of S. typhi reference strain
The phenol-chloroform method was used to isolate the DNA fromS. typhi. The tissue was homogenized, then incubated with 1 mg (100 μl) lysozyme, 100 μl SDS (10%, pH 7.2) and 100 μl TBE (1×) and incubated at 37 ℃ for 60 minutes. One milliliter 0.1% Triton-X100 and 5 μl proteinase-K were added and incubated again at 65 ℃for 120 minutes. To this, an equal volume of chloroform∶IAA (24∶1) was added and mixed by vortexing for 15 seconds and then centrifuged at 10 000 rpm for 10 minutes, when the aqueous phase was collected. One hundredforty microliters phenol∶chloroform∶IAA (25∶24∶1) was added and mixed by vortexing for 15 seconds and then centrifuged at 10 000 rpm for 10 minutes, when the aqueous phase was collected. An equal volume of phenol∶chloroform∶IAA (25∶24∶1) was added and mixed by vortexing for 15 seconds and then centrifuged at 10 000 rpm for 10 minutes, when the aqueous phase was collected. Then an equal volume of isopropanol was added and the solution was kept at room temperature for 5 minutes and centrifuged at 10 000 rpm for 10 minutes, when the supernatant was discarded. The pellet was washed in 200 μl of 70% ethanol and centrifuged at 10 000 rpm for 10 minutes. The tubes containing the DNA were dried at 37 ℃ (in inverted condition) for 30 minutes and resuspended in 30 μl of TE buffer (pH 8).
PCR for detection of S. typhi in test samples
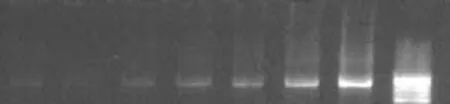
Fig. 1. Sensitivity of PCR with serially diluted DNA from culture strain of S. typhi. Amplification products of 343bp from nested PCR were analyzed in lanes 7, 6, 5 and 4. The dilution factor was 4 times (0.6 PCR product +0.4 D water), six times (0.4 PCR product +0.6 D water), eight times (0.2 PCR product +0.8 D water) and ten times (0.1 PCR product +0.9 D water) of primary reaction in lanes 7, 6, 5, 4 respectively. The estimated DNA amount was 2.0, 1.5, 1.1, and 0.6 ng in lanes 7, 6, 5, 4 respectively. PCR: polymerase chain reaction. Mmr: the 100bp molecular marker.
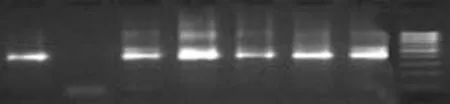
Fig. 2. Representative amplification pattern with nested primers of PCR-positive cases, i.e. patients with GBC. Mmr is the 100bp molecular marker; lane 1 shows amplification of reference DNA whereas lane 2 is negative control DNA of healthy human. Lanes 3 to 7 were amplified with sample DNA. PCR: polymerase chain reaction.
First, DNA isolated from the reference strain ofS. typhiand other organisms was amplified to test the specificity of the PCR products. Second, the minimum detectable level by PCR was established by amplification of serially diluted DNA fromS. typhiATCC 19430 (Fig. 1). Primers were synthesized from two conserved regions of the gene ofS. typhi(AE014613) located between 38 and 679 (642bp) for external amplification. Sequences of sop E are given in Table 1. TheS. typhigenome was evaluated by nested PCR (Fig. 2). The flagellin gene primer (ST) was tested (http://www.ncbi.nlm.gov/BLAST) and used as designed by Song et al[3]and modified by Frankel.[6]
Two μl of loading buffer and 8 μl of PCR product were analyzed by agarose gel electrophoresis (1.2% agarose gel in 1× TBE). This was run at 75 V and made visible by ethidium bromide staining and UV transillumination.
Culture
Each specimen was inoculated onto Petri dishes containing blood agar, MacConkey agar, deoxycholate citrate agar, and also in enrichment Selinite F broth. The culture specimens in Selnite broth culture bottles were incubated at 37 ℃ for 7 days. First sub-culture was done on blood agar and MacConkey agar plates. The subculture plates were incubated overnight and examined for the presence of bacterial colonies. Suspected pure non-lactose fermenting colonies were subjected to Gramstaining and motility testing after incubation in peptone water for 1 hour. The Gram-negative motile bacilli were put on different biochemical substrates for identification. Biochemical characterization of isolated strains was done with a battery of biochemical tests (triple sugar iron, methyl red, Voges-Proskauer, Simmons citrate, indole, urease, lysine, arginine, ornithine, glucose, lactose, sucrose, mannitol, and adonitol). The Salmonella isolated was confirmed by both primers, along with Widal and IHA.
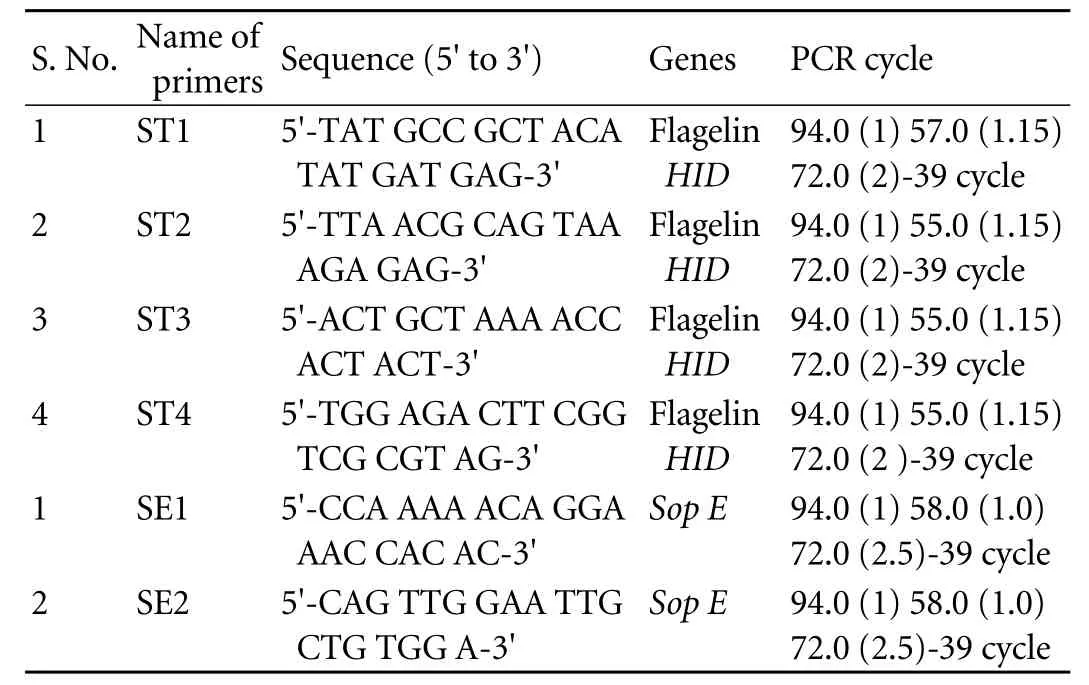
Table 3. Primer names, nucleotide sequences, PCR amplification cycles
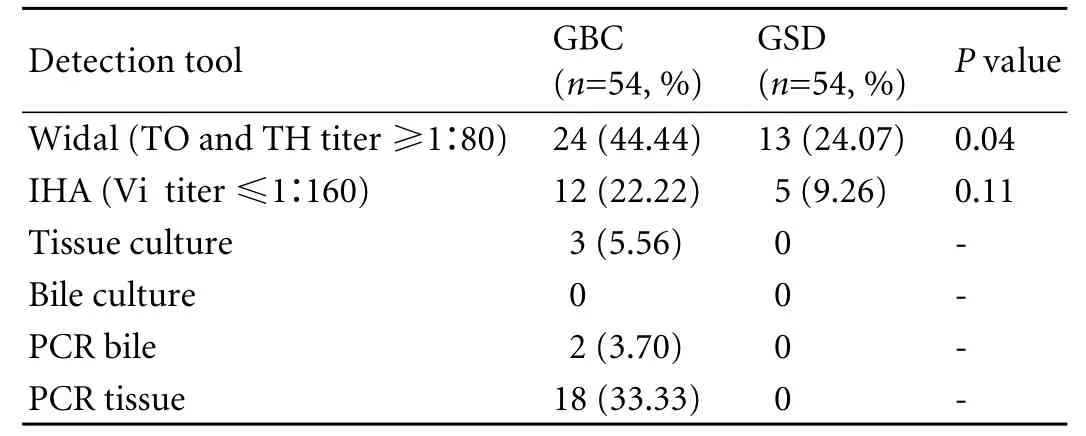
Table 4. Results of Widal, IHA, bile culture and PCR in cases and controls
Statistical analysis
MEDCALC statistical software was used for analysis. The Chi-square test was applied for contingency tables and proportions. APvalue of ≤0.05 was taken as significant.
Results
The primer names, nucleotide sequences and PCR amplification cycles used for detection ofS. typhiby PCR are presented in Table 3. Twenty-four (44.44%) cases were positive on Widal test vs. 13 (24.07%) controls (P=0.04), and 12 (22.22%) on IHA vs. 5 (9.26%) incontrols (P=NS) (Table 4). Thus, our results are contrary to previously published reports that show a highly significant difference on IHA between GBC and GSD regardingS. typhi.

Table 5. Bacterial isolates present in cases (GBC)
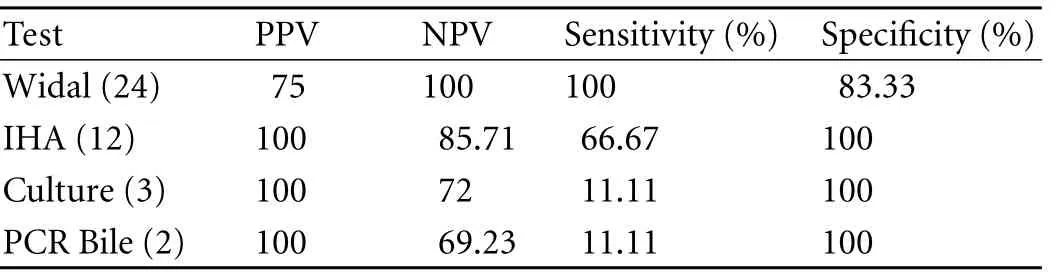
Table 6. Comparative evaluation of tests (Widal, IHA, tissue culture and PCR bile) used in detection of Salmonella taking PCR (tissue) as the standard in GBC cases
However, 18 (33.33%) cases showed positive results on PCR (tissue) with ST primer and 2 on PCR (bile) vs. none in the controls. Of the 18 GBC patients, only 12 were positive by both IHA and Widal tests (Table 4). These 12 cases were also positive on PCR with SE primer specific for S. enterica subsp. enterica serovar Typhi (Ty2). No amplification was evident on PCR (with both ST and SE) in the controls positive on the Widal and IHA tests. In addition, bile culture failed to grow any Salmonella in either cases or controls. The bacterial flora grown in culture from GBC samples are depicted in Table 4. Only 3 tissue cultures of cases were positive for Salmonella compared to none in the controls (Tables 4, 5). With PCR (tissue) as the standard, Widal, IHA, culture (tissue and bile) and PCR (bile) had a sensitivity 100% vs. 66.67%, 11.11%, and 11.11% and a specificity 83.33% vs. 100%, 100%, and 100% respectively (Table 6 ).
Discussion
GBC is a dismal disease, the etiology of which is still poorly understood.[7]It has high incidence pockets around the globe and is the third most common cancer of our region.[1,8]The interplay of various suspectedetiological factors and infections in gallbladder carcinogenesis (S. typhi,H. pylori,H. billis) is now strongly suspected to cause GBC.[9,10]Bacteria (likeH. pyloriin gastric cancer and MALT lymphoma) producing site-specific persistent infections by colonization may initiate or promote neoplasia in susceptible individuals. There is however a long time gap, often years, between infection and development of cancer, and not all those who are infected develop cancer.
Infection withS. typhiis transmitted by the fecaloral route through contaminated food and water. The bacteria attach themselves to the surface of the epithelial cells of the intestinal villi. The virulent bacilli resist intracellular killing by neutrophils and macrophages and eventually appear in the blood stream. In a cell culture study, Haghjoo and Galán[11]found thatS. typhisubsequent to internalization reaches an unusual membrane-bound compartment where it can survive and replicate. It is possible that a unique cytolethal distending toxin may be involved in some aspects of the ability ofS. typhito cause long, persistent infections in humans, because, at least in other bacteria, this toxin has been shown to possess immunomodulatory activity. From the blood, some organisms localize in the gallbladder producing a chronic bacterial carrier state there.[12]The gallbladder shows catarrhal inflammation and bacilli multiply in bile and are discharged into the intestine. Mostly, the infection is cleared, but about 3% turns into chronic carriers. They harbor the organisms in the gallbladder and biliary tract and may continue to excrete the bacilli for several years or throughout life.
Epidemiological studies have shown that those who become carriers ofS. typhihave 8.47 times increased risk of developing GBC compared with the people who have had acute typhoid and have cleared the infection.[10]These findings agree with earlier investigations by Caygill et al[12]and Welton et al.[13]
A case-control study by Welton et al[13]compared those who experienced acute infection withS. typhito those who subsequently became chronic carriers following the 1922 typhoid outbreak in New York. Carriers were six times more likely to die of hepatobiliary carcinoma than matched controls. Additional evidence was found in an analysis of the 1964 typhoid outbreak in Aberdeen,[12]also suggesting a strong association between chronic carrier status and hepatobiliary carcinoma. These studies also agreed that the people who contract typhoid but do not become carriers are not at higher risk of cancer.[10-14]
Conversely, several studies from regions with a high incidence of GBC do question the role of Salmonella in the etiopathogenesis of GBC. Roa et al[15]carried out a microbiological study of gallbladder bile in 608 patients from Chile.E. coliwas isolated in 51% of positive cases,Streptococci-Enterococciin 24%,Enterobacter sp.in 9%, andKlebsiellaandProteusin lower proportions.Salmonella sp.was isolated in only 4 cases, of whom were women with chronic cholecystitis. The authors thus opined that the role of Salmonella in the pathogenesis of GBC must be reassessed. Another study revealed positive results of bile culture in 47/58 (81%) patients with GBC although Salmonella was rare at 4/58 (8.5%).[16]
We also carried out gallstone culture for 100 consecutive patients in a prospective study between December 1997 and March 1999 at our Institute.[17]Cultures were obtained from the core of the gallstones after breaking a freshly removed stone on a culture plate. Only enteric organisms were cultured from 11 patients with GBC. No Salmonella was cultured in any of the samples.
So far, various studies have used serological tests and culture for detecting the presence of Salmonella in blood, bile, urine and stool specimens. All studies thus far have demonstrated indirect evidence of Salmonella by serological tests, which are often not specific forS. typhi. For example, even Vi antigen is present in strains of S. paratyphi C, S. dublin and Citrobacter besides most strains ofS. typhi. Itah and Akpan[18]found in their study that of 39 patients suspected with typhoid fever who tested positive on Widal test with titers ranging from 1∶80 to 1∶320, no Salmonella organism was encountered in some cultures (statistically significant). This prompted the authors to suggest that serological investigations alone may not be a reliable index for the diagnosis of Salmonella infections. The Widal test is of little help in detection of carriers in endemic countries like India and is always associated with significant false positivity and negativity. The Widal test revealed 24 GBC patients positive for Salmonella infection compared to 13 controls and this difference was statistically significant (P=0.04) in our study. But none of these 13 controls showed amplification on PCR withS. typhispecific primers.
Antibody to Vi antigen is present in serum from most carriers and is thought to be of great value although subject to confirmation by culture. We also found 5 patients with GSD having positive titers on IHA although again none showed amplification on PCR. Moreover, IHA detected only 12 of 18 patients with GBC ,who were positive on PCR. This thereby questions the sensitivity and specificity of both serological tests in detecting the presence ofS. typhi.
Culture is often sterile, especially in endemic zones with inadequate exposure to prior antibiotic therapy.[3]Sometimes even patients with positive titers are negative on culture. As in our study, no positive bile culture was obtained for Salmonella. Hence, the accuracy of these tests in categorically documenting the presence ofS. typhican be debated.
Song et al[3]were among the first to show PCR to be helpful in successfully detecting amplification products in blood specimens of suspected but culture-negative patients with typhoid fever. Since then, several reports have appeared in the literature suggesting PCR be the gold standard for the diagnosis of typhoid fever.
PCR with sequences of Vi antigen is not feasible because the nucleotide sequence of this antigen has not been fully investigated. A PCR-based assay detectingS. typhiDNA by amplification of its flagellin gene is feasible and has been worked out in this study. The flagellar antigen ofS. typhi(Hld) is encoded by a 1530-bp DNA sequence.[19,20]Although flagellar antigen is not structurally specific toSalmonella sp.and d antigen is also present in manySalmonellaspecies other thanS. typhi,[21]the flagellin gene ofS. typhihas unique nucleotide sequences in the hypervariable region.[19]The nucleotide sequences and predicted amino acid sequences of region VI (corresponding to nucleotides 969 to 1077) of the HJ-d flagellin gene ofS. typhiare different from those of other Salmonella species. The nucleotide sequences are highly homologous withS. typhi.[19,20]These findings suggested that the PCR test, based on the unique sequence in the flagellin gene ofS. typhi, could be used to detectS. typhispecifically in clinical specimens.
We also used designed primers of the Sop E gene that are specific forS. typhisubspecies viz.,S. entericasubsp. enterica serovar Typhi (Ty2). Twelve patients with GBC showed amplification with SE in our study. Interestingly, none of the controls proved to be positive onS. typhispecific PCR analysis. In addition, only 2 cases showed amplification on PCR of bile specimens. Salmonella is known to be susceptible to bile salts and the low positivity rate could be attributed to this. The 2 positive cases may be the result of the presence of dead bacteria or those in circulation. This means that bile of both cases and controls was devoid of a significant presence ofS. typhi. It may even be hypothesized thatS. typhicolonized the diseased gallbladder after the development of GBC due to biliary tract obstruction and lowered patient immunity. It is also our experience that GBC often clinically presents as empyema of the gallbladder.[22]If this had not been true we would have had a few positive results in controls as well. Thus it raises a doubt thatS. typhiis really an initiator of carcinogenesis in the gallbladder in this region of India with a high prevalence of GBC, and its presence might just be a coincidental finding. This study also questions the importance given to studies based solely on serological tests from the region in documentingS. typhias a factor responsible for the high incidence of GBC.
In conclusion, the present study is one of the first of its kind from an endemic region that provides direct evidence for the presence ofS. typhi, especially Ty2, in GBC tissue samples from a large number of GBC patients using a highly sensitive and specific PCR test. No positive bile culture and only 3 positive tissue cultures for Salmonella indicate that the bacteria present in the circulation are virtually dead and incapable of causing serious damage. Thus, the present study opens a forum for further studies looking into the role ofS. typhiin gallbladder carcinogenesis.
Funding:None.
Ethical approval:Not needed.
Contributors:TM and SHS proposed the study. TM wrote the first draft. TM and MRR analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. SHS is the guarantor.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Shukla HS, Awasthi K, Naithani YP, Gupta SC. A clinicopathological study of carcinoma of the gall bladder. Indian J Cancer 1981;18:198-201.
2 Kumar S, Kumar S, Kumar S. Infection as a risk factor for gallbladder cancer. J Surg Oncol 2006;93:633-639.
3 Song JH, Cho H, Park MY, Na DS, Moon HB, Pai CH. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J Clin Microbiol 1993;31:1439-1443.
4 Sleigh JD, Duguid JP. Salmonella. In: Colle JG and Duguid Jp, Fraser AG and Marmion BP (Eds). Practical Medical Microbiology. 13th Edn. Churchill Livingstone: Edinburgh; 1989:456.
5 Barrett TJ. Improvement of the indirect hemagglutination assay for Salmonella typhi Vi antibodies by use of glutaraldehyde-fixed erythrocytes. J Clin Microbiol 1985;22: 662-663.
6 Frankel G. Detection of Salmonella typhi by PCR. J Clin Microbiol 1994;32:1415.
7 Shukla HS. Gallbladder cancer. J Surg Oncol 2006;93:604-606.
8 Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer 2006;118:1591-1602.
9 Matsukura N, Yokomuro S, Yamada S, Tajiri T, Sundo T, Hadama T, et al. Association between Helicobacter bilis in bile and biliary tract malignancies: H. bilis in bile from Japanese and Thai patients with benign and malignant diseases in the biliary tract. Jpn J Cancer Res 2002;93:842-847.
10 Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin 2001;51:349-364.
11 Haghjoo E, Galán JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 2004;101:4614-4619.
12 Caygill CP, Hill MJ, Braddick M, Sharp JC. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 1994; 343:83-84.
13 Welton JC, Marr JS, Friedman SM. Association between hepatobiliary cancer and typhoid carrier state. Lancet 1979;1: 791-794.
14 Lax AJ, Thomas W. How bacteria could cause cancer: one step at a time. Trends Microbiol 2002;10:293-299.
15 Roa I, Ibacache G, Carvallo J, Melo A, Araya J, De Aretxabala X, et al. Microbiological study of gallbladder bile in a high risk zone for gallbladder cancer. Rev Med Chil 1999;127:1049-1055.
16 Csendes A, Becerra M, Burdiles P, Demian I, Bancalari K, Csendes P. Bacteriological studies of bile from the gallbladder in patients with carcinoma of the gallbladder, cholelithiasis, common bile duct stones and no gallstones disease. Eur J Surg 1994;160:363-367.
17 Hazrah P, Oahn K, Tewari M, et al. The frequency of live bacteria in gallstones. HPB (Oxford) 2004;6:28-32.
18 Itah AY, Akpan CJ. Correlation studies on Widal agglutination reaction and diagnosis of typhoid fever. Southeast Asian J Trop Med Public Health 2004;35:88-91.
19 Frankel G, Newton SM, Schoolnik GK, Stocker BA. Unique sequences in region VI of the flagellin gene of Salmonella typhi. Mol Microbiol 1989;3:1379-1383.
20 Wei LN, Joys TM. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J Mol Biol 1985;186: 791-803.
21 Ewing WH. Antigenic scheme for Salmonella. In W. H. Ewing (ed.), Identification of Enterobacteriaceae. Elsevier, New York;1986:243-318.
22 Tewari M, Kumar V, Mishra RR, Kumar M, Shukla HS. Is there a role for cholecystectomy in gallbladder carcinoma discovered to be unresectable for cure at laparotomy? World J Surg 2008;32:2683-2687.
April 14, 2010
Accepted after revision August 20, 2010
Author Affiliations: Department of Surgical Oncology, Institute of Medical Sciences, Banaras Hindu University, Varanasi-221005, U. P., India (Tewari M, Mishra RR and Shukla HS)
Prof. Hari S Shukla, MS, FRCSEd, PhD, 7 SKG Colony, Lanka, Varanasi-221005, U. P., India (Tel: 0091-542-2367718; Fax: 0091-542-2368856; Email: harishukla@usa.net)
© 2010, Hepatobiliary Pancreat Dis Int. All rights reserved.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- News
- Advances in prognostic factors in acute pancreatitis: a mini-review
- Hepatocellular carcinoma metastatic to the kidney mimicking renal oncocytoma
- Simultaneous breast and ovarian metastasis from gallbladder carcinoma
- An eight-year journey of Hepatobiliary & Pancreatic Diseases International
- Interferon and lamivudine combination therapy versus lamivudine monotherapy for hepatitis B e antigen-negative hepatitis B treatment: a meta-analysis of randomized controlled trials
