植物干旱和盐胁迫响应相关miRNA研究进展
2024-09-12宋子荷甄艳
摘要:非编码RNA(non-coding RNA,ncRNA)是一类由生物基因组转录产生但不编码蛋白质的遗传信息分子,作为表观遗传学研究的主要内容,曾一度被视为基因组中的“暗物质”或“转录噪音”。最广为人知的微小RNA (microRNA,miRNA)是在进化上高度保守的一类长20~24个核苷酸的短链非编码小分子RNA,通过与靶位点碱基互补配对切割降解靶基因转录本或抑制其翻译,从而实现对生物体生长发育等过程的调控。随着小RNA测序和降解组测序等miRNA研究手段的发展,越来越多的miRNA及其生物学功能在动植物中被相继报道,揭示了miRNA在逆境响应过程中的重要调控作用。笔者较为系统地论述了植物miRNA的特征、合成过程、作用方式及其在抗旱耐盐方面的研究进展,以期揭示植物抗旱耐盐的miRNA调控机理,为创制抗旱耐盐新种质提供依据。
关键词:非编码RNA;盐胁迫;干旱胁迫;小RNA测序;降解组
中图分类号:S718;Q522"""" 文献标志码:A开放科学(资源服务)标识码(OSID):
文章编号:1000-2006(2024)04-0001-11
Advancements in the research of miRNAs associated with
plant" drought and salt stress responses
SONG Zihe, ZHEN Yan*
(Co-Innovation Center for"" Sustainable Forestry in Southern China, College of Forestry and Grassland, Nanjing Forestry University, Nanjing 210037, China)
Abstract:
China, as a maritime power with an extensive coastline and abundant coastal resources, also faces challenges due to vast areas of saline-alkali land exacerbated by global warming-induced seasonal droughts. These extreme conditions result in low survival rates for most plants, making research on molecular mechanisms of drought resistance and salt tolerance crucial. Enhancing plant survival in arid and saline-alkali regions can yield significant ecological benefits and economic value. Non-coding RNA (ncRNA) represents a class of genetic information molecules transcribed from the genome that do not encode proteins. Once considered genomic “dark matter” or “transcriptional noise,” ncRNAs, particularly microRNAs (miRNAs), have emerged as pivotal in epigenetic research. miRNAs are short (20-24 nucleotides), highly conserved non-coding small RNA molecules that regulate organismal growth and development by cleaving, degrading, or inhibiting translation of target gene transcripts via complementary base pairing. Advancements in miRNA research methods such as small RNA sequencing and degradome sequencing have unveiled numerous miRNAs and their target genes across animals and plants. Insights into their synthesis, processing, maturation, and functional impacts have expanded. In plants, miRNAs predominantly target genes in the open reading frame, employing a straightforward recognition pattern with full or nearly full complementarity to target site sequences. This simplicity has fueled rapid advancements in plant miRNA research, revealing their pivotal regulatory roles in growth, development, and stress responses. This article systematically reviews plant miRNA features, synthesis processes, modes of action, and recent research progress in drought and salt resistance. It summarizes key techniques and strategies in plant miRNA research, addresses current challenges and future prospects, and aims to deepen understanding of miRNA regulatory mechanisms in plant drought and salt resistance. Such insights provide a foundation for developing new drought- and salt-resistant plant varieties.
China is a maritime power with a long coastline and abundant coastal resources. However, this long costline" brings vast areas of saline-alkali land. Additionally, global warming has led to more" frequent seasonal drought. Under such extreme conditions, the survival rate of most plants is very low. Therefore, research on the molecular mechanisms of drought resistance and salt tolerance is particularly important. Improving the survival rate of plants in arid and saline-alkali areas can bring significant ecological benefits and economic value. Non-coding RNA (ncRNA) is a class of genetic information molecules transcribed from the genome that do not encode proteins. As a major focus of epigenetic research, ncRNAs were once considered the “dark matter” or “transcriptional noise” of the genome. The most well-known type of ncRNA is microRNA (miRNA), a highly conserved class of short non-coding small RNA molecules that are 20-24 nucleotides in length. They regulate the growth and development of organisms by cleaving and degrading target gene transcripts or inhibiting the" translation of target genes through complementary base pairing with target sites. With the development of miRNA research methods such as small RNA sequencing and degradome sequencing, an increasing number of miRNAs and their target genes has been reported in animals and plants. Their biological synthesis, processing, maturation, and functional effects have been elucidated. Plant miRNAs complementarily pair with their target genes mostly in the open reading frame, with a"" full or nearly full complementarity to the target" sequences. These characteristics stimulated the" rapid development in plant miRNA research, and a large amount of research has revealed"" important regulatory roles of miRNAs in plant growth, development, and stress responses. This article provides a systematic review of the features, synthesis process, mode of action, and research progress of plant miRNAs in drought and salt resistance. We summarizes the main techniques and strategies for plant miRNA research in recent years, discuss the existing problems and prospects, and" reveals the" regulatory mechanisms of miRNA in" plant drought and salt resistance, providing a basis for" generating" new varieties of drought and salt resistance.
China, as a maritime power with an extensive coastline and abundant coastal resources, also faces challenges due to vast areas of saline-alkali land exacerbated by global warming-induced seasonal droughts. These extreme conditions result in low survival rates for most plants, making research on molecular mechanisms of drought resistance and salt tolerance crucial. Enhancing plant survival in arid and saline-alkali regions can yield significant ecological benefits and economic value. Non-coding RNA (ncRNA) represents a class of genetic information molecules transcribed from the genome that do not encode proteins. Once considered genomic “dark matter” or \"transcriptional noise,\" ncRNAs, particularly microRNAs (miRNAs), have emerged as pivotal in epigenetic research. miRNAs are short (20-24 nucleotides), highly conserved non-coding small RNA molecules that regulate organismal growth and development by cleaving, degrading, or inhibiting translation of target gene transcripts via complementary base pairing. Advancements in miRNA research methods such as small RNA sequencing and degradome sequencing have unveiled numerous miRNAs and their target genes across animals and plants. Insights into their synthesis, processing, maturation, and functional impacts have expanded. In plants, miRNAs predominantly target genes in the open reading frame, employing a straightforward recognition pattern with full or nearly full complementarity to target site sequences. This simplicity has fueled rapid advancements in plant miRNA research, revealing their pivotal regulatory roles in growth, development, and stress responses. This article systematically reviews plant miRNA features, synthesis processes, modes of action, and recent research progress in drought and salt resistance. It summarizes key techniques and strategies in plant miRNA research, addresses current challenges and future prospects, and aims to deepen understanding of miRNA regulatory mechanisms in plant drought and salt resistance. Such insights provide a foundation for developing new drought- and salt-resistant plant varieties.
Keywords:non-coding RNA; salt stress; drought stress; small RNA-Seq; degradome
土壤盐渍化以及周期性干旱是全球生态以及农业面临的日益严重的问题[1-2]。据联合国粮农组织报道,全球范围内干旱地区覆盖约61亿hm2土地(占地球陆地面积约41%),盐渍土壤面积逾8.33亿hm2(占地球陆地面积约5.6%)。干旱和土壤盐渍化都是影响植物生长的重要非生物胁迫,气候干旱、地面蒸发、地下径流汇集和地下水位接近地表都会促使土壤盐渍化,因此,干旱和盐渍两种胁迫往往会伴随出现[3]。干旱胁迫时土壤水分含量降低以及盐胁迫时土壤盐离子浓度升高,都会使土壤水势降低,从而导致植物根系吸水困难,因此两者在一定意义上都属于渗透胁迫。区别在于盐胁迫对植物而言不是单一的逆境信号,其初级胁迫除了渗透胁迫还包含细胞吸收了过多无机离子后引发的离子毒害[4]。干旱和盐胁迫均可以进一步诱发次级氧化胁迫及营养胁迫,从而使植物的呼吸作用、光合作用、代谢以及生长受到严重影响[5-7]。对两种胁迫同时开展研究,更有利于深入了解植物逆境下的调控机制。近年来,微小RNA(miRNA)通过调控关键基因在植物生长发育以及胁迫响应中发挥重要的作用,逐渐成为当前研究的热点内容。笔者从miRNA介导植物响应干旱及盐胁迫的角度切入,总结国内外最新研究进展,为阐明植物抗旱耐盐机理、提高植物在极端环境下的适应能力提供参考。
1 植物miRNA来源及作用机制
1.1 植物miRNA的发现及特征
miRNA是真核生物中广泛存在的一类长20~24个核苷酸的单链非编码小分子RNA,可以通过识别并剪切特定的mRNA,实现其调控功能。miRNA的发现可以追溯到1993年,Lee等[8]从秀丽隐杆线虫(Caenorhabditis elegans)中鉴定出2个不编码蛋白质但可以结合靶基因并抑制其翻译的小分子RNA(miRNA)。直到2001年这类小分子RNA被正式命名为microRNA。2002年Reinhart等[9]从拟南芥(Arabidopsis thaliana)中发现了植物中的首个miRNA。随着生物信息学的发展以及高通量测序技术的迭代更新,越来越多的miRNA被鉴定出来。miRBase数据库(https://www.mirbase.org/)收录了包括已发表的miRNA序列数据、注释、预测基因靶标等信息以供查询和研究,是最主要的miRNA公共数据库之一[10]。
与mRNA相比,miRNA不具有编码蛋白质的能力,本身不含有开放阅读框(open reading frame,ORF);其次,miRNA 的5′端具有高度保守的特殊“种子”区,因而大多数miRNA在不同植物体中的序列相对比较保守[11]。此外,miRNA 倾向于靶向一类mRNA,而不是特定的某一个mRNA。动物miRNA与靶基因互补主要在3′端非翻译区(3′-untranslated region,3′-UTR),植物miRNA与靶基因的互补区多在ORF区,部分在3′-UTR区,很少在5′-UTR区[12-14]。
1.2 植物miRNA的合成
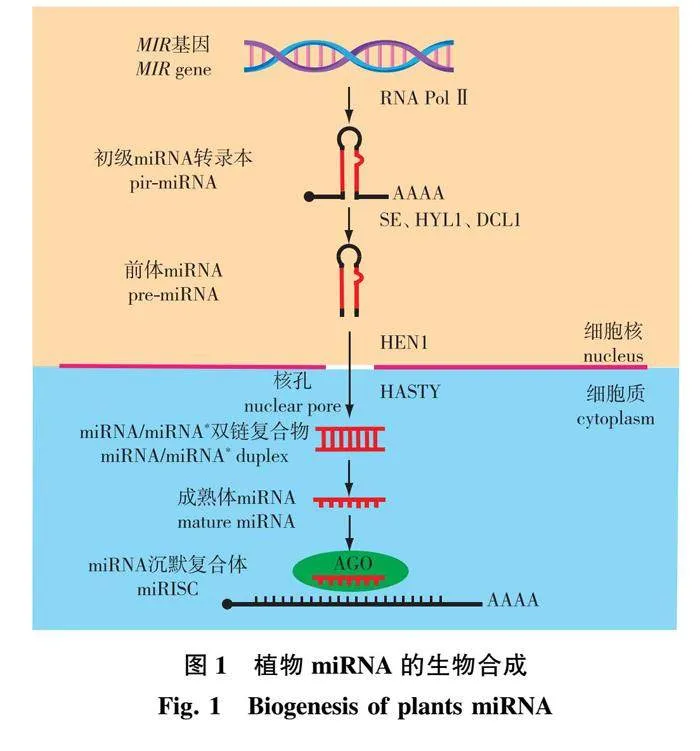
动植物中miRNA生物合成均起始于microRNA基因(MIR)(图1),其表达需要多种蛋白精准且有序的调控。与蛋白质编码基因类似,大多数MIR基因的启动子中含有转录因子结合元件,其中也包括植物激素应答元件,表明部分MIR受转录因子的调控并且可能参与植物激素信号传导[15-16]。首先,在RNA聚合酶Ⅱ(Pol Ⅱ)的作用下MIR转录形成长度为几百个核苷酸的miRNA初级转录产物(primary transcript,pri-miRNA)[17-18]。紧接着由RNA结合蛋白DAWDLE(DDL)结合并稳定pri-miRNA,使其在SmD3/SmB核小体中加工剪切成为具有茎环结构的前体miRNA(precursor miRNA,pre-miRNA)。这一过程需要C2H2型锌指蛋白SERRATE(SE)、双链RNA结合蛋白Hyponastic" Leaves" 1(HYL1)、DICER的同源蛋白Dicer-Like 1(DCL1)和核帽结合复合物(cap-binding complex,CBC)的协同作用来完成[19]。DCL是植物pri-miRNA的剪切加工过程中的关键角色,它分两步缩短pri-miRNA,首先DCL1剪切去除pri-miRNA不完全折叠的末端,形成pre-miRNA的发夹二级结构,之后进一步切割pre-miRNA,生成miRNA/miRNA*双链复合体。在剪切加工过程中SE和HYL1可以提高DCL1切割效率和准确性[20]。随后HUA enhancer" 1(HEN1)将miRNA/miRNA*双链复合体甲基化以保护其不被miRNA降解酶(small rna degrading nuclease,SDN)降解[21-22]。甲基化的miRNA/miRNA*双链复合体被核转运蛋白Hasty(HST)输送到细胞质,随后miRNA作为引导链加载到Argonaute(AGO)蛋白中以形成miRNA诱导沉默复合物miRISC(miRNA-containing RNA induced silencing coplex)[23-24],miRNA*链作为随从链稳定性较差,通常被降解[25],但有研究表明miRNA*也被发现可以富集并加载到AGO蛋白中,以抑制特定植物组织或胁迫条件下的基因表达[26]。
1.3 植物miRNA的作用机制
在植物中miRNA作用的主要方式就是降解靶向mRNA[27],阻止进一步的翻译从而发挥沉默效应。典型的植物miRNA靶位点存在于5′-UTR、ORF和3′-UTR以及非蛋白质编码转录本中。AGO蛋白的P-element induced wimpy testis(PIWI)结构域形成类似于核糖核酸酶H(RNase H)的折叠,构成催化中心[28-29],miRNA与AGO1形成的miRNA诱导的沉默复合物(miRISC)在与目标mRNA配对后,在配对区域进行切割,产生5′端和3′端的裂解片段[30-31]。然而,并非所有植物miRNA与靶基因配对都会导致AGO催化的剪切。一些植物miRNA靶标在5′和3′端的完美碱基配对区域内嵌入了保守的中心错配,允许miRISC复合物结合但阻止剪切的发生[32-33]。
miRNA也可以通过翻译抑制发挥作用[34]。该现象最早在拟南芥中被发现,apetala 2(AP2)是参与花器官发育调控基因,miR172靶向该基因使其蛋白的积累受到影响,但是AP2转录产生的mRNA水平却无明显变化[35]。翻译抑制主要是由AGO1、AGO7和AGO10组装成的miRISC复合体介导[36]。AGO1-miRISC执行的翻译抑制机制取决于miRNA靶位点的位置。该miRISC靶向5′-UTRs,从而阻断核糖体募集和翻译起始,而靶向开放阅读框架的AGO1-miRISC可阻断核糖体运动和翻译延长。由miRNA介导的蛋白质翻译抑制作用可能是由空间位阻引起的,AGO1-miRISCs也可以通过与3′UTR结合影响帽子依赖性翻译中的一个特定环节。有研究表明与5′UTR结合的AGO1-miRISC同时抑制帽子依赖性翻译和非帽子依赖性翻译,而3′UTR结合的AGO1-miRISC只抑制帽子依赖性翻译[37]。
miRNA还可以介导DNA甲基化调控基因表达,在拟南芥的研究中发现,phabulosa(PHB)和phavoluta(PHV)编码序列在miRNA互补位点下游被严重甲基化,并且在相关基因突变体中甲基化程度较低[38]。在水稻(Oryza sativa)中,部分pri-miRNA被DCL3加工成24个核苷酸的长miRNA。这些长miRNA被分类到效应器AGO4中,通过与靶基因进行碱基配对介导DNA甲基化[39]。
2 植物miRNA研究的主要方法与策略
2.1 miRNA鉴定——小RNA测序
目前广为研究的小RNA(small RNA)主要是miRNA、short interfering RNA(siRNA)和 PIWI-interacting RNA(piRNA),其中miRNA的研究最为深入。小RNA测序是目前主流的miRNA检测手段,技术流程主要包含建库测序和生物信息分析两个部分。与大多数RNA测序方法相似,小RNA测序同样需要构建cDNA文库。文库构建的第1步则是将所有RNA片段的3′和5′两端连接接头,接头上包含用于后续测序的引物以及区分样品的独特index,可以在分析测序数据时识别单个reads的文库来源,从而使同时对大量样本进行测序成为可能。然后逆转录得到第1链的cDNA,经过PCR扩增,PAGE胶电泳分离目标DNA片段,切胶回收得到cDNA文库,在库检合格后进行上机测序[40]。高通量测序技术可以对样本中所有小RNA家族进行测序和表达定量,从而解析miRNA、siRNA、piRNA和其他非编码RNA以及相应靶序列。测序原始数据质控后,进行clean reads长度分布统计,小RNA注释,已知miRNA注释,已知piRNA注释,ncRNA分析,新miRNA预测(有参考基因组),差异基因分析,miRNA家族分析,差异表达miRNA聚类分析,miRNA靶基因预测,靶基因GO、KEGG富集分析,协助研究者从海量数据中找出与条件相关的特异性基因,然后进一步分析这些特异性基因的生物学意义[41]。Chen等[42]将无瓣海桑(Sonneratia apetala)进行盐胁迫处理,共构建9个小RNA文库进行测序,鉴定出114个已知miRNA和24个新型miRNA,其中40个miRNA在胁迫处理1 d后相较于对照组差异表达,72个miRNA在胁迫处理28 d后相较于对照差异表达。Wu等[43]进行柑橘(Citrus reticulata)的晚熟突变体和野生型的对比研究,构建4个小RNA文库进行测序,鉴定出107个已知miRNA和21个新型miRNA,其中有24个miRNA在两个类型之间差异表达。
2.2 miRNA靶基因检测——降解组测序
植物miRNA通常与mRNA进行完全或接近完全的配对引起靶基因转录本的剪切,从而调控基因的表达,因此鉴定miRNA的靶基因是了解其功能的关键。普通的生物信息学方法预测准确率较低,而5′-RLM-RACE(5′-RNA ligase mediated rapid amplification of cDNA ends)技术虽然结果精准,但需要了解靶标mRNA的序列,且在同一实验中不能检测多个靶标,对低表达的靶基因不敏感等诸多问题也局限了其应用范围。随即一种结合下一代测序(next-generation sequencing,NGS)和5′-RLM-RACE验证的高通量检测方法降解组测序(degradome sequencing)应运而生[44]。首先靶基因经(RLM-RACE)剪切产生5′端剪切片段和3′端剪切片段,3′剪切片段由于包含有自由的5′单磷酸和3′polyA尾巴,在RNA连接酶的作用下与5′端接头序列连接。随后利用oligo(DT)引物将新形成的单链RNA序列反转录成cDNA序列,并进行PCR扩增。未经处理的cDNA过长无法满足高通量测序的要求,所以利用限制性内切酶Mme I特异性识别5′接头上的酶切位点进行切割,产生20~21个核苷酸的产物,接着与3′端DNA接头连接后进行RCR扩增并上机测序[45]。最后分析降解组文库的数据,删除接头序列后利用相关生物信息学工具例如PAREsnip[46]、CleaveLand4[47]或sPARTA[48]进行分析。Zhang等[49]利用小RNA和降解组测序鉴定出21个miRNA-mRNA作用对参与芝麻盐胁迫响应。Candar等[50]对干旱胁迫下番茄(Solanum lycopersicum)不同组织进行小RNA和降解组测序测序,并利用CleaveLand4软件预测出与59个降解位点相关联的115个特异的miRNA-mRNA作用对。
2.3 miRNA功能研究——TM和STTM
TM(target mimicry)技术近来在植物中被用于miRNA的功能研究,由于miRNA可以靶向多个靶基因,TM技术相比于将靶基因沉默的传统研究方法更能全面且准确地探究miRNA的作用。早期研究人员在拟南芥中首次发现了TM机制,miR399通常会在植物处于磷饥饿状态时表达,但其靶基因Phosphate2(PHO2)的表达水平并未显著下降,同时非蛋白编码基因induced by phosphate starvation 1(IPS1)也被磷饥饿诱导表达,IPS1与miR399 5′端的第10和11位序列之间形成了一个2~4个碱基的错配环结构(CUA),其与miR399的结合能力较PHO2更强,IPS1通过与PHO2竞争性结合miR399从而上调PHO2的表达[33]。在拟南芥中分别超表达TM-MIM156、TM-MIM319和TM-MIM858,检测到靶基因不同程度上调且植株均有表型变化[33]。Wang等[51]研究发现水稻中的一种eTM(endogenous target mimic)(osa-eTM160)显著降低了osa-miR160在花药发育早期对auxin" response factor18(osa-ARF18)的抑制作用,从而调节水稻种子结实和种子大小。Li等[52]在烟草(Nicotiana tabacum)中发现了一种eTM(nta-eTMX27),过量表达nta-eTMX27的转基因植株中nta-miRX27的表达量显著降低,同时其靶基因quinolinate phosphoribosyltaransferase2(QPT2)的表达量显著升高,利用RNAi沉默nta-eTMX27的转基因植株相关基因表达量变化趋势相反。然而,并非所有的miRNA都能够被TM显著抑制[53]。此后研究人员在此基础上改良出一种更加高效的miRNA沉默新方法,短串联模拟靶标 (short tandem target mimic,STTM) 技术,STTM由一段长48、88或96个核苷酸的间隔序列以及两个miRNA结合位点组成[54]。STTM技术能对靶向的miRNA进行有效降解,而这种降解需要SDN的活性。STTM相较于TM有着更高的效率、更强的特异性,并且转基因植株的表型也更加稳定[55]。
3 miRNA广泛参与植物干旱和盐胁迫应答过程
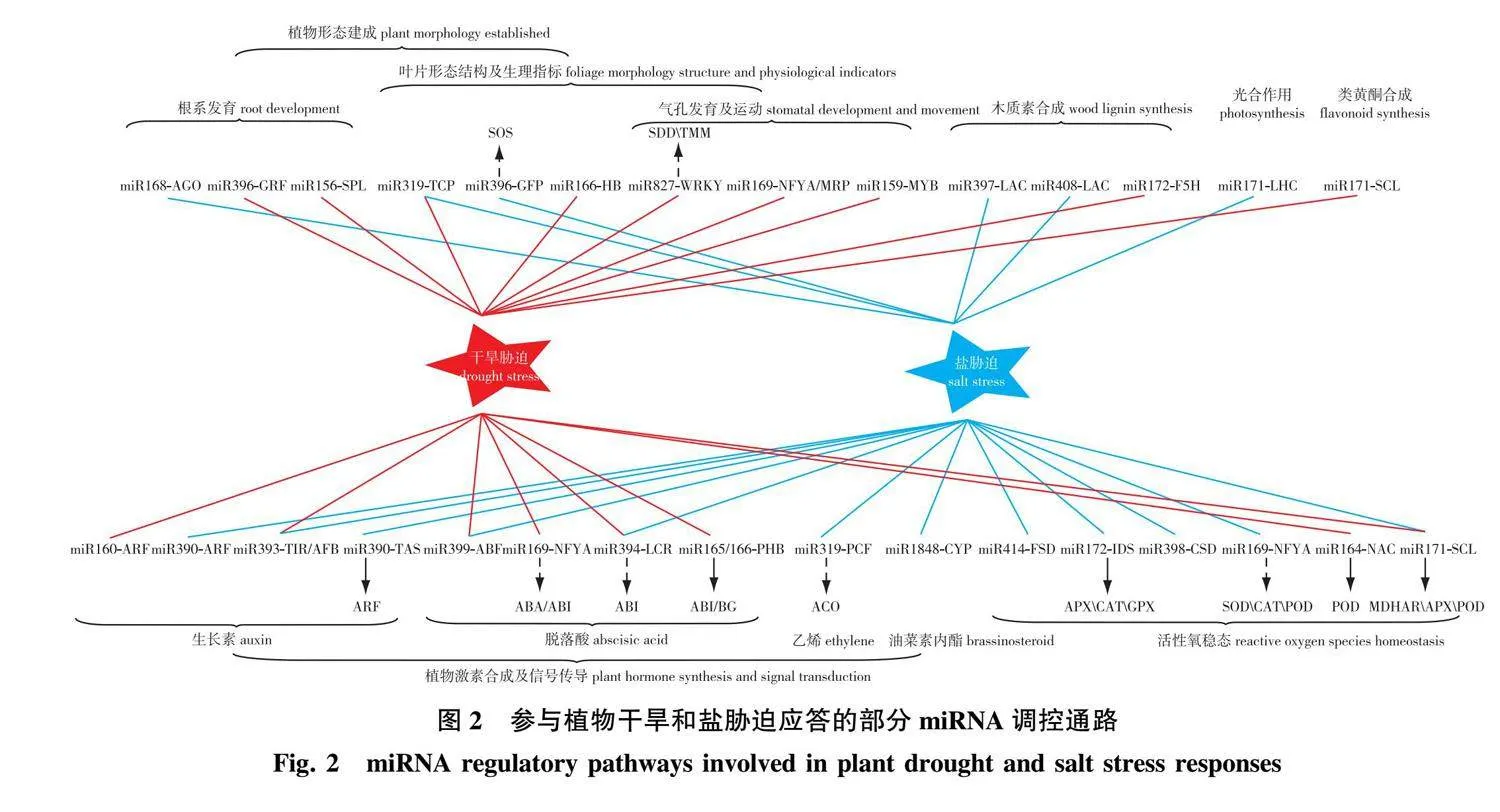
植物在复杂的环境中繁衍生息,受到各种生物或非生物因子的胁迫,非生物因子包括空气质量和流量、光强和光质、温度、水分、矿质营养和微量元素含量、盐分及土壤的pH和氧化还原电势,当这些环境因子的变化波动超出正常范围,就会对植物的生长发育造成负面影响,包括活性氧(ROS)产生、膜稳定性降低、蛋白质变性增加、离子平衡化、新陈代谢紊乱及物理损伤等[56-57]。植物对逆境的应答分为环境适应性(adaptation)和表型可塑性(phenotypic plasticity),环境适应性是经过多代自然环境选择之后形成的能适应环境的稳定遗传变化,表型可塑性则是植物暴露在新环境中产生生理和形态上的可逆变化,无需遗传修饰[58-59]。植物逆境响应涉及复杂且精密的调控过程,miRNA作为重要的转录后调控的一类小RNA,通过直接靶向功能基因或通过靶向转录因子间接调控下游功能基因等模式,在植物生长发育和响应非生物和生物胁迫等过程中扮演了极为重要的角色。近年来,高通量测序技术快速发展,研究者们通过小RNA测序和降解组测序在不同植物种中发现许多响应干旱和盐胁迫的miRNA-mRNA模块并进一步探究验证其调控功能,相关模块主要介导植物形态建成、激素合成与信号传导和活性氧稳态调节等途径来响应干旱和盐胁迫(图2)。
3.1 miRNA介导干旱和盐胁迫下的植物形态建成
在干旱和盐胁迫下植物发达根系对应着较大的吸收面积,提高营养吸收能力,缓解水分以及营养胁迫对植物生长的抑制,miR396-GRF、miR168-AGO和miR156-SPL等模块通过影响植物根系发育来响应干旱或盐胁迫。Zhang等[60]研究发现,SimiR396d靶向growth-regulating factor 1(SiGRF1)正调控谷子(Setaria italica)根系生长和抗旱性,过表达SimiR396d和沉默SiGRF1促进了突变植株的根系生长,增强了抗旱能力,过表达SiGRF1和对SimiR396d进行靶标模拟(TM)的突变植株中根系生长受到抑制,抗旱能力下降。Wan等[61]研究发现Osa-miR168-OsAGO1模块参与调控水稻盐胁迫耐受性,研究人员利用STTM技术沉默miR168提高了水稻盐胁迫耐受性,突变植株在盐胁迫下根的数量以及根和茎的长度均显著高于野生型。Arshad等[62]研究发现苜蓿(Medicago sativa)中MsmiR156靶向squamosa" promoterbinding-like13(MsSPL13)通过促进根系生长来提高植物抗旱性,过表达MsmiR156的植株干旱胁迫下根系生物量的积累显著增加,利用RNA interference(RNAi)沉默SPL13的植株在干旱胁迫下根长显著增加,抗旱能力提高。
叶片是植物光合作用和蒸腾作用的主要器官,在干旱和盐胁迫下植物叶片的生理、形态以及结构特征均会做出响应,miR319-TCP、miR396-GRF和miR166-HB等模块通过影响叶片形态结构以及生理指标响应干旱或盐胁迫。Li等[63]研究发现在拟南芥中过表达Mtr-miR319a,其靶基因teosinte branched1/cycloidea/proliferating" cell factors4(AtTCP4)表达量显著下降,相比于野生型,转基因植株表现为叶片卷曲,在盐胁迫下含水量和脯氨酸含量显著升高,一定程度上提高了植株的耐盐性。Zhou等[64]将Osa-miR319在西伯利亚翦股颖(Agrostis stolonifera)中过量表达,TCP家族的相关靶基因表达量下降,突变植株叶片角质层、蜡质覆盖率和厚度以及茎的粗细显著增加,在干旱和盐胁迫下突变植株保水能力增强且细胞膜完整性提高,表现出更强的抗旱耐盐能力。Yuan等[65]研究发现Osa-miR396c-GRF模块对匍匐翦股颖盐胁迫耐受性的调控起重要作用,过量表达Osa-miR396c的匍匐翦股颖中分蘖数量和长度显著减少,且叶片变短变窄,在盐胁迫下突变植株叶片的含水量和叶绿素含量增加,叶片的电解质泄露减少,值得注意的突变植株中salt" overly" sensitive1(OsSOS1)表达量升高,从而导致盐胁迫处理后植株Na+相对含量降低,K+/Na+升高,表现出更强的耐盐性。Zhang等[66]在水稻中利用STTM技术沉默Osa-miR166,转基因植株叶表皮泡状细胞宽度减小导致近轴叶片卷曲,抗旱能力增强,将抗miR166降解形式的靶基因homeodomain containing protein4(OsHB4)过量表达也出现相似表型。
气孔是植物器官上皮的特殊结构,由保卫细胞、副卫细胞及中间的小孔构成,是植物地上部分与外界环境之间气体和水分交换的通道,影响着光合、呼吸以及蒸腾作用等生理活动,在植物抵御干旱胁迫中发挥着重要作用。近年来研究表明,miR827-WRKY、miR169-NFYA/MRP和miR159-MYB等模块可能通过影响下游气孔发育相关基因调控气孔发育和运动进而响应干旱胁迫。Yang等[67]研究发现马铃薯(Solanum tuberosum)Stu-miR827受干旱胁迫显著诱导,利用STTM技术沉默Stu-miR827,突变植株中Stu-miR827的靶基因StWRKY48表达量显著升高,同时调控气孔发育的基因stomatal density and" distribution1(StSDD1)和toomanymouths(StTMM)表达受到抑制,导致突变植株中气孔密度显著增加,抗旱性显著降低。有研究表明multidrug resistance-associated proteins(MPRs)参与调节植物气孔运动,Zhang等[68]在番茄中过表达Sly-miR169c,突变植株中miR169c的靶基因SlNF-YA1/2/3和SlMRP1表达量显著下调,突变植株气孔孔径、气孔导度和蒸腾速率都显著降低,水分流失减少,抗旱能力增强。Jiang等[69]研究发现拟南芥在干旱胁迫下Ath-miR159表达量降低,其靶基因AtMYB33表达量升高,miR159ab功能丧失突变体在干旱胁迫下气孔开放的比例显著降低,从而降低了失水率,突变植株胁迫下的存活率显著高于野生型,抗旱能力增强。
3.2 miRNA参与干旱和盐胁迫下的植物激素合成与信号传导
植物激素是植物生长发育乃至胁迫响应中重要的信号分子[70]。植物细胞生长素(IAA)可以通过调控植物生长发育来参与胁迫响应,当植物细胞内生长素含量较低时,Aux/indoleacetic" acids" proteins(IAAs)与auxin" response factors(ARFs)形成异源二聚体抑制其转录调控功能,生长素含量升高后AUX/IAAs与transport inhibitor 11(TIR1)/auxin-signaling F-box(AFB)复合物结合,随后被泛素化降解,从而解除对ARF的抑制作用,ARF调控下游相关基因表达,完成生长素信号传导[71]。miR160-ARF、miR390-ARF和miR393-TIR/AFB等模块通过调控生长素信号传导响应干旱和盐胁迫。Shen等[72]研究发现过表达Mdm-miR160和RNAi沉默其靶基因MdARF17的转基因苹果(Malus domestica)抗旱能力显著增加,下游基因MdHYL1正调控苹果不定根发育和抗旱能力,MdARF17通过与MdHYL1的启动子结合来抑制MdHYL1的表达,结果表明Mdm-miR160-ARF17-HYL1模块在苹果干旱胁迫响应中发挥重要作用。Zhang等[73]将棉花(Gossypium hirsutum)中的Ghr-miR160b在拟南芥中过表达,其靶基因AtARF17/18表达量显著降低,同时突变植株在盐胁迫下种子的萌发率显著提高,增强了植株的耐盐性。He等[74]研究发现毛白杨(Populus tomentosa)中Ptr-miR390靶向trans-acting short interfering" RNA 3(PtTAS3)产生tasiARFs,tasiARFs靶向PtARF4从而调控杨树的盐胁迫响应,过表达Ptr-miR390的植株在盐胁迫下侧根的长度和密度以及嫩枝和根的干质量都显著升高,表现出更强的耐盐性,过表达STTM390的植株则在盐胁迫下表现出完全相反的表型,对盐胁迫更为敏感,有趣的是,在盐胁迫下引入PtIAA17通过阻断生长素信号传导抑制了Ptr-miR390-PtARF4调控的侧根伸长。Zhao等[75]研究发现Osa-miR393靶向AsTIR1/AsAFB2介导匍匐翦股颖的盐胁迫响应,过量表达Osa-miR393的植株,AsTIR1和AsAFB2的表达量显著降低,突变植株在盐胁迫下含水量和叶绿素含量增加,MDA含量和电解质泄露率降低,耐盐性增强,在干旱胁迫下,叶片含水率增加,叶片电解质泄露率降低,抗旱性增强。
脱落酸(ABA)是一种重要的植物激素,在植物受到环境胁迫时发挥作用,不仅可以调节植物的生长和气孔关闭还可以调节种子的成熟和休眠,miR399-ABF、miR169-NFYA、miR394-LCR和miR165/166-PHB等模块通过调节ABA的信号传导来响应干旱和盐胁迫。Baek等[76]研究发现随着NaCl和ABA浓度的增加,拟南芥中Ath-miR399f前体的表达量显著增加,过量表达Ath-miR399f的转基因植株中靶基因abscisic acid-responsive element binding factors3(AtABF3)的表达量显著降低,对盐胁迫和ABA处理的耐受性强于野生型植株,但在干旱胁迫下水分损失更为严重,抗旱能力降低。Ni等[77]发现大豆(Glycine max)中GmmiR169c靶向nuclear transcription factory subunit alpha 3(GmNFYA3),过表达GmNFYA3的转基因拟南芥ABA生物合成的相关基因aba deficient1(AtABA1)和AtABA2以及ABA信号传导的相关基因abl interactor1(AtABI1)和AtABI2表达量显著上调,突变植株叶片水分损失减少,耐旱性增强,但对外源性ABA的敏感性增加。Song等[78]研究发现拟南芥中Ath-miR394受ABA处理以及干旱和盐胁迫诱导,过表达Ath-miR394和靶基因ligase chain reaction(AtLCR)功能丧失突变植株抗旱能力增强,但对盐胁迫更为敏感,突变植株中ABA响应基因ABI3/4/5表达量升高,在种子萌发、子叶发育和根系伸长等方面表现出对ABA超敏感,而过量表达抗miR394降解的LCR的突变植株耐盐能力增强,但对干旱胁迫更为敏感,ABA响应基因表达量降低,对ABA不敏感。Yan等[79]利用STTM技术沉默Ath-miR165/166,转基因拟南芥在干旱胁迫下存活率以及含水量增加,抗旱能力增强,但在种子萌发和子叶绿化方面表现出对ABA敏感,进一步研究发现miR165/166的靶基因Phbulosa(AtPHB)通过直接调控AtABI4和beta-1,3-glucanase1(AtBG1)的表达来调节ABA稳态。
miRNA还可以通过调控乙烯和油菜素内酯的生物合成来响应胁迫。Liu等[80]研究发现在柳枝稷(Panicum virgatum)中过量表达Osa-miR319b以及抑制其靶基因proliferating cell factor5(PvPCF5)表达都可以通过促进乙烯合成基因1-aminocyclopropane-1-carboxylate" oxidase(PvACO)的表达从而提高突变体植株的耐盐性。Xia等[81]研究发现水稻中Osa-miR1848靶向cytochrome p450 subfamily" 51(OsCYP51G3)介导植物甾醇和油菜素内酯的生物合成来响应盐胁迫,过表达osa-miR1848和沉默OsCYP51G3的突变植株相比野生型有更多的电解质泄露,对盐胁迫更为敏感。
3.3 miRNA调节干旱和盐胁迫下的植物体内活性氧稳态
植物活性氧的清除能力与抗旱耐盐能力息息相关,干旱和盐胁迫时植物细胞会增加活性氧的产出和积累,导致活性氧代谢平衡被打破,诱发膜脂过氧化作用,损伤生物大分子,破坏细胞结构和功能[82]。植物体内的酶促防御系统,包括超氧化物歧化酶(superoxide dismutase,SOD)、过氧化物酶(peroxidase,POD)、谷胱甘肽过氧化物酶(glutathione peroxidase,GSH-Px)、抗坏血酸过氧化物酶 (ascorbate peroxidase,APX)和过氧化氢酶(catalase,CAT)等[83]。miR414-FSD、miR172-IDS、miR398-CSD、miR169-NFYA、miR164-NAC和miR171-SCL等模块可以通过调节植物体内相关抗氧化酶的活性来响应干旱和盐胁迫。Wang等[84]研究发现棉花中Ghr-miR414c通过靶向GhFSD1(Fe-SOD)参与植物对盐胁迫响应的调节,过表达Ghr-miR414c与沉默GhFSD1的转基因拟南芥呈现相似的表型,都对盐胁迫更为敏感。Cheng等[85]发现水稻中miR172a/b通过切割indeterminate spikelet1(OsIDS1)的转录本,解除OsIDS1对下游ROS清除相关基因的抑制,从而介导盐胁迫的响应,miR172a/b在水稻盐胁迫0.5和4.0 h时被显著诱导,过量表达miR172a的水稻在盐胁迫下表现出更强的耐受性,相反过量表达STTM172a的水稻对盐胁迫更为敏感。He等[86]研究发现番茄(Solanum lycopersicum)中Sly-miR398b靶向copper/zinc superoxide dismutase 1(Slycsd1),在盐胁迫下,过量表达Sly-miR398b抑制突变植株的生长,活性氧和MDA含量显著增加,光合速率显著降低。Xing等[87]研究发现玉米(Zea mays)中ZmmiR169q表达受到盐胁迫的抑制,且其表达水平与植株体内活性氧水平高度相关,ZmmiR169q的过表达株系对盐胁迫高度敏感,而其沉默株系表现耐盐表型,过量表达miR169q的靶基因ZmNF-YA8可以诱导过氧化物酶的表达提升,降低活性氧水平,提高玉米对盐胁迫的耐受性。Wang等[88]研究发现小麦(Triticum aestivum)中Tae-miR9674a在盐和干旱胁迫初期表达显著上调,将Tae-miR9674a在烟草中过量表达,突变植株生长状况显著优于野生型,有更强的耐盐性,并且过量表达株系中脯氨酸合成基因pyrroline-5-carboxylate" synthase1(NtP5CS1)和部分抗氧化酶基因如NtFeSOD、NtCAT1和NtPOD4在盐和干旱胁迫下表达上调,相反这类基因在沉默突变株系中表达下调,沉默株系也表现出对盐和干旱胁迫更为敏感。Peng等[89]研究发现苹果中Msi-miR164g在干旱胁迫下被显著诱导,Msi-miR164g靶向NAM/ATAF/CUC(NAC)家族的成员MsNAC022,MsNAC022通过与MsPOD启动子特定区域结合来激活其转录,从而维持植物体内活性氧平衡,过表达MsNAC022的转基因苹果表现出更强的抗旱性,而过表达Msi-miR164g的拟南芥植株则对干旱胁迫更敏感。Wang等[90]证实苹果中mdm-miR171i靶向Scarecrow-like 26(MsSCL26),MsSCL26与下游抗氧化基因Monodehydroascorbate" reductase(MsMDHAR)、MsPOD和MsAPX启动子结合促进其表达,过表达miR171i的转基因苹果对干旱胁迫敏感,过表达MsSCL26和敲除mdm-miR171i的转基因植株抗旱耐盐能力显著增强。
3.4 植物干旱和盐胁迫应答的其他miRNA调控途径
miRNA还可以通过调控木质素合成、类黄酮合成及光合作用等模块响应干旱和盐胁迫。Nguyen等[91]在拟南芥中过表达Ath-miR397和Sv-miR397,其靶基因Laccase(LAC)家族基因成员AtLAC2/4/17在突变植株中表达水平降低导致木质素积累减少,突变株对盐胁迫更为敏感,胁迫下植株鲜质量和叶绿素含量显著降低。Qin等[92]研究发现玉米中ZmmiR408靶向ZmLAC9,过表达ZmmiR408的植株中木质素积累减少从而表现为对盐胁迫敏感,相反敲除ZmmiR408a/b和过表达ZmLAC9的转基因植株中木质素积累增加,细胞壁增厚,耐盐性提高。Jeena等[93]研究发现假马齿苋(Bacopa monnieri)中Bm-miR172c-5p通过靶向苯丙烷代谢途径上的关键基因ferulate" 5-hydroxylase(BmF5H)调节木质素生物合成,过表达Bm-miR172-5p的转基因假马齿苋中木质素的含量降低,表现出对干旱敏感的表型,内源性靶标模拟沉默Bm-miR172-5p的转基因植株中木质素含量升高,抗旱能力增强。Fan等[94]将研究发现百慕大草 (Cynodon dactylon)中CdmiR171f靶向编码光合系统中的电子转运蛋白的基因light-harvesting" complex1(CdLHC1),在蒺藜苜蓿(Medicago truncatula)中异源过表达CdmiR171f的植株在盐胁迫下相比野生型表现出干物质的积累增加和光合作用性能的提升。Um等[95]研究发现水稻中Osa-miR171f在干旱胁迫下被显著诱导,过表达Osa-miR171f的突变植株抗旱能力显著增强,敲除Osa-miR171f的突变植株对干旱胁迫更为敏感,值得注意的是chalcone synthase(OsCHS1)、chalcone isomerase(OsCHI)、flavanone 3-hydroxylase(OsF3H)等类黄酮合成相关基因在突变植株中表达量受到Osa-miR171f-OsSCL6模块调控,Osa-miR171f-OsSCL6可能通过调控水稻类黄酮合成来增强抗旱能力。
4 展 望
干旱和土壤盐渍化严重制约了农业的发展,随之带来的粮食减产问题也迫在眉睫。大多数非盐生和旱生植物在胁迫初期可以通过形态和生理变化来适应环境,但随着胁迫的持续,大都受到严重的影响甚至死亡。了解植物干旱和盐胁迫响应的分子机制,对培育抗逆作物育种具有重要意义。近年来,随着高通量测序技术的发展,许多miRNA的功能被逐步揭晓,其已被证明参与调控植物生长发育、生物与非生物胁迫等方面。miRNA的研究也为审视植物抗旱耐盐的机制提供了全新的角度。但如今大量的研究仍停留在对不同植物种中miRNA的鉴定层面,对于miRNA在植物抗逆途径中的详细机制仍了解不足,需要更深入的研究揭示其精准的调控网络,但并非局限于miRNA对靶基因的调控,对此不仅可以通过对MIR启动子的研究鉴定miRNA上游的调控关系,还可以发掘竞争性内源RNA(competing endogenous RNA,ceRNA)网络,并验证ceRNA分子之间的竞争关系。在植物胁迫响应中存在许多发挥关键作用的基因,但其中大部分并没有发现被miRNA靶向,因此可以设计并利用靶向植物胁迫响应中关键mRNA的人工miRNA(artificial miRNAs,amiRNA),利用人工miRNA对目的基因实现精准切割,为开发抗逆品种提供了新的思路。
参考文献(reference):
[1]CAI W J,LENGAIGNE M,BORLACE S,et al.More extreme swings of the South Pacific convergence zone due to greenhouse warming[J].Nature,2012,488(7411):365-369.DOI: 10.1038/nature11358.
[2]QADIR M,QUILLROU E,NANGIA V,et al.Economics of salt-induced land degradation and restoration[J].Nat Resour Forum,2014,38(4):282-295.DOI: 10.1111/1477-8947.12054.
[3]ZHAO S,ZHANG Q,LIU M,et al.Regulation of plant responses to salt stress[J].Int J Mol Sci,2021,22(9):4609.DOI: 10.3390/ijms22094609.
[4]DEINLEIN U,STEPHAN A B,HORIE T,et al.Plant salt-tolerance mechanisms[J].Trends Plant Sci,2014,19(6):371-379.DOI: 10.1016/j.tplants.2014.02.001.
[5]FOYER C H,RASOOL B,DAVEY J W,et al.Cross-tolerance to biotic and abiotic stresses in plants:a focus on resistance to aphid infestation[J].J Exp Bot,2016,67(7):2025-2037.DOI: 10.1093/jxb/erw079.
[6]MILLER G,SUZUKI N,CIFTCI-YILMAZ S,et al.Reactive oxygen species homeostasis and signalling during drought and salinity stresses[J].Plant Cell Environ,2010,33(4):453-467.DOI: 10.1111/j.1365-3040.2009.02041.x.
[7]MUNNS R,GILLIHAM M.Salinity tolerance of crops-what is the cost?[J].New Phytol,2015,208(3):668-673.DOI: 10.1111/nph.13519.
[8]LEE R C,FEINBAUM R L,AMBROS V.The C.elegans heterochronic gene Lin-4 encodes small RNAs with antisense complementarity to Lin-14[J].Cell,1993,75(5):843-854.DOI: 10.1016/0092-8674(93)90529-y.
[9]REINHART B J,WEINSTEIN E G,RHOADES M W,et al.MicroRNAs in plants[J].Genes Dev,2002,16(13):1616-1626.DOI: 10.1101/gad.1004402.
[10]AMBROS V,BARTEL B,BARTEL D P,et al.A uniform system for microRNA annotation[J].RNA,2003,9(3):277-279.DOI: 10.1261/rna.2183803.
[11]AXTELL M J.Classification and comparison of small RNAs from plants[J].Annu Rev Plant Biol,2013,64:137-159.DOI: 10.1146/annurev-arplant-050312-120043.
[12]BONNET E,WUYTS J,ROUZ P,et al.Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes[J].Proc Natl Acad Sci USA,2004,101(31):11511-11516.DOI: 10.1073/pnas.0404025101.
[13]WANG X J,REYES J L,CHUA N H,et al.Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets[J].Genome Biol,2004,5(9):R65.DOI: 10.1186/gb-2004-5-9-r65.
[14]LYTLE J R,YARIO T A,STEITZ J A.Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR[J].Proc Natl Acad Sci USA,2007,104(23):9667-9672.DOI: 10.1073/pnas.0703820104.
[15]XIE Z X,ALLEN E,FAHLGREN N,et al.Expression of Arabidopsis MIRNA genes[J].Plant Physiol,2005,138(4):2145-2154.DOI: 10.1104/pp.105.062943.
[16]MEGRAW M,BAEV V,RUSINOV V,et al.MicroRNA promoter element discovery in Arabidopsis[J].RNA,2006,12(9):1612-1619.DOI: 10.1261/rna.130506.
[17]KIM Y J,ZHENG B L,YU Y,et al.The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana[J].EMBO J,2011,30(5):814-822.DOI: 10.1038/emboj.2011.3.
[18]SONG X W,LI Y,CAO X F,et al.MicroRNAs and their regulatory roles in plant-environment interactions[J].Annu Rev Plant Biol,2019,70:489-525.DOI: 10.1146/annurev-arplant-050718-100334.
[19]HAN M H,GOUD S,SONG L,et al.The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation[J].Proc Natl Acad Sci USA,2004,101(4):1093-1098.DOI: 10.1073/pnas.0307969100.
[20]YANG L,LIU Z Q,LU F,et al.SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis[J].Plant J,2006,47(6):841-850.DOI: 10.1111/j.1365-313X.2006.02835.x.
[21]LI J J,YANG Z Y,YU B,et al.Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis[J].Curr Biol,2005,15(16):1501-1507.DOI: 10.1016/j.cub.2005.07.029.
[22]YANG Z Y,EBRIGHT Y W,YU B,et al.HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide[J].Nucleic Acids Res,2006,34(2):667-675.DOI: 10.1093/nar/gkj474.
[23]AXTELL M J,WESTHOLM J O,LAI E C.Vive la différence:biogenesis and evolution of microRNAs in plants and animals[J].Genome Biol,2011,12(4):221.DOI: 10.1186/gb-2011-12-4-221.
[24]EAMENS A L,SMITH N A,CURTIN S J,et al.The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes[J].RNA,2009,15(12):2219-2235.DOI: 10.1261/rna.1646909.
[25]TOMARI Y,MATRANGA C,HALEY B,et al.A protein sensor for siRNA asymmetry[J].Science,2004,306(5700):1377-1380.DOI: 10.1126/science.1102755.
[26]XU L,HU Y G,CAO Y,et al.An expression atlas of miRNAs in Arabidopsis thaliana[J].Sci China Life Sci,2018,61(2):178-189.DOI: 10.1007/s11427-017-9199-1.
[27]JONES-RHOADES M W,BARTEL D P,BARTEL B.MicroRNAS and their regulatory roles in plants[J].Annu Rev Plant Biol,2006,57:19-53.DOI: 10.1146/annurev.arplant.57.032905.105218.
[28]SONG J J,SMITH S K,HANNON G J,et al.Crystal structure of Argonaute and its implications for RISC slicer activity[J].Science,2004,305(5689):1434-1437.DOI: 10.1126/science.1102514.
[29]YUAN YR,PEI Y,MA J B,et al.Crystal structure of A. aeolicus argonaute,a site-specific DNA-guided endoribonuclease,provides insights into RISC-mediated mRNA cleavage[J].Mol Cell,2005,19(3):405-419.DOI: 10.1016/j.molcel.2005.07.011.
[30]ADDO-QUAYE C,ESHOO T W,BARTEL D P,et al.Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome[J].Curr Biol,2008,18(10):758-762.DOI: 10.1016/j.cub.2008.04.042.
[31]GERMAN M A,PILLAY M,JEONG D H,et al.Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends[J].Nat Biotechnol,2008,26(8):941-946.DOI: 10.1038/nbt1417.
[32]AXTELL M J,JAN C,RAJAGOPALAN R,et al.A two-hit trigger for siRNA biogenesis in plants[J].Cell,2006,127(3):565-577.DOI: 10.1016/j.cell.2006.09.032.
[33]FRANCO-ZORRILLA J M,VALLI A,TODESCO M,et al.Target mimicry provides a new mechanism for regulation of microRNA activity[J].Nat Genet,2007,39(8):1033-1037.DOI: 10.1038/ng2079.
[34]BRODERSEN P,SAKVARELIDZE-ACHARD L,BRUUN-RASMUSSEN M,et al.Widespread translational inhibition by plant miRNAs and siRNAs[J].Science,2008,320(5880):1185-1190.DOI: 10.1126/science.1159151.
[35]SCHWAB R,PALATNIK J F,RIESTER M,et al.Specific effects of microRNAs on the plant transcriptome[J].Dev Cell,2005,8(4):517-527.DOI: 10.1016/j.devcel.2005.01.018.
[36]HOU C Y,LEE W C,CHOU H C,et al.Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants[J].Plant Cell,2016,28(10):2398-2416.DOI: 10.1105/tpc.16.00295.
[37]IWAKAWA HO,TOMARI Y.Molecular insights into microRNA-mediated translational repression in plants[J].Mol Cell,2013,52(4):591-601.DOI: 10.1016/j.molcel.2013.10.033.
[38]BAO N,LYE K W,BARTON M K.MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome[J].Dev Cell,2004,7(5):653-662.DOI: 10.1016/j.devcel.2004.10.003.
[39]WU L,ZHOU H,ZHANG Q,et al.DNA methylation mediated by a microRNA pathway[J].Mol Cell,2010,38(3):465-475.DOI: 10.1016/j.molcel.2010.03.008.
[40]VAN GOETHEM A,MESTDAGH P,VAN MAERKEN T,et al.MicroRNA expression analysis using small RNA sequencing discovery and RT-qPCR-based validation[J].Methods Mol Biol,2017,1654:197-208.DOI: 10.1007/978-1-4939-7231-9_13.
[41]CHVEZ MONTES RA,JAIMES-MIRANDA F,DE FOLTER S.Bioinformatic analysis of small RNA sequencing libraries[J].Methods Mol Biol,2019,1932:51-63.DOI: 10.1007/978-1-4939-9042-9_4.
[42]CHEN B B,DING Z Y,ZHOU X,et al.Integrated full-length transcriptome and microRNA sequencing approaches provide insights into salt tolerance in mangrove (Sonneratia apetala Buch.-Ham.)[J].Front Genet,2022,13:932832.DOI: 10.3389/fgene.2022.932832.
[43]WU J X,ZHENG S S,FENG G Z,et al.Comparative analysis of miRNAs and their target transcripts between a spontaneous late-ripening sweet orange mutant and its wild-type using small RNA and degradome sequencing[J].Front Plant Sci,2016,7:1416.DOI: 10.3389/fpls.2016.01416.
[44]董淼,黄越,陈文铎,等.降解组测序技术在植物miRNA研究中的应用[J].植物学报,2013,48(3):344-353.DONG M,HUANG Y,CHEN W D,et al.Use of degradome sequencing in study of plant microRNAs[J].Chin Bull Bot,2013,48(3):344-353.DOI: 10.3724/SP.J.1259.2013.00344.
[45]BAKSA I,SZITTYA G.Identification of ARGONAUTE/small RNA cleavage sites by degradome sequencing[J].Methods Mol Biol,2017,1640:113-128.DOI: 10.1007/978-1-4939-7165-7_7.
[46]FOLKES L,MOXON S,WOOLFENDEN H C,et al.PAREsnip:a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing[J].Nucleic Acids Res,2012,40(13):e103.DOI: 10.1093/nar/gks277.
[47]ADDO-QUAYE C,MILLER W,AXTELL M J.CleaveLand:a pipeline for using degradome data to find cleaved small RNA targets[J].Bioinformatics,2009,25(1):130-131.DOI: 10.1093/bioinformatics/btn604.
[48]KAKRANA A,HAMMOND R,PATEL P,et al.SPARTA:A parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets,including new miRNA target-identification software[J].Nucleic Acids Res,2014,42(18):e139.DOI: 10.1093/nar/gku693.
[49]ZHANG Y J,GONG H H,LI D H,et al.Integrated small RNA and Degradome sequencing provide insights into salt tolerance in sesame (Sesamum indicum L.)[J].BMC Genomics,2020,21(1):494.DOI: 10.1186/s12864-020-06913-3.
[50]CANDAR C B,ARICAN E,ZHANG B H.Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes[J].Plant Biotechnol J,2016,14(8):1727-1746.DOI: 10.1111/pbi.12533.
[51]WANG M,WU H J,FANG J,et al.A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160[J].Sci Bull,2017,62(7):470-475.DOI: 10.1016/j.scib.2017.03.013.
[52]LI F F,WANG W D,ZHAO N,et al.Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco[J].Plant Physiol,2015,169(2):1062-1071.DOI: 10.1104/pp.15.00649.
[53]TODESCO M,RUBIO-SOMOZA I,PAZ-ARES J,et al.A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana[J].PLoS Genet,2010,6(7):e1001031.DOI: 10.1371/journal.pgen.1001031.
[54]YAN J,GU Y Y,JIA X Y,et al.Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis[J].Plant Cell,2012,24(2):415-427.DOI: 10.1105/tpc.111.094144.
[55]郑文清,张倩,杜亮.短串联靶标模拟技术及其在植物miRNA功能研究中的应用[J].生物技术通报,2020,36(12):256-264.ZHENG W Q,ZHANG Q,DU L.Short tandem target mimic and its application in analyzing plant miRNA functions[J].Biotechnol Bull,2020,36(12):256-264.DOI: 10.13560/j.cnki.biotech.bull.1985.2020-0257.
[56]SHARMA M,KUMAR P,VERMA V,et al.Understanding plant stress memory response for abiotic stress resilience:molecular insights and prospects[J].Plant Physiol Biochem,2022,179:10-24.DOI: 10.1016/j.plaphy.2022.03.004.
[57]SUZUKI N,RIVERO R M,SHULAEV V,et al.Abiotic and biotic stress combinations[J].New Phytol,2014,203(1):32-43.DOI: 10.1111/nph.12797.
[58]WANG Z R,BASKIN J M,BASKIN C C,et al.Great granny still ruling from the grave:phenotypical response of plant performance and seed functional traits to salt stress affects multiple generations of a halophyte[J].J Ecol,2022,110(1):117-128.DOI: 10.1111/1365-2745.13789.
[59]LMKE J,BURLE I.Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants[J].Genome Biol,2017,18(1):124.DOI: 10.1186/s13059-017-1263-6.
[60]ZHANG Y F,XIAO T,YI F,et al.SimiR396d targets SiGRF1 to regulate drought tolerance and root growth in foxtail millet[J].Plant Sci,2023,326:111492.DOI: 10.1016/j.plantsci.2022.111492.
[61]WAN J,MENG S J,WANG Q Y,et al.Suppression of microRNA168 enhances salt tolerance in rice (Oryza sativa L.)[J].BMC Plant Biol,2022,22(1):563.DOI: 10.1186/s12870-022-03959-1.
[62]ARSHAD M,FEYISSA B A,AMYOT L,et al.MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13[J].Plant Sci,2017,258:122-136.DOI: 10.1016/j.plantsci.2017.01.018.
[63]LI M N,XU L,ZHANG L X,et al.Overexpression of Mtr-miR319a contributes to leaf curl and salt stress adaptation in Arabidopsis thaliana and Medicago truncatula[J].Int J Mol Sci,2022,24(1):429.DOI: 10.3390/ijms24010429.
[64]ZHOU M,LI D Y,LI Z G,et al.Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass[J].Plant Physiol,2013,161(3):1375-1391.DOI: 10.1104/pp.112.208702.
[65]YUAN S R,ZHAO J M,LI Z G,et al.MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass[J].Hortic Res,2019,6:48.DOI: 10.1038/s41438-019-0130-x.
[66]ZHANG J S,ZHANG H,SRIVASTAVA A K,et al.Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development[J].Plant Physiol,2018,176(3):2082-2094.DOI: 10.1104/pp.17.01432.
[67]YANG J W,ZHANG N,BAI J P,et al.Stu-miR827-targeted StWRKY48 transcription factor negatively regulates drought tolerance of potato by increasing leaf stomatal density[J].Int J Mol Sci,2022,23(23):14805.DOI: 10.3390/ijms232314805.
[68]ZHANG X H,ZOU Z,GONG P J,et al.Over-expression of microRNA169 confers enhanced drought tolerance to tomato[J].Biotechnol Lett,2011,33(2):403-409.DOI: 10.1007/s10529-010-0436-0.
[69]JIANG Y Q,WU X,SHI M,et al.The miR159-MYB33-ABI5 module regulates seed germination in Arabidopsis[J].Physiol Plant,2022,174(2):e13659.DOI: 10.1111/ppl.13659.
[70]黎家,李传友.新中国成立70年来植物激素研究进展[J].中国科学:生命科学,2019,49(10):1227-1281.LI J,LI C Y.Seventy-year major research progress in plant hormones by Chinese scholars[J].Sci Sin (Vitae),2019,49(10):1227-1281.DOI: 10.1360/SSV-2019-0197.
[71]PAQUE S,WEIJERS D Q A:Auxin:the plant molecule that influences almost anything[J].BMC Biol,2016,14(1):67.DOI: 10.1186/s12915-016-0291-0.
[72]SHEN X X,HE J Q,PING Y K,et al.The positive feedback regulatory loop of miR160-Auxin Response Factor 17-HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees[J].Plant Physiol,2022,188(3):1686-1708.DOI: 10.1093/plphys/kiab565.
[73]ZHANG X P,SHEN J,XU Q J,et al.Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton[J].Plant Cell Environ,2021,44(10):3302-3321.DOI: 10.1111/pce.14133.
[74]HE F,XU C Z,FU X K,et al.The MICRORNA390/TRANS-ACTING" SHORT" INTERFERING"" RNA3 module mediates lateral root growth under salt stress via the auxin pathway[J].Plant Physiol,2018,177(2):775-791.DOI: 10.1104/pp.17.01559.
[75]ZHAO J M,YUAN S R,ZHOU M,et al.Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance[J].Plant Biotechnol J,2019,17(1):233-251.DOI: 10.1111/pbi.12960.
[76]BAEK D,CHUN H J,KANG S,et al.A role for Arabidopsis miR399f in salt,drought,and ABA signaling[J].Mol Cells,2016,39(2):111-118.DOI: 10.14348/molcells.2016.2188.
[77]NI Z Y,HU Z,JIANG Q Y,et al.GmNFYA3,a target gene of miR169,is a positive regulator of plant tolerance to drought stress[J].Plant Mol Biol,2013,82(1/2):113-129.DOI: 10.1007/s11103-013-0040-5.
[78]SONG J B,GAO S,SUN D,et al.miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner[J].BMC Plant Biol,2013,13:210.DOI: 10.1186/1471-2229-13-210.
[79]YAN J,ZHAO C Z,ZHOU J P,et al.The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana[J].PLoS Genet,2016,12(11):e1006416.DOI: 10.1371/journal.pgen.1006416.
[80]LIU Y, LI D, YAN J, et al. MiR319-mediated ethylene biosynthesis, signalling and salt stress response in switchgrass[J]. Plant Biotechnology Journal, 2019, 17(12): 2370-2383. DOI: 10.1111/pbi.13154.
[81]XIA K F,OU X J,TANG H D,et al.Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress[J].New Phytol,2015,208(3):790-802.DOI: 10.1111/nph.13513.
[82]GOMEZ J M,JIMENEZ A,OLMOS E,et al.Location and effects of long-term NaCl stress on superoxide dismutase and ascorbate peroxidase isoenzymes of pea (Pisum sativum cv.Puget) chloroplasts[J].J Exp Bot,2004,55(394):119-130.DOI: 10.1093/jxb/erh013.
[83]LEE D H,KIM Y S,LEE C B.The inductive responses of the antioxidant enzymes by salt stress in the rice (Oryza sativa L.)[J].J Plant Physiol,2001,158(6):737-745.DOI: 10.1078/0176-1617-00174.
[84]WANG W,LIU D,CHEN D D,et al.MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress[J].RNA Biol,2019,16(3):362-375.DOI: 10.1080/15476286.2019.1574163.
[85]CHENG X L,HE Q,TANG S,et al.The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops[J].New Phytol,2021,230(3):1017-1033.DOI: 10.1111/nph.17211.
[86]HE Y,ZHOU J X,HU Y F,et al.Overexpression of sly-miR398b increased salt sensitivity likely via regulating antioxidant system and photosynthesis in tomato[J].Environ Exp Bot,2021,181:104273.DOI: 10.1016/j.envexpbot.2020.104273.
[87]XING L J,ZHU M,LUAN M D,et al.miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS[J].Plant Physiol,2022,188(1):608-623.DOI: 10.1093/plphys/kiab498.
[88]WANG L,BAI X Y,QIAO Y,et al.Tae-miR9674a,a microRNA member of wheat,confers plant drought and salt tolerance through modulating the stomata movement and ROS homeostasis[J].Plant Biotechnol Rep,2023,17(4):471-488.DOI: 10.1007/s11816-022-00787-5.
[89]PENG X,FENG C,WANG Y T,et al.miR164g-MsNAC022 acts as a novel module mediating drought response by transcriptional regulation of reactive oxygen species scavenging systems in apple[J].Hortic Res,2022,9:uhac192.DOI: 10.1093/hr/uhac192.
[90]WANG Y T,FENG C,ZHAI Z F,et al.The apple microR171i-SCARECROW-LIKE PROTEINS26.1 module enhances drought stress tolerance by integrating ascorbic acid metabolism[J].Plant Physiol,2020,184(1):194-211.DOI: 10.1104/pp.20.00476.
[91]NGUYEN D Q,BROWN C W,PEGLER J L,et al.Molecular manipulation of microRNA397 abundance influences the development and salt stress response of Arabidopsis thaliana[J].Int J Mol Sci,2020,21(21):7879.DOI: 10.3390/ijms21217879.
[92]QIN R D,HU Y M,CHEN H,et al.MicroRNA408 negatively regulates salt tolerance by affecting secondary cell wall development in maize[J].Plant Physiol,2023,192(2):1569-1583.DOI: 10.1093/plphys/kiad135.
[93]JEENA G S,JOSHI A,SHUKLA R K.Bm-miR172c-5p regulates lignin biosynthesis and secondary xylem thickness by altering the ferulate 5 hydroxylase gene in Bacopa monnieri[J].Plant Cell Physiol,2021,62(5):894-912.DOI: 10.1093/pcp/pcab054.
[94]FAN S G,AMOMBO E,AVOGA S,et al.Salt-responsive bermudagrass microRNAs and insights into light reaction photosynthetic performance[J].Front Plant Sci,2023,14:1141295.DOI: 10.3389/fpls.2023.1141295.
[95]UM T,CHOI J,PARK T,et al.Rice microRNA171f/SCL6 module enhances drought tolerance by regulation of flavonoid biosynthesis genes[J].Plant Direct,2022,6(1):e374.DOI: 10.1002/pld3.374.
(责任编辑 吴祝华)
收稿日期Received:2023-05-16""" 修回日期Accepted:2024-01-10
基金项目:国家自然科学基金项目(31000287)。
第一作者:宋子荷(zihesong_njfu@outlook.com),博士生。*通信作者:甄艳(zhenyongni30@aliyun.com),副教授。
引文格式:宋子荷,甄艳.
植物干旱和盐胁迫响应相关miRNA研究进展[J]. 南京林业大学学报(自然科学版),2024,48(4):1-11.
SONG Z H, ZHEN Y.
Advancements in the research of miRNAs associated with plant" drought and salt stress responses
[J]. Journal of Nanjing Forestry University (Natural Sciences Edition),2024,48(4):1-11.
DOI:10.12302/j.issn.1000-2006.202305016.
