Comamonas kerstersii细菌致病性的基因组分析(英文)
2024-05-29王慧明德松王明席
王慧 明德松 王明席

Genomic Insights of Pathogenicity of Comamonas kerstersii
WANG Hui1,MING Desong2,WANG Mingxi1
( 1. School of Medicine,Huaqiao University,Xiamen 361021,China;2. Quanzhou First Hospital Affiliated to Fujian Medical University,Quanzhou 362000,China )
Abstract:To achieve better management of Comamonas kerstersii(C. kerstersii) infections,the aim was to understand its virulence and pathogenicity by analyzing the virulence factor genes (VFGs) in the genome of C. kerstersii 121606. The genome was sequenced previously, and the VFGs were predicted by BLAST searching through the VFDB database. The results showed that C. kerstersii contained a large number of VFGs,some of which were exemplified to correlate to the survival and growth in humans,and the pathogenicity of C. kerstersii in the gastrointestinal tract and lungs. Genomic analysis revealed that C. kerstersii might be an intracellular pathogen. The harbored VFGs were conducive to explaining its virulence and pathogenicity in gastrointestinal tract and lung infections reported in the literature or encountered in clinic. The data also provided ideal targets for novel antibiotic and vaccine development for the therapy of C. kerstersii infections.
Keywords:Comamonas kerstersii;virulence factor gene;pathogenicity;intracellular pathogen;gastrointestinal tract infection;lung infection
CLC Number:中图分类号:Q 939.48;R 378文献标志码:Document Code:AArticle Number:文章編号:1000-5013(2024)03-0394-23
摘要:为了更好地管理Comamonas kerstersii (C. kerstersii)感染,通过分析C. kerstersii菌株121606基因组中的毒力因子基因(VFGs)来了解其毒力和致病性。 对已经完成测序的C. kerstersii 121606基因组,通过BLAST搜索VFDB数据库来预测其VFGs。结果表明:C. kerstersii含有大量VFGs,其中一些与其在人体内的生存和生长,以及在胃肠道和肺部的致病性有关。基因组分析表明:C. kerstersii可能是一种细胞内病原体。C. kerstersii所携带的VFGs有助于解释其在文献中所报道的胃肠道感染和临床工作中所遇到的肺部感染中的毒力和致病性,这些数据也为新型抗生素和疫苗开发提供了理想的靶点,以用于治疗C. kerstersii感染。
关键词:Comamonas kerstersii;毒力因子基因;致病性;细胞内病原体;胃肠道感染;肺部感染中图分类号:
Comamonas kerstersii (C. kerstersii) is a Gram-negative,aerobic,motile rod bacterium[1],which is found ubiquitously in water,soil,and plants[1]. As summarized in our previously submitted work,we knew that,it primarily causes intra-abdominal infections,such as peritonitis and appendicitis,as well as extra-abdominal infections,including lung infection encountered in our clinical settings. Due to difficulties in identification using conventional phenotypic methods or even 16S rRNA gene sequencing,the actual prevalence of its infections are underestimated. While it is rarely resistant to antibiotics and is closely associated with a favorable clinical prognosis,we have also isolated a pan-drug resistant C. kerstersii strain 121606 from an 82-year-old male patient. Given the potential health threat posed by pan-drug resistance,we have conducted genomic studies to elucidate the mechanisms of pan-drug resistance at the genomic level (submitted). In this article,we aim to further understand the survival ability,virulence,and pathogenicity of C. kerstersii,particularly in the gastrointestinal tract and lungs,by analyzing the virulence factor genes (VFGs) in the genome of the exemplified C. kerstersii 121606 strain. This strain shares nearly identical VFGs with our two other C. kerstersii strains: one (strain 12322-1) isolated from the sputum of a patient with a fatal lung infection,to whom it promoted the patient′s death,and another (strain 202149) isolated from peritoneal pus of a patient with acute perforated appendicitis.
1 Materials and Methods
1.1 Genome Sequencing,Assembly,Annotation for C. kerstersii 121606
These works had been completed as described in previously submitted article. The 16S rRNA gene sequence (1 460 bp,GenBank No. KY014106) and the genome sequences of C. kerstersii strain 121606 (GenBank assembly accession No. GCA_002002445.1,ASM200244v1) had been deposited into GenBank.
1.2 Prediction of VFGs in C. kerstersii 121606 Genome
The VFGs in C. kerstersii 121606 were predicted by conducting a BLAST search against the VFDB protein Set B database[2]in collaboration with Beijing Novogene Bioinformatics Technology Co.,Ltd.. The identity parameter was set at >40%. No stringent cut-off parameters were applied to further filter the initially predicted VFGs because doing so would exclude a large number of potentially unknown VFGs and result in an inability to correlate the remaining VFGs to the infectivity of C. kerstersii in patients.
2 Experimental Results and Analysis
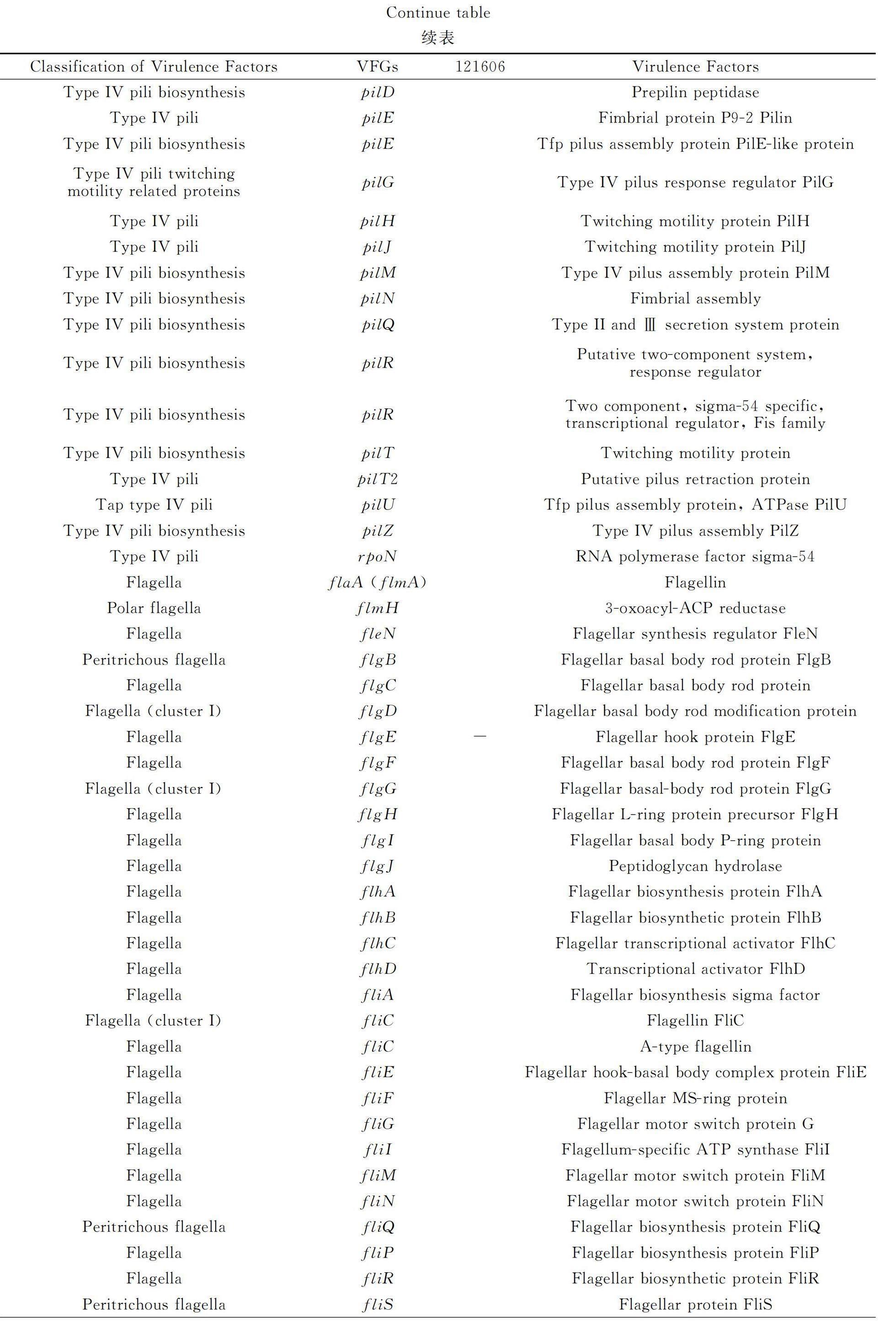
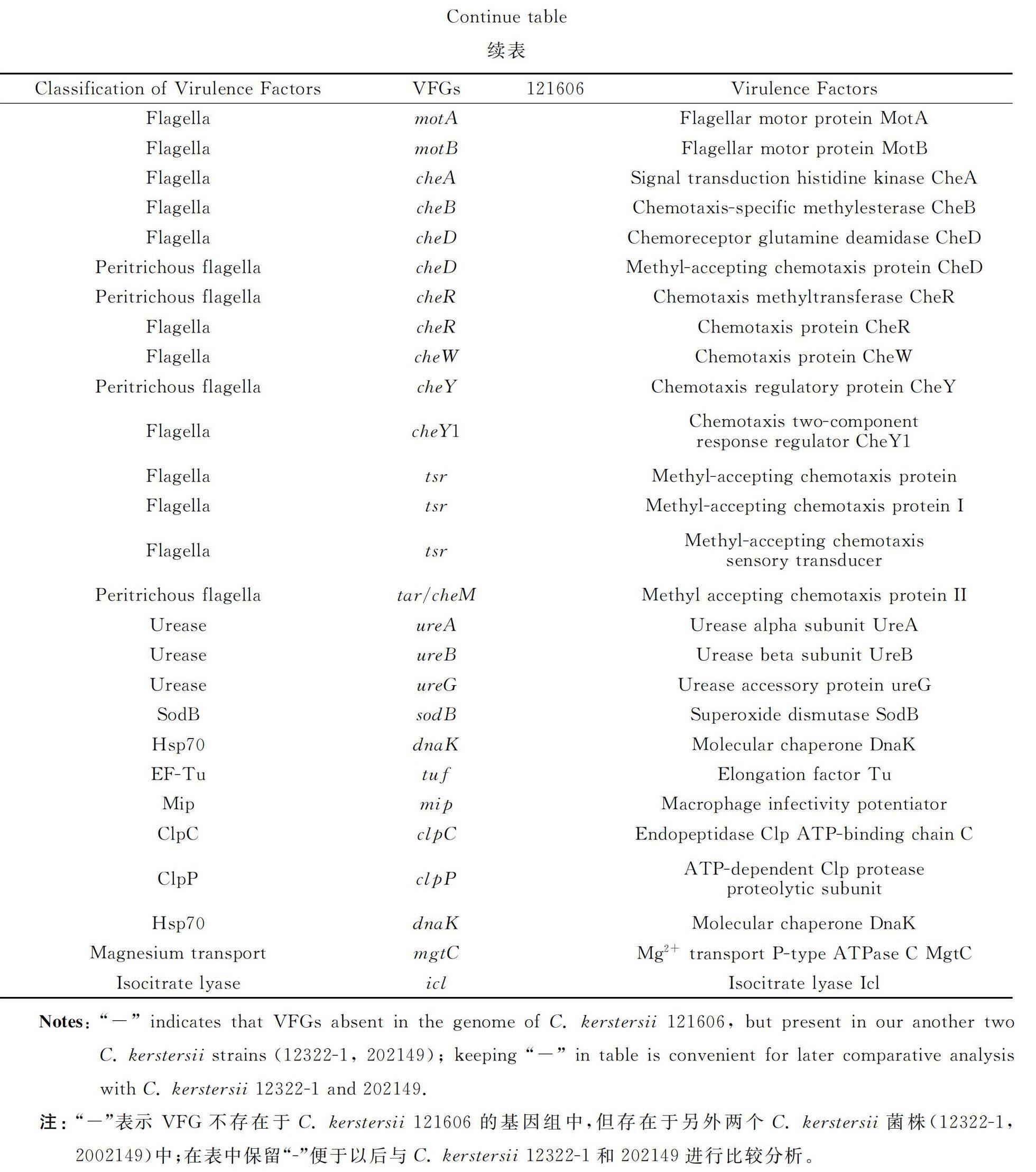
The VFGs inC. kerstersii 121606 were listed in table 1 (discussed) and table 2 (not discussed),such as the genes involved in formation/biosynthesis/regulation of capsular polysaccharide (rmlA,rmlB,rmlC,wbfV/wcvB,wbjD/wecB,wecC,gtaB),LOS (galE,kdsB,kdtA/waaA,lpxA,lpxB,lpxC,orfM),lipopolysaccharide (acpXL,fabZ,gtrB,hisF,kdsA,lpxD,kdtB),motility/attachment apparatuses,fimbriae (cupA1,cupA3,cupD2,cupD4,cupD5),type IV pili (pilA,pilB,pilC,pilD,pilE,pile,pilG,pilH,pilJ,pilM,pilN,pilQ,pilR,pilT,pilT2,pilU,pilZ,rpoN),flagella (motility: flaA/flmA,flmH,fleN,flgB,flgC,flgD,flgE,flgF,flgG,flgH,flgI,flgJ,flhA,flhB,flhC,flhD,fliA,fliC,fliE,fliF,fliG,fliI,fliM,fliN,fliQ,flip,fliR,fliS,motA,motB;chemotaxis: cheA,cheB,cheD,cheR,cheW,cheY,cheY1,tsr,tar/cheM),yersiniabactin siderophore (fyuA,ybtQ,ybtP,irp2,ECVR50_2112),Haemophilus iron transport system HitABC (hitA,hitC),molecular chaperones (groEL,dnaK),hemolysin (hlyⅢ,hlyA),heme uptake system (hutC,hutD,bhuT,hmuV,phuS),HasA-type hemophore-mediated heme uptake system (hasD,hasE),heme biosynthesis pathway (hemB,hemC,hemE,hemH,hemL,hemN),heme utilization (AB57_0992),the ATP-binding cassette (ABC) transporter-FbpABC (fbpC) and other iorn uptake systems (fatD,vctC,irgA,fur).
Several conclusions can be drawn from table 1:
1) C. kerstersii possesses multiple iron acquisition systems vital for the bacterial survival and virulence.
(a) The yersiniabactin siderophore system sequesters irons from transferrin and lactoferrin and delivers them to the cytosol and is an essential virulence factor;the yersiniabactin siderophore system also acts as chelator of Cu2+,Zn2+,Ni2+and plays the corresponding important role in bacterial survival in enviroment and virulence in human;
(b) Haemophilus Iron Transport system functions in assimilation of iron from human transferrin;
(c) FbpABCtransporter functions in the periplasm-to-cytosol transport of iron;
(d) Molecular chaperones are involved in iron uptake;
(e) Hemolysins lyse erythrocyte cells and release heme into blood and act as vital virulence factors;
(f) Heme uptake pathways;
(g) Heme biosynthesis pathway.
2) C. kerstersii possesses many virulence factors crucial for their growth,survival and pathogenicity in gastrointestinal tract.
(a) Type IV pili are essential for bacterial colonization and pathogenicity in gastrointestinal tract;
(b) Flagella are vital for bacterial pathogenicity to gastrointestinal tract;flagella are involved in the motility and chemotaxis functions in intestinal colonization and pathogenicity in bacterial gastroenteritis;flagellar structure,assembly,biosynthesis regulation,gene clustering;constituents and the signal pathway of flagellar chemotaxis;
(c) The virulence factors urease,SodB,DnaK,Elongation factor Tu vital for bacterial growth,survival and pathogenicity in the gastrointestinal tract.
3) The virulence factors Mip,ClpP,ClpC,DnaK,GroEL,MgtC and Icl are vital for intracellular survival and infectivity of C. kerstersii to lung.
3 Discussion
As described above,C. kerstersii mainly causes intra-abdominal infections,and the gastrointestinal tract is its natural habitat for survival and virulence. Since we also isolated a C. kerstersii strain (12322-1) from the sputum of our patient,it is possible that the respiratory tract is also a habitat for its survival and virulence. The correlation of VFGs with bacterial survival,growth in humans,and infectivity of C. kerstersii in the gastrointestinal tract and lungs was exemplified by analyzing the following VFGs.
3.1 C. kerstersii Possesses Multiple Iron Acquisition Systems Vital for Bacterial Survival and Virulence
3.1.1 Iron Sources in Host are Vital for Bacterial Survival
Iron is essential for almost all organisms[3- 4]. In the human body,iron exists as ferric (Fe3+) and ferrous (Fe2+) ions,as well as iron-heme coordinated with protoporphyrin IX[5]. The concentrations of free iron (mostly Fe2+) and insoluble iron (Fe3+,with extremely low solubility of 10-18mol·L-1) are negligible[5-6]. Heme-iron (Fe2+) incorporated into hemoglobin (Hb) in red blood cells is the main form of circulating iron,accounting for approximately 80% of all human iron[5]. The main circulating non-heme iron in the human bloodstream includes transferrin (Fe3+) and lactoferrin (Fe3+)[5]. C. kerstersii has several iron acquisition mechanisms,which may confer higher survival ability and pathogenicity[7].
3.1.2 Yersiniabactin Siderophore System Sequesters Irons From Transferrin and Lactoferrin and Delivers Them to Cytosol and is an Essential Virulence Factor
The Yersiniabactin siderophore is made up of one salicylate,one thiazoline,and two thiazolidine rings[4]. It has an extremely high affinity for ferric (Fe3+) ions (Ka=4×10-36L·mol-1)[8]. Yersiniabactin efficiently removes Fe3+ions from transferrin and lactoferrin[4],then binds to the TonB-dependent outer membrane receptor-Psn in Yersinia pestis[4]and FyuA in Y. enterocolitica[9]to pass through the outer membrane into the periplasm[4]. It subsequently delivers Fe3+ions to the cytosol via the inner membrane ABC transporter,which is composed of two inner membrane fused-function permease/ATP-binding proteins,YbtP and YbtQ[4],followed by the reduction of Fe3+to Fe2+.
The yersiniabactin siderophore promotes the growth of Y. enfevocolitica and Escherichia coli under iron-restricted conditions[9]. It is also a virulence factor for other highly pathogenic bacteria,such as Y. pseudotuberculosis,Y. enterocolitica biotype IB,and extraintestinal pathogenic E. coli (ExPEC)[10]. Additionally,the yersiniabactin siderophore-based iron transport system is essential for the pathogenesis of bubonic and pneumonic plague[4,11],which is a flea-borne zoonosis caused by Y. pestis[12].
Yersiniabactin is synthesized througha nonribosomal peptide/polyketide mechanism involving several proteins,including high-molecular-weight protein 1 (HMWP1),HMWP2,YbtD,YbtT,YbtE,YbtS,and YbtU. Its secretion requires an inner membrane exporter encoded by a four-gene operon (ybtPQXS)[4]. Most of the genes required for yersiniabactin biosynthesis (ybtT,ybtE,ybtS),transport (fyuA,ybtP,ybtQ) and the regulation (ybtA) are located within a Yersinia high-pathogenicity island (HPI)[4,13],This island is also crucial for the virulence of uropathogenic E. coli (UPEC),the main causative agents of non-nosocomial urinary tract infections,particularly in the highly inflammatory infection environment of high-titer mouse cystitis[14].
C. kerstersii possesses the yersiniabactin siderophore system,as it ontains genes encoding for the yersiniabactin siderophore system: (a) fyuA;(b) ybtQ/ybtP,which are overlapping genes,with each encoding half of an inner membrane ABC transporter containing an amino-terminal membrane-spanning permease domain and a carboxy-terminal ATPases domain[15];and (c) irp2,which encodes the putative peptide synthetase-HMWP2 for the siderophore yersiniabactin[16]. Laboratory verification of the presence of these yersiniabactin biosynthesis genes contained in HPI in C. kerstersii genome would help clarify its pathogenicity to patients.
3.1.3 Haemophilus Iron Transport System Functions in Assimilation of Iron From Human Transferrin
C. kerstersii may be able to utilize another classic high-affinity,TonB-independent iron acquisition system-called Hit (Haemophilus Iron Transport) ABC. This system is composed of a periplasmic iron-binding protein (HitA),a cytoplasmic permease (HitB),and a nucleotide-binding protein (HitC) found in Haemophilus influenza[17]. These components could potentially allow C. kerstersii to assimilate iron from human transferrin,as hitA and hitC were predicted in the C. kerstersii 121606 genome.
3.1.4 FbpABC Transporter Functions in Periplasm-to-Cytosol Transport of Iron
Together with hitABC,the prediction of fbpC in the C. kerstersii 121606 genome suggests that it has an additional periplasmic protein-dependent iron ABC transporter,FbpABC. This transporter consists of the ferric binding protein (FbpA),a homologue of HitA,FbpB,a cytoplasmic permease,and FbpC protein,a nucleotide binding protein[18]. It is required for iron uptake from the host ferric binding proteins[19].
Additionally,C. kerstersii 121606 contains: (a) iron compound ABC transporter permease protein gene,fatD;(b) Fe3+ABC transporter ATP-binding protein gene,vctC;(c) iron-negatively regulated virulence gene,irgA,and d) its negative regulator gene,fur[20]. Fur was reported to sense intracellular iron amount and maintain iron homeostasis in Bacillus subtilis[21]. Fur might also be regulator of hitA as it was a regulator of afuA,which encodes a homologue of HitA[22].
3.1.5 Molecular Chaperones are Involved in Iron Uptake
Helicobacterpylori is the major pathogen of gastritis[23]and requires iron for growth[24]. Bacterial GroEL,a homologue of human heat shock protein 60 (Hsp60),is a well-characterized molecular chaperone that prevents aggregation and facilitates proper protein folding in bacteria[25]. It,along with urease (discussed below),are the major surface-exposed proteins of H. pylori[26]. GroEL can assist in iron acquisition for H. pylori through specific binding with iron extracellularly and scavenging iron from heme[27],as well as in the maturation of metalloenzymes by supporting the insertion of a metal cluster into an apoprotein,such as Molybdenum-iron protein[28]. GroEL and another bacterial molecular chaperone,DnaK,the main homologue of human Hsp70 in bacteria[29],were also shown to bind to lactoferrin[30]. The prediction of groEL and dnaK in C. kerstersii 121606 genome indicated that this bacterium had these iron acquisition mechanisms.
3.1.6 Hemolysins Lyse Erythrocyte Cells and Release Heme Into Blood and Act as Vital Virulence Factors
C. kerstersii 121606 possesses the genes (hlyⅢ,hlyA) that encode hemolysins capable of lysing erythrocyte cells,releasing Hb into the blood serum,and converting it into oxidized Hb (metHb),in which ferric heme is loosely bound and can be utilized by bacterial heme uptake systems[5]. It has been reported that haemolysin Ⅲ (encoded by hlyⅢ) from B. cereus can form 3–3.5 nm transmembrane pores in diameter on the erythrocyte membrane and lyse them[31]. HlyA (110 ku,listeriolysin O)[32]can also lyse erythrocytes[33]and a wide spectrum of cells,such as granulocytes,monocytes,and endothelial cells[33]. When produced by the intramacrophage Listeria monocytogenes,a food-borne pathogen causing listeriosis with a mortality rate of approximately 20%[32],HlyA can break the endosome membrane and help this bacterium escape the hostile endosome environment[34]. HlyA is verified as a unique and essential virulence factor for virulent L. monocytogenes[32]and plays a vital role in E. coli-caused extraintestinal diseases[35].
3.1.7 Heme Uptake Pathways
These findings of the genes hutC,hutD,bhuT,hmuV in C. kerstersii 121606 indicated that C. kerstersii had multiple heme uptake systems.
Gram-negative bacteria are found to have two heme uptake mechanisms[5]to transport of host heme,heme proteins (Hb,hemopexin) into cytosol,one is through the heme scavenging proteins-hemophores,another is by binding outer membrane receptors,then heme is transported across the bacterial outer membrane via a TonB-dependent outer membrane receptor into the periplasm,then transferred to the periplasmic heme binding protein (PBP),which transmits heme to the inner membrane ABC transporter complex,finaly translocated into the cytoplasm[5,36-37].
The prediction of the genes,hutC,encoding a putative inner membrane permease,and hutD,encoding a putative ABC-transporter ATPase,which form the heme/Hb ABC transporter-hutCD[36-38]in C. kerstersii 121606,and bhuT,encoding the periplasmic heme-binding protein which functions as HutB[37],indicated that they might own the TonB dependent heme uptake mechanism[36],but whether another component-the outer membrane heme receptor encoding gene hutA is in C. kerstersii strains requires experimental evidence.
The presence of hmuV in C. kerstersii 121606,which encode a ATPase,suggested the possible presence of another heme ABC transport system-HmuTUV,if the genes encoding other two components-the periplasmic binding protein (hmuT),and the permease (hmuU) are verified to be present in C. kerstersii by experiment[39].
C. kerstersii may also possess HasA-type hemophore-mediated heme uptake system,a two component of a ABC exporter-HasDEF,which exports a heme acquisition protein,HasA[40],as C. kerstersii 121606 contained the genes hasD,encoding ABC transporter ATP-binding protein,and hasE,encoding ABC transporter periplasmic substrate-binding protein[40].
3.1.8 Heme Biosynthesis Pathway
In addition to heme uptake,C. kerstersii may also have the ability to biosynthesize heme. The heme biosynthetic pathway starts from GltX-catalyzed conversion of L-glutamate to charged glutamyl-tRNAGlu,which acts as substrate of glutamyl-tRNA reductase (HemA),the latter works sequentially with glutamate-1-semialdehyde aminotransferase (HemL) to synthesize 5-aminolevulinic acid (ALA). Subsequently,ALA is used to synthesize uroporphyrinogen Ⅲ by porphobilinogen synthase (HemB),then heme by uroporphyrinogen decarboxylase (HemE) and protoporphyrin ferrochelatase (HemH)[41]. Prediction of the genes endcoding HemB,HemC (porphobilinogen deaminase),HemE,HemH (ferrochelatase),HemL and HemN (coproporphyrinogen Ⅲ oxidase) in C. kerstersii 121606 suggests that it possesses this heme biosynthesis pathway[42].
3.1.9 Iron Acquisition Proteins are Potential Drug and Vaccine Targets
The iron acquisition proteins play an essential role in the pathogenesis of causative bacteria. They have been identified as potentially novel antibiotic and vaccine targets to combat infections. For example,FyuA[43],HitA[44],GroEL and DnaK[45]are the potential vaccine candidates against bacteria.
3.2 Yersiniabactin Siderophore System Also Acts as Chelator of Cu2+,Zn2+,Ni2+and Plays Corresponding Important Role in Bacterial Survival in Enviroment and Virulence in Human
Yersiniabactin is also a chelator of Cu2+ [46],Zn2+[16]. Copper serves as both a nutrient and a toxin during bacterial infections. Yersiniabactin efficiently scavenges extracellular Cu2+in the form of a Cu2+-Yersiniabactin complex. It then uses the same Fe3+-Yersiniabactin transport proteins to facilitate copper import and release in UPEC. This supports copper-dependent enzyme activity in low copper conditions,such as cuproenzymes[47],and helps UPEC resist copper toxicity in higher copper conditions[47]by preventing Cu2+reduction to the more bactericidal Cu+[46]. Additionally,it protects UPEC against phagocytic killing by catalyzing superoxide dismutation intraphagocytically with the superoxide dismutase (SOD)-like activity of the Cu2+-Yersiniabactin complex[46].
Zn2+is a necessary micronutrient during bacterial infections. The yersiniabactin siderophore also plays a crucial role in acquiring Zn2+and causing lethal pathogenesis in mouse pneumonic plague[16]. In addition to Fe3+,Cu2+,and Zn2+,yersiniabactin also has a high affinity forchelating Ni2+,Cr3+,Co3+,and Ga3+ [14],as well as Co2+and Pd2+[48]. This promotes the survival of bacteria in heavy metal-polluted environments.
3.3 C. kerstersii Possesses Many Virulence Factors Crucial for Their Growth,Survival and Pathogenicity in Gastrointestinal Tract
3.3.1 Type IV Pili are Essential for Bacterial Colonization and Pathogenicity in Gastrointestinal Tract
As proteinaceous hair-like appendages,pili are essential for bacterial adherence to human gut cells[49]. Type IV pili (T4P),which are approximately 5-8 nm in diameter and several micrometers in length,have three subtypes: Type IVa pili (T4aP),Type IVb pili (T4bP),and Type IVc pili (T4cP)[50]. They are predicted to be produced by about 30% of the known gut microbiome[49]. T4P promotes the intestinal colonization of enterohemorrhagic E. coli (EHEC) O157:H7,a food-borne pathogen of hemorrhagic colitis[51],enterotoxigenic E. coli (ETEC),the most common bacterial pathogen of diarrhea in developing countries and travelers diarrhea[52],and enteropathogenic E. coli (EPEC)[53]. It also promotes the survival,virulence,and transmission of EHEC O157:H7[51],invasion of epithelial cells,hemagglutination of erythrocytes,biofilm formation,and twitching motility of EHEC O157:H7[51].
The biogenesis process of Type IV pili in Gram-negative bacteria begins with the attachment of an ATPase-PilB to the cytoplasmic ring formed by PilM and the platform protein PilC[49]. PilM,along with the subunits PilN,PilO,and PilP,forms an inner membrane alignment complex called PilMNOP. This complex has a cage-like ring in the inner membrane and periplasm,which is formed by conformational changes in PilN and PilO resulting from the attachment of PilB to PilM. Then,the platform protein PilC incorporates the major pilin subunit PilA after the removal of the leader peptide by the prepilin peptidase PilD. PilD also methylates PilA or minor pilins,such as pilE,into the growing pilus based on the PilMNOP alignment complex. Afterward,PilA is translocated through the outer membrane pore formed by PilQ to elongate the pilus[49]. The cytoplasmic ATPase PilB drives pilin assembly,while the PilT ATPase drives disassembly or retraction. Both of them can reversibly associate with the PilC protein[49]. In the final step,the minor pilins are synthesized in a similar way to PilA and create a priming complex for pilin formation at the pilus tip and along the pilus[49].
The presence of the Type IV pili biogenesis genes encoding PilA,PilB[54],PilC,PilD,PilE,PilJ {a minor pilin and a methyl-accepting chemotaxis protein (Mcp)-like chemosensor,which directly interacts with the major pilin subunit PilA and regulates surface-induced gene expression and pathogenicity[54]},PilG {a polytopic membrane protein dedicated to assist transformation of bacteria by binding DNA and interacting with the N-terminal region of PilQ,which can also bind DNA[55]and essential for twitching motility)[56]},PilH {an ABC transporter homologue required for the biogenesis of any bacterial pilus type[57]},PilM,PilN,PilQ {a secretin working in concert with PilG to bind DNA during T4P-mediated transformation[55]},PilR {the response regulator of a two (PilS and PilR)-component transcriptional regulatory system dedicated to activate transcription of pilA[58]},PilT,PilT2,PilU {also a retraction ATPase motor[54]},PilZ {which can stimulate T4P formation or regulate T4P-dependent motility)[59]} in C. kerstersii 121606 indicated that C. kerstersii possessed the virulence factor T4aP possibly invovled in the pathogenicity of C. kerstersii in intesintal diseases[49],such as peritonitis,appendicitis,as mentioned above.
3.3.2 Flagella are Vital for Bacterial Pathogenicity to Gastrointestinal Tract
3.3.2.1 Flagella are Ivolved in Motility and Chemotaxis Functions in Intestinal Colonization and Pathogenicity in Bacterial Gastroenteritis
Flagella are approximately 20 nm in diameter[60]. The possession of rapid,darting motility in viscous milieus conferred by the polar flagella is an exclusive characteristic of virulent Campylobacter jejuni and E. coli[61],the major pathogen of acute human bacterial gastroenteritis worldwide[62- 63],and are required for intestinal mucus colonization[61],a key pathogenicity determinant in intestinal infection by C. jejuni[64-65]and H. pylori[66]. Additionally,they function in secreting virulence proteins,microcolony formation,biofilm formation and escape of the innate immune response[61]. Chemotaxis conferred by polar flagella is also critical for colonization and pathogenicity in the establishment of gastrointestinal infections by C. jejuni[61- 62]and H. pylori[66].
3.3.2.2 Flagellar Structure,Assembly,Biosynthesis Regulation,Gene clustering
Flagella are composed of approximately 20 000-30 000 protein subunits,consisting of over 20 different types of proteins[67]. They comprise: (a) the base located in the cytoplasm and inner membrane,which is composed of the flagellar type Ⅲ secretion system (T3SS),a membrane-embedded export gate composed of six transmembrane proteins-FlhA,FlhB,FliO,FliP,FliQ,and FliR within the central membrane patch of the ring,the water-soluble ATPase ring complex composed of three cytoplasmic proteins,FliH,FliI,and FliJ[68-71],the inner membrane MS ring (a homomultimer of flagellar motor switch protein FliF)[68,72],the cytoplasmic C ring or switch complex {which is formed by two motor/switch proteins FliM and FliN[68,73]and functions as the flagellar switch and also aids in secretion[66]},and the motor {formed by multimers of the motor/switch protein FliG[68,72]} attached to MS ring[68];(b) the coaxial periplasmic rod {a helical cylinder cotaining four flagellin proteins-FliE,FlgB,FlgC,and FlgF in the proximal part and FlgG in the distal part[66-67]} and the associated ring structures;(c) the surface-localized hook structure (a helical assembly of about 120 FlgE subunits) spanning the periplasm and outer membrane;and (d) the extracellular filament {composed primarily by the major flagellin,FlaA,with the minor flagellin,FlaB[61,66]or FliC[67]}. The rod is surrounded by: (a) three disk structures,consisting of the basal (composed of FlgP),the medial paralyzed flagellum protein A (PflA),and the proximal disk (composed of PflB);(b) the motor components of the base including MotA and MotB (encoded by motA,motB),which forms a proton channel[74];(c) the stators of flagellum in periplasm that interact with FliG and function to link proton flow and generation of flagellar rotation torque[66,68,72];(d) the periplasmic P ring (FlgI) in peptidoglycan;and (e) the L ring (FlgH) in outer membrane[66,68].
Flagellar assembly begins at the base,then progresses to the hook,and finally to the filament[70,75]. The peptidoglycan-hydrolyzing activity of the FlgJ protein (peptidoglycan hydrolase) is necessary for flagellar rod formation[76]. To assemble the hook and filament from the cell surface,their component proteins are transported from the cytoplasm to the distal end of the growing structure by T3SS[66]. Chaperones FlgN,FliS,and FliT assist in the transfer of flagellin subunits to T3SS from the cytoplasm[69]. The initial entry of the flagellin subunits into the narrow pore of the export gate is facilitated by the ATPase ring complex and powered by proton motive force,which is controlled via the specific interaction of FlhA with FliJ[69]. The scaffolding protein FlgD is also essential for flagellar hook assembly in Salmonella typhimurium[77]. Its export requires FliE,a flagellar basal body protein,and an adaptor protein located between the MS ring and rod substructures[68,78]. The flagellar number is controlled by the antiactivator FleN through binding to the enhancer of the flagellar gene,fleQ,a multidomain sigma-54 (σ54) factor[79].
Flagellar biogenesis is tightly controlled by the activities of σ54and σ28[66]. For instance,the two-component signal transduction system,FlgSR,is composed of the FlgS sensor kinase and the FlgR response regulator. This system recognizes a regulatory checkpoint in the MS ring and rotor to activate the σ54-dependent expression of flagellar rod and hook genes,and ultimately the σ28-dependent flagellins and fed gene expression[72].
In the genome,a number of flagellar genes are structurally organized as operons,with a master and several secondary levels of gene expression regulation[80]. Examples include the flgGHIJKL operon[81],the flgBCDEF operon,whose genes are transcribed as a single mRNA in a σ54-dependent way in Rhodobacter sphaeroides[82],the flhBAE operon in Y. enterocolitica[83],the flmA(flaA)B operon,and the flmGH operon in Caulobacter crescentus[84]. Another example of a master regulator in flagellum biogenesis is the flhD operon,which encodes two genes,flhD and flhC in E. coli. This operon produces the FlhD/FlhC complex,which is a transcriptional activator required for the transcription of the three class II operons-fliA,flhB,and fliL[85]. Additionally,as a subunit of RNA polymerase and a homologue of σ28,the FliA protein activates flagellin synthesis by transcribing the fliC gene[86].
The prediction of the flagellar components and biogenesis genes in C. kerstersii 121606 indicates that C. kerstersii possesses flagella with intact motility function.
3.3.2.3 Constituents and Signal Pathway of Flagellar Chemotaxis
The signal transduction proteins of chemotaxis include the core components. These consist of chemoreceptors,which sense environmental stimuli and transduce this signal to mediate rotation of flagella. The two-component system is composed of the histidine kinase CheA and its cognate response regulator CheY[87]. The phosphorylated form of CheY,regulated by ligand binding to the chemoreceptors,interacts with the FliM component of the flagellar motor switch to cause clockwise motor rotation[88]. The CheW coupling protein physically conjugates chemoreceptors to CheA[66]. Other components include the phosphatases CheZ and FliY,which promote CheY dephosphorylation,the chemoreceptor modification enzymes CheR (a methyltransferase that transfers methyl groups from S-adenosylmethionine to glutamate residues on the cytoplasmic domains of the chemoreceptors Mcps),and CheB (a methylesterase),that governs adaptation responses to sustained ligand levels by methylating or demethylating glutamyl residues of chemoreceptor respecitively[66]. The chemoreceptor glutamine deamidase CheD deamidates the Mcps essential for chemoreceptors to effectively transduce signals to the CheA kinase in B. subtilis[87]and increases the receptor kinase activity or enhances CheC phosphatase activity to regulate the CheY levels in B. subtilis[89]. Some bacteria have multiple homologues of bacterial chemotaxis and chemosensing genes,such as tsr,tar/cheM,and have a great ability to respond to a wide variety of compounds,like Chromobacterium violaceum[74],an extremely virulent opportunistic pathogen ubiquitously distributed in water and soil[90].
The prediction of cheA,cheB,cheD,cheR,cheW,cheY,cheY1,tsr and tar/cheM in our C. kerstersii 121606 suggested that their flagella have full chemotaxis function.
As a whole,the prediction of almost all of the flagellar components,biogenesis,motility,and chemotaxis genes in C. kerstersi 121606 indicates that they have fully functional flagella,which contributes to their high pathogenicity in the gastrointestinal tract.
3.3.3 Virulence Factors Urease,SodB,DnaK,Elongation Factor Tu Vital for Bacterial Growth,Survival and Pathogenicity in Gastrointestinal Tract
Survival,colonization and pathogenicity of H. pylori in gastric acidic environment depends on the abundantly produced nickel-dependent urease,which hydrolyses urea into ammonia to create a pH neutral microenvironment[26]and block the phagosome-lysosome fusion to resist against the host immune system[91]. Most bacterial ureases are made of a (αβγ)3(encoded by ureA,ureB,ureC) trimer[92]. Urease also requires different sets of accessory proteins,such as UreE,UreF,UreG,UreH and UreI[93]or UreD,UreE,UreF and UreG[94-95],to coordinate nickel ions into the UreABC subunits. Considering that the urease encoding genes predicted through VFDB database might be not complete,we manually searched the annotated genome of C. kerstersii 121606,and found that it contained the genes encoding the subunit α,β,γ,and the accessory proteins UreD,UreE,UreF,UreG,meaning that C. kerstersii could express functional urease.
The survival,colonization,and pathogenicity of H. pylori in the gastric acidic environment depend on the abundantly produced nickel-dependent urease. This enzyme hydrolyzes urea into ammonia to create a pH-neutral microenvironment[26]and blocks the phagosome-lysosome fusion to resist the host immune system[91]. Most bacterial ureases are composed of a (αβγ)3trimer (encoded by ureA,ureB,ureC)[92]. Urease also requires different sets of accessory proteins,such as UreE,UreF,UreG,UreH,and UreI[93],or UreD,UreE,UreF,and UreG[94-95],to coordinate nickel ions into the UreABC subunits. Considering that the urease-encoding genes predicted through the VFDB database might not be complete,we manually searched the annotated genome of C. kerstersii 121606 and found that it contained the genes encoding the subunits α,β,γ,and the accessory proteins UreD,UreE,UreF,UreG,indicating that C. kerstersii could express functional urease.
SodB,an iron-cofactored superoxide dismutase,is necessary for H. pylori to combat the oxidative stress produced by neutrophils and macrophages[96]while colonizing the host gastric mucosa[97].
DnaK is found on the cell surface ofseveral pathogens,such as human probiotic intestinal microbiota Lactobacillus salivarius,and Bifidobacterium animalis subsp. lactis[98]. It can be upregulated by bile salts in the gastrointestinal tract of mammals to enhance the adaptation and colonization of B. animalis subsp. lactis in the gut bile environment by acting as a high affinity receptor of host plasminogen[98].
The elongation factor thermal unstable Tu (EF-Tu,encoded by tuf) is a translational GTPase (G protein),that delivers aminoacyl-tRNAs (aa-tRNAs) to the A-site of the ribosome[99]and has additional adhesive moonlighting functions[100]. EF-Tu on bacterial cell surfaces can bind to membrane receptors and fibronectin,a key component of the extracellular matrix (ECM) proteins on the surface of host cells[100]. It can also bind to human intestinal cells and mucins,indicating the important role of EF-Tu in gut colonization for the probiotic bacterium,L. johnsonii[101]. The secreted EF-Tu from H. pylori also promotes adherence and invasion to host cells during pathogenesis[102].
The above analyses collectively indicate the potential of survival and pathogenicity of C. kerstersi in the gastrointestinal tract. These data help to explain the fact that nearly all reported C. kerstersi infections are gut diseases.
3.4 Virulence Factors Mip,ClpP,ClpC,DnaK,GroEL,MgtC and Icl are Vital for Intracellular Survival and Infectivity of C. kerstersii to Lung
Legionella pneumophilais a Gram-negative,facultative intracellular bacterium[103]and the causative agent of Legionnaires disease,which presents as pneumonia[104]. It is able to survive and multiply in lung macrophages[103]and Type I and II pneumocytes[105]. The Legionella Mip (macrophage infectivity potentiator) protein,a homodimeric lipoprotein of 24 ku located on the bacterial surface and on host membranes[106-107],is necessary for the early intracellular infection step of L. pneumophila to alveolar macrophages[108]and the human phagocytic cell line U937[109],intracellular multiplication[110],and the full virulence of L. pneumophila in the lungs of guinea pigs and the spread to the spleen after intratracheal inoculation[108]. It also facilitates L. pneumophila to pass through the collagen IV-containing ECMs or lung epithelial cell lines,and disseminate within the lung tissue and spread to the spleen[111].
ClpP is a 21.6 ku serine-type protease in E. coli that is highly conserved in prokaryotes and eukaryotes[112]. It plays a similar role in bacterial pathogenicity in the lungs. ClpP is necessary for the colonization of Streptococcus pneumoniae in the nasopharynx,survival in the lungs of mice after intranasal challenge,and intracellular survival in murine macrophages[113]. ClpP must bind with the Hsp100/Clp family of molecular chaperones,ClpA,or ClpX,or ClpC,to form the Clp protease ClpAP,ClpXP,or ClpCP complex in order to degrade aggregated protein[112]. Therefore,it is understandable that ClpC ATPase is also found to promote the intracellular survival of the facultative intracellular pathogen L. monocytogenes in macrophages and survival in host tissues[114].
Similar to the ClpC ATPase,two other chaperones,DnaK and GroEL,play important roles in the survival and pathogenicity of bacteria. The stress-induced DnaK homologue-Hsp70 is involved in protein folding,proteostasis control[29],and removal of aggregated proteins[115]. It has been verified to be essential for intramacrophage survival of the intracellular pathogen S. enterica serovar Typhimurium,invasion of epithelial cells,leading to systemic infection[116],and intramacrophagic replication of Brucella suis[117].
MgtC is also a vital virulence factor for several intracellular pathogens,such as Burkholderia cenocepacia[118],M. abscessus[119],both of them being important respiratory tract pathogens in patients with cystic fibrosis,Mycobacterium tuberculosis[120],S. enterica serovar Typhimurium[121],and Bordetella pertussis,the causative agent of whooping cough[122]. It also promote bacterial survival of B. cenocepacia[118],M. abscessus[119],and M. tuberculosis[120]within macrophages through postponing the phagolysosomal fusion of bacteria[118]. These functions of mgtC are also observed in P. aeruginosa,a highly lethal extracellular pathogen in cystic fibrosis patients with persistent pulmonary infections[123],during the intramacrophage phase or when there is a limitation of Mg2+,impairing host phagocytic functions[124].
Isocitrate lyase (Icl),an enzyme indispensible for the metabolism of fatty acids,has been to promote the persistence of M. tuberculosis in activated macrophages and its virulence in mice[125]. It is also indispensable for the pathogenesis of Pseudomonas aeruginosa in the rat lung infection model[126].
These predicted VFGs in C. kerstersii 121606 indicate that C. kerstersii might be an intracellular pathogen,especially forr lung macrophages and Type I and II pneumocytes. This helps to explain the infectivety of C. kerstersii in our patient′s lung.
In summary,these exemplified VFGs helped us understand the ability of intracellular survival,growth in humans,and the infectivity of C. kerstersii in the gastrointestinal tract and lungs of the reported cases and our patients. Some of them might act as novel drug and vaccine targets.
4 Conclusion
C. kerstersii possessed a wide range of VFGs to demonstrate its status as an intracellular pathogen and to elucidate its pathogenicity in the reported gastrointestinal tract infection and a case of lung infection encountered in our clinical work. These findings also offer potential targets for antibiotic innovation and vaccine development to address the rising incidence of C. kerstersii infections.
Acknowledgment:We express gratitude towards the Beijing Novogene Bioinformatics Technology Co.,Ltd.,for providing technical support.
Ethics Statement:This article does not contain any studies with humans or animals performed by any of the authors. This work was approved by the Ethics Committee of Quanzhou First Hospital.
References:
[1]WAUTERS G,DE BAERE T,WILLEMS A,et al.Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena[J].Int J Syst Evol Microbiol,2003,53(3):859-62.DOI:10.1099/ijs.0.02450-0
[2]CHEN Lihong,XIONG Zhaohui,SUN Lilian,et al.VFDB 2012 update:toward the genetic diversity and molecular evolution of bacterial virulence factors[J].Nucleic Acids Res,2012,40(D1):D641-D645.DOI:10.1093/nar/gkr989.
[3]MILLER M C,PARKIN S,FETHERSTON J D,et al.Crystal structure of ferric-yersiniabactin,a virulence factor of Yersinia pestis[J].J Inorg Biochem,2006,100(9):1495-500.DOI:10.1016/j.jinorgbio.2006.04.007.
[4]FETHERSTON J D,KIRILLINA O,BOBROV A G,et al.The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague[J].Infect Immun,2010,78(5):2045-2052.DOI:10.1128/IAI.01236-09.
[5]CHAO A,SIEMINSKI P J,OWENS C P,et al.Iron acquisition in Mycobacterium tuberculosis[J].Chem Rev,2019,119(2):1193-1220.DOI:10.1021/acs.chemrev.8b00285.
[6]BUTT A T,THOMAS M S.Iron acquisition mechanisms and their role in the virulence of Burkholderia species[J].Front Cell Infect Microbiol,2017,7:460(1-21).DOI:10.3389/fcimb.2017.00460.
[7]KOCZURA R,KAZNOWSKI A.The Yersinia high-pathogenicity island and iron-uptake systems in clinical isolates of Escherichia coli[J].J Med Microbiol,2003,52(8):637-642.DOI:10.1099/jmm.0.05219-0.
[8]PERRY R D,BALBO P B,JONES H A,et al.Yersiniabactin from Yersinia pestis: Biochemical characterization of the siderophore and its role in iron transport and regulation[J],Microbiology (Reading),1999,145 (5):1181-1190.DOI:10.1099/13500872-145-5-1181.
[9]HAAG H,HANTKE K,DRECHSEL H,et al.Purification of yersiniabactin: A siderophore and possible virulence factor of Yersinia enterocolitica[J].J Gen Microbiol,1993,139(9):2159-2165.DOI:10.1099/00221287-139-9-2159.
[10]SCHUBERT S,PICARD B,GOURIOU S,et al.Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections[J].Infect Immun,2002,70(9):5335-5337.DOI:10.1128/IAI.70.9.5335-5337.2002.
[11]SEBBANE F,JARRETT C,GARDNER D,et al.Role of the Yersinia pestis yersiniabactin iron acquisition system in the incidence of flea-borne plague[J].PLoS One,2010,5(12):e14379.DOI:10.1371/journal.pone.0014379.
[12]PERRY R D,BOBROV A G,FETHERSTON J D.The role of transition metal transporters for iron,zinc,manganese,and copper in the pathogenesis of Yersinia pestis[J].Metallomics,2015,7(6):965-978.DOI:10.1039/c4mt00332b.
[13]CARNIEL E.The Yersinia high-pathogenicity island[J].Int Microbiol,1999,2(3):161-167.
[14]KOH E I,HUNG C S,HENDERSON J P.The yersiniabactin-associated ATP binding cassette proteins YbtP and YbtQ enhance Escherichia coli fitness during high-titer cystitis[J].Infect Immun,2016,84(5):1312-1319.DOI:10.1128/IAI.01211-15.
[15]FETHERSTON J D,BERTOLINO V J,PERRY R D.YbtP and YbtQ: Two ABC transporters required for iron uptake in Yersinia pestis[J].Mol Microbiol,1999,32(2):289-299.DOI:10.1046/j.1365-2958.1999.01348.x.
[16]BOBROV A G,KIRILLINA O,FETHERSTON J D,et al.The Yersinia pestis siderophore,yersiniabactin,and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice[J].Mol Microbiol,2014,93(4):759-775.DOI:10.1111/mmi.12693.
[17]ADHIKARI P,KIRBY S D,NOWALK A J,et al.Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon[J].J Biol Chem,1995,270(42):25142-25149.DOI:10.1074/jbc.270.42.25142.
[18]FERREIR?S C,CRIADO M T,G?MEZ J A.The neisserial 37 kDa ferric binding protein (FbpA)[J].Comp Biochem Physiol B Biochem Mol Biol,1999,123(1):1-7.DOI:10.1016/s0305-0491(99)00044-9.
[19]LAU G H,MACGILLIVRAY R T,MURPHY M E.Characterization of a nucleotide-binding domain associated with neisserial iron transport[J].J Bacteriol,2004,186(10):3266-3269.DOI:10.1128/JB.186.10.3266-3269.2004.
[20]GOLDBERG M B,BOYKO S A,CALDERWOOD S B.Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae[J].Proc Natl Acad Sci USA,1991,88(4):1125-1129.DOI:10.1073/pnas.88.4.1125.
[21]PI Hualiang,HELMANN J D.Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis[J].Proc Natl Acad Sci USA,2017,114(48):12785-12790.DOI:10.1073/pnas.1713008114.
[22]WILLEMSEN P T,VULTO I,BOXEM M,et al.Characterization of a periplasmic protein involved in iron utilization of Actinobacillus actinomycetemcomitans[J].J Bacteriol,1997,179(15):4949-4952.DOI:10.1128/jb.179.15.4949-4952.1997.
[23]GENTA R M. Helicobacter pylori,mucosal damage,and apoptosis:pathogenesis and definition of gastric atrophy[J].Gastroenterology,1997,113(Suppl l):S51-S55.DOI:10.1016/s0016-5085(97)80012-1.
[24]SENKOVICH O,CEASER S,MCGEE D J,et al.Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media[J].Infect Immun,2010,78(5):1841-1849.DOI:10.1128/IAI.01258-09.
[25]HAYER-HARTL M,BRACHER A,HARTL F U.The GroEL-GroES Chaperonin machine: A nano-cage for protein folding[J].Trends Biochem Sci,2016,41(1):62-76.DOI:10.1016/j.tibs.2015.07.009.
[26]DUNN B E,PHADNIS S H.Structure,function and localization of Helicobacter pyloriurease[J]. Yale J Biol Med,1998,71(2):63-73.
[27]GONZ?LEZ-L?PEZ M A,VEL?ZQUEZ-GUADARRAMA N,REMERO-ESPEJEL M E,et al.Helicobacter pylori secretes the chaperonin GroEL (HSP60),which binds iron[J].FEBS Lett,2013,587(12):1823-1828.DOI:10.1016/j.febslet.2013.04.048.
[28]RIBBE M W,BURGESS B K.The chaperone GroEL is required for the final assembly of the molybdenum-iron protein of nitrogenase[J].Proc Natl Acad Sci USA,2001,98(10):5521-5525.DOI:10.1073/pnas.101119498.
[29]HARTL F U,BRACHER A,HAYER-HARTL M.Molecular chaperones in protein folding and proteostasis[J].Nature,2011,475(7356):324-332.DOI:10.1038/nature10317.
[30]HAGEMANN L,GR?NDEL A,JACOBS E,et al.The surface-displayed chaperones GroEL and DnaK of Mycoplasma pneumoniae interact with human plasminogen and components of the extracellular matrix[J].Pathog Dis,2017,75(3):1-12.DOI:10.1093/femspd/ftx017.
[31]BAIDA G E,KUZMIN N P.Mechanism of action of hemolysin Ⅲ from Bacillus cereus[J].Biochim Biophys Acta,1996,1284(2):122-124.DOI:10.1016/s0005-2736(96)00168-x.
[32]CHURCHILL R L,LEE H,HALL J C.Detection of Listeria monocytogenes and the toxin listeriolysin O in food[J].J Microbiol Methods,2006,64(2):141-170.DOI:10.1016/j.mimet.2005.10.007.
[33]STANLEY P,KORONAKIS V,HUGHES C.Acylation of Escherichia coli hemolysin: A unique protein lipidation mechanism underlying toxin function[J].Microbiol Mol Biol Rev,1998,62(2):309-333.DOI:10.1128/MMBR.62.2.309-333.1998.
[34]HILTBOLD E M,ZIEGLER H K.Mechanisms of processing and presentation of the antigens of Listeria monocytogenes[J].Infect Agents Dis,1993,2(5):314-323.
[35]MENESTRINA G,MOSER C,PELLET S,et al.Pore-formation by Escherichia coli hemolysin (HlyA) and other members of the RTX toxins family[J].Toxicology,1994,87(1/2/3):249-267.DOI:10.1016/0300-483x(94)90254-2.
[36]R?O S J,OSORIO C R,LEMOS M L.Heme uptake genes in human and fish isolates of Photobacterium damselae: Existence of hutA pseudogenes[J].Arch Microbiol,2005,183(5):347-358.DOI:10.1007/s00203-005-0779-4.
[37]NAOE Y,NAKAMURA N,DOI A,et al.Crystal structure of bacterial haem importer complex in the inward-facing conformation[J].Nat Commun,2016,7:13411.DOI:10.1038/ncomms13411.
[38]OSORIO C R,JUIZ-RO ?S,LEMOS M L.The ABC-transporter hutCD genes of Photobacterium damselae subsp. piscicida are essential for haem utilization as iron source and are expressed during infection in fish[J].J Fish Dis,2010,33(8):649-655.DOI:10.1111/j.1365-2761.2010.01169.x.
[39]CU?V P O,KEOGH D,CLARKE P,et al.The hmuUV genes of Sinorhizobium meliloti 2011 encode the permease and ATPase components of an ABC transport system for the utilization of both haem and the hydroxamate siderophores,ferrichrome and ferrioxamine B[J].Mol Microbiol,2008,70(5):1261-1273.DOI:10.1111/j.1365-2958.2008.06479.x.
[40]IDEI A,KAWAI E,AKATSUKA H,et al.Cloning and characterization of the Pseudomonas fluorescens ATP-binding cassette exporter,HasDEF,for the heme acquisition protein HasA[J].J Bacteriol,1999,181(24):7545-7551.DOI:10.1128/JB.181.24.7545-7551.1999.
[41]WANG Liying,BROWN L,ELLIOTT M,et al.Regulation of heme biosynthesis in Salmonella typhimurium: Activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein[J].J Bacteriol,1997,179(9):2907-2914.DOI:10.1128/jb.179.9.2907-2914.1997.
[42]DAILEY H A,DAILEY T A,GERDES S,et al.Prokaryotic heme biosynthesis: Multiple pathways to a common essential product[J].Microbiol Mol Biol Rev,2017,81(1):e00048-16.DOI:10.1128/MMBR.00048-16.
[43]BRUMBAUGH A R,SMITH S N,SUBASHCHANDRABOSE S,et al.Blocking yersiniabactin import attenuates extraintestinal pathogenic Escherichia coli in cystitis and pyelonephritis and represents a novel target to prevent urinary tract infection[J].Infect Immun,2015,83(4):1443-1450.DOI:10.1128/IAI.02904-14.
[44]ELHOSARY M A,BAHEY-EL-DIN M,ABDELBARY A,et al.Immunization with the ferric iron-binding periplasmic protein HitA provides protection against Pseudomonas aeruginosa in the murine infection model[J].Microb Pathog,2019,131:181-185.DOI:10.1016/j.micpath.2019.04.014.
[45]FOURIE K R,WILSON H L.Understanding GroEL and DnaK stress response proteins as antigens for bacterial diseases[J].Vaccines (Basel),2020,8(4):773.DOI:10.3390/vaccines8040773.
[46]CHATURVEDI K S,HUNG C S,GIBLIN D E,et al.Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic[J].ACS Chem Biol,2014,9(2):551-561.DOI:10.1021/cb400658k.
[47]KOH E I,ROBINSON A E,BANDARA N,et al.Copper import in Escherichia coli by the yersiniabactin metallophore system[J].Nat Chem Biol,2017,13(9):1016-1021.DOI:10.1038/nchembio.2441.
[48]MOSCATELLO N J,PFEIFER B A.Yersiniabactin metal binding characterization and removal of nickel from industrial wastewater[J].Biotechnol Prog,2017,33(6):1548-1554.DOI:10.1002/btpr.2542.
[49]LIGTHART K,BELZER C,DE VOS W M,et al.Bridging bacteria and the gut: Functional aspects of type IV pili[J].Trends Microbiol,2020,28(5):340-348.DOI:10.1016/j.tim.2020.02.003.
[50]CRAIG L,PIQUE M E,TAINER J A.Type IV pilus structure and bacterial pathogenicity[J].Nat Rev Microbiol,2004,2(5):363-378.DOI:10.1038/nrmicro885.
[51]XICOHTENCATL-CORTES J,MONTERIRO-NETO V,SALDA?A Z,et al.The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes[J].J Bacteriol,2009,191(1):411-421.DOI:10.1128/JB.01306-08.
[52]MAZARIEGO-ESPINOSA K,CRUZ A,LEDESMA M A,et al.Longus,a type IV pilus of enterotoxigenic Escherichia coli,is involved in adherence to intestinal epithelial cells[J].J Bacteriol,2010,192(11):2791-2800.DOI:10.1128/JB.01595-09.
[53]MUNDY R,PICKARD D,WILSON R K,et al.Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium[J].Mol Microbiol,2003,48(3):795-809.DOI:10.1046/j.1365-2958.2003.03470.x.
[54]PERSAT A,INCLAM Y F,ENGEL J N,et al.Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa[J].Proc Natl Acad Sci USA,2015,112(24):7563-7568.DOI:10.1073/pnas.1502025112.
[55]FRYE S A,L?NG E L,BEYENE G T,et al.The inner membrane protein PilG interacts with DNA and the secretin PilQ in transformation[J].PLoS One,2015,10(8):e0134954(1-25).DOI:10.1371/journal.pone.0134954.
[56]DARZINS A.The pilG gene product,required for Pseudomonas aeruginosa pilus production and twitching motility,is homologous to the enteric, single-domain response regulator CheY[J].J Bacteriol,1993,175(18):5934-5944.DOI:10.1128/jb.175.18.5934-5944.1993.
[57]WU S S,WU Jie,CHENG Y L,et al.The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus[J].Mol Microbiol,1998,29(5):1249-1261.DOI:10.1046/j.1365-2958.1998.01013.x.
[58]HOBBS M,COLLIE E S,FREE P D,et al.PilS and PilR,a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa[J].Mol Microbiol,1993,7(5):669-682.DOI:10.1111/j.1365-2958.1993.tb01158.x.
[59]KUZMICH S,SKOTNICKA D,SZADKOWSKI D,et al.Three pilZ domain proteins,PlpA,PixA,and PixB,have distinct functions in regulation of motility and development in Myxococcus xanthus[J].J Bacteriol,2021,203(13):e00126-21.DOI:10.1128/JB.00126-21.
[60]MACNAB R M.How bacteria assemble flagella[J].Annu Rev Microbiol,2003,57:77-100.DOI:10.1146/annurev.micro.57.030502.09083.
[61]GUERRY P.Campylobacter flagella: Not just for motility[J].Trends Microbiol,2007,15(10):456-461.DOI:10.1016/j.tim.2007.09.006.
[62]VAN VLIET A H,KETLEY J M.Pathogenesis of enteric Campylobacterinfection[J].Symp Ser Soc Appl Microbiol,2001,(30):45S-56S.DOI:10.1046/j.1365-2672.2001.01353.x.
[63]WALLIS M R.The pathogenesis of Campylobacter jejuni[J].Br J Biomed Sci,1994,51(1):57-64.
[64]FERRERO R L,LEE A.Motility of Campylobacter jejuni in a viscous environment: Comparison with conventional rod-shaped bacteria[J].J Gen Microbiol,1988,134(1):53-59.DOI:10.1099/00221287-134-1-53.
[65]LEE A,O′ROURKE J L,BARRINGTON P J,et al.Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: A mouse cecal model[J].Infect Immun,1986,51(2):536-546.DOI:10.1128/iai.51.2.536-546.1986.
[66]LERTSETHTAKARN P,OTTEMANN K M,HENDRIXSON D R.Motility and chemotaxis in Campylobacter and Helicobacte[J].Annu Rev Microbiol,2011,65:389-410.DOI:10.1146/annurev-micro-090110-102908.
[67]SAIJO-HAMANO Y,MATSUNAMI H,NAMBA K,et al.Architecture of the bacterial flagellar distal rod and hook of Salmonella[J].Biomolecules,2019,9(7):260-271.DOI:10.3390/biom9070260.
[68]MINAMINO T,YAMAGUCHI S,MACNAB R M.Interaction between FliE and FlgB,a proximal rod component of the flagellar basal body of Salmonella[J].J Bacteriol,2000,182(11):3029-3036.DOI:10.1128/JB.182.11.3029-3036.2000.
[69]MINAMINO T.Protein export through the bacterial flagellar type Ⅲ export pathway[J].Biochim Biophys Acta,2014,1843(8):1642-1648.DOI:10.1016/j.bbamcr.2013.09.005.
[70]MINAMINO T,INOUE Y,KINOSHITA M,et al.FliK-driven conformational rearrangements of FlhA and FlhB are required for export switching of the flagellar protein export apparatus[J].J Bacteriol,2020,202(3):e00637-19.DOI:10.1128/JB.00637-19.
[71]BARKER C S,MESHCHERYAKOVA I V,INOUE T,et al.Assembling flagella in Salmonella mutant strains producing a type Ⅲ export apparatus without FliO[J].J Bacteriol,2014,196(23):4001-4011.DOI:10.1128/JB.02184-14.
[72]BURNHAM P M,HENDRIXSON D R.Campylobacter jejuni: Collective components promoting a successful enteric lifestyle[J].Nat Rev Microbiol,2018,16(9):551-565.DOI:10.1038/s41579-018-0037-9.
[73]ZHAO Ronghao,PATHAK N,JAFFE H,et al.FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body[J].J Mol Biol,1996,261(2):195-208.DOI:10.1006/jmbi.1996.0452.
[74]PEREIRA M,PAENTE J A,BATAUS L A,et al.Chemotaxis and flagellar genes of Chromobacterium violaceum[J].Genet Mol Res,2004,3(1):92-101.
[75]NAKAMURA S,MINAMINO T.Flagella-driven motility of Bacteria[J].Biomolecules,2019,9(7):279-302.DOI:10.3390/biom9070279.
[76]NAMBU T,MINAMINA T,MACNAB R M,et al.Peptidoglycan-hydrolyzing activity of the FlgJ protein,essential for flagellar rod formation in Salmonella typhimurium[J].J Bacteriol,1999,181(5):1555-1561.DOI:10.1128/JB.181.5.1555-1561.1999.
[77]OHNISHI K,OHTO Y,AIZAWA S,et al.FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium[J].J Bacteriol,1994,176(8):2272-2281.DOI:10.1128/jb.176.8.2272-2281.1994.
[78]M?LLER V,JONES C J,KAWAGISHI I,et al.Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex[J].J Bacteriol,1992,174(7):2298-2304.DOI:10.1128/jb.174.7.2298-2304.199.
[79]DASGUPTA N,RAMPHAL R.Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa[J].J Bacteriol,2001,183(22):6636-6644.DOI:10.1128/JB.183.22.6636-6644.2001.
[80]JONES C J,HOMMA M,MACNAB R M.L-,P-,and M-ring proteins of the flagellar basal body of Salmonella typhimurium: Gene sequences and deduced protein sequences[J].J Bacteriol,1989,171(7):3890-3900.DOI:10.1128/jb.171.7.3890-3900.1989.
[81]GONZ?LEZ-PEDRAJO B,DE LA MORA J,BALLADO T,et al.Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis[J].Biochim Biophys Acta,2002,1579(1):55-63.DOI:10.1016/s0167-4781(02)00504-3.
[82]BALLADO T,CAMARENA L,GONZ?LEZ-PEDRAJO B,et al.The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: Study of an flgE mutant[J].J Bacteriol,2001,183(5):1680-1687.DOI:10.1128/JB.183.5.1680-1687.2001.
[83]FAUCONNIER A,ALLAOUI A,CAMPOS A,et al.Flagellar flhA,flhB and flhE genes,organized in an operon,cluster upstream from the inv locus in Yersinia enterocolitica[J].Microbiology (Reading),1997,143 ( 11):3461-3471.DOI:10.1099/00221287-143-11-3461.
[84]LECLERC G,WANG Shuiping,ELY B.A new class of Caulobacter crescentus flagellar genes[J].J Bacteriol,1998,180(19):5010-5019.DOI:10.1128/JB.180.19.5010-5019.1998.
[85]LIU Xiaoying,MATSUMURA P.The FlhD/FlhC complex,a transcriptional a ctivator of the Escherichia coli flagellar class II operons[J].J Bacteriol,1994,176(23):7345-7351.DOI:10.1128/jb.176.23.7345-7351.
[86]OHNISHI K,KUTSUKAKE K,SUZUKI H,et al.Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium[J].Mol Gen Genet,1990,221(2):139-147.DOI:10.1007/BF00261713.
[87]KRISTICH C J,ORDAL G W.Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis[J].J Biol Chem,2002,277(28):25356-25362.DOI:10.1074/jbc.M201334200.
[88]WELCH M,OOSAWA K,AIZAWA S I,et al.Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria[J].Proc Natl Acad Sci USA,1993,90(19):8787-8791.DOI:10.1073/pnas.90.19.8787.
[89]MOON K H,HOBBS G,MOTALEB M A.Borrelia burgdorferi CheD promotes various functions in chemotaxis and the pathogenic life cycle of the spirochete[J].Infect Immun,2016,84(6):1743-1752.DOI:10.1128/IAI.01347-15.
[90]DUR?N N,MENCK C F M.Chromobacterium violaceum: A review of pharmacological and industiral perspectives[J].Crit Rev Microbiol,2001,27(3):201-222.DOI:10.1080/20014091096747.
[91]GORDON A H,HART P D A,YOUNG M R.Ammonia inhibits phagosome-lysosome fusion inmacrophages[J].Nature,1980,286(5768):79-80.DOI:10.1038/286079a0.
[92]MAZZEI L,MUSIANIL F,CIURLI S.The structure-based reaction mechanism of urease,a nickel dependent enzyme:tale of a long debate[J].J Biol Inorg Chem,2020,25(6):829-845.DOI:10.1007/s00775-020-01808-w.
[93]MOBLEY H L.The role of Helicobacter pylori urease in the pathogenesis of gastritis and peptic ulceration[J].Aliment Pharmacol Ther,1996,10(Suppl 1):57-64.DOI:10.1046/j.1365-2036.1996.22164006.x.
[94]RIOT B,BERCHE P,SIMONET M.Urease is not involved in the virulence of Yersinia pseudotuberculosis in mice[J].Infect Immun,1997,65(5):1985-90.DOI:10.1128/iai.65.5.1985-1990.1997.
[95]NOLDEN L,BECKERS G,M?CKEL B,et al.Urease of Corynebacterium glutamicum: Organization of corresponding genes and investigation of activity[J].FEMS Microbiol Lett,2000,189(2):305-310.DOI:10.1111/j.1574-6968.2000.tb09248.x.
[96]BAGCHI D,BHAITACHARYA G,STOHS S J.Production of reactive oxygen species by gastric cells in association with Helicobacter pylori[J].Free Radic Res,1996,24(6):439-450.DOI:10.3109/10715769609088043.
[97]TSUGAWA H,MORI H,MATSUZAKI J,et al.Nordihydroguaiaretic acid disrupts the antioxidant ability of Helicobacter pylori through the repression of SodB activity in vitro[J].Biomed Res Int,2015,2015:734548(1-8).DOI:10.1155/2015/734548.
[98]CANDELA M,CENTANNI M,FIORI J,et al.DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts[J].Microbiology (Reading),2010,156(6):1609-1618.DOI:10.1099/mic.0.038307-0.
[99]SPRINZL M.Elongation factor Tu: A regulatory GTPase with an integrated effector[J].Trends Biochem Sci,1994,19(6):245-250.DOI:10.1016/0968-0004(94)90149-x.
[100]HARVEY K L,JAROCKI V M,CHARLES I G,et al.The diverse functional roles of elongation factor Tu (EF-Tu) in microbial pathogenesis[J].Front Microbiol,2019,10:2351(1-19).DOI:10.3389/fmicb.2019.02351.
[101]GRANATO D,BERGONZALLI G E,PRIDMORE R D,et al.Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins[J].Infect Immun,2004,72(4):2160-2169.DOI:10.1128/IAI.72.4.2160-2169.2004.
[102]CHIU K H,WANG Linghui,TSAI T T,et al.Secretomic analysis of host-pathogen interactions reveals that elongation factor-Tu is a potential adherence factor of Helicobacter pylori during pathogenesis[J].J Proteome Res,2017,16(1):264-273.DOI:10.1021/acs.jproteome.6b00584.
[103]WINN W C JR.Legionnaires disease: Historical perspective[J].Clin Microbiol Rev,1988,1(1):60-81.DOI:10.1128/CMR.1.1.60.
[104]MC DADE J E,SHEPARD C C,FRASER D W,et al.Legionnaires disease: Isolation of a bacterium and demonstration of its role in other respiratory disease[J].N Engl J Med,1977,297(22):1197-1203.DOI:10.1056/NEJM197712012972202.
[105]CIANCIOTTO N P,STAMOS J K,KAMP D W.Infectivity of Legionella pneumophila mip mutant for alveolar epithelialcells[J].Curr Microbiol,1995,30(4):247-250.DOI:10.1007/BF00293641.
[106]HELBIG J H,L?CK P C,STEINERT M,et al.Immunolocalization of the Mip protein of intracellularly and extracellularly grown Legionella pneumophila[J].Lett Appl Microbiol,2001,32(2):83-88.DOI:10.1046/j.1472-765x.2001.00861.x..
[107]K?HLER R,FANGHA?NEL J,K?NIG B,et al.Biochemical and functional analyses of the Mip protein: Influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila[J].Infect Immun,2003,71(8):4389-4397.DOI:10.1128/IAI.71.8.4389-4397.2003.
[108]CIANCIOTTO N P,EISENSTEIN B I,MODY C H,et al.A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence[J].J Infect Dis,1990,162(1):121-126.DOI:10.1093/infdis/162.1.121.PMID:2355188.
[109]HELBIG J H,K?NIG B,KNOSPE H,et al.The PPIase active site of Legionella pneumophila Mip protein is involved in the infection of eukaryotic host cells[J].Biol Chem,2003,384(1):125-137.DOI:10.1515/BC.2003.013.
[110]SUSA M,HACKER J,MARRE R.De novo synthesis of Legionella pneumophila antigens during intracellular growth in phagocytic cells[J].Infect Immun,1996,64(5):1679-1684.DOI:10.1128/iai.64.5.1679-1684.1996.
[111]WAGNER C,KHAN A S,KAMPHAUSEN T,et al.Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix[J].Cell Microbiol,2007,9(2):450-462.DOI:10.1111/j.1462-5822.2006.00802.x.
[112]PORANKIEWICZ J,WANG Jimin,CLARKE A K.New insights into the ATP-dependent Clp protease: Escherichia coli and beyond[J].Mol Microbiol,1999,32(3):449-458.DOI:10.1046/j.1365-2958.1999.01357.x.
[113]KWON H Y,OGUNNIYI A D,CHOI M H,et al.The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge[J].Infect Immun,2004,72(10):5646-5653.DOI:10.1128/IAI.72.10.5646-5653.2004.
[114]ROUQUETTE C,RIPIO M T,PELLEGRINI E,et al.Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes[J].Mol Microbiol,1996,21(5):977-987.DOI:10.1046/j.1365-2958.1996.641432.x.
[115]TOMOYASU T,TABATA A,IMAKI H,et al.Role of Streptococcus intermedius DnaK chaperone system in stress tolerance and pathogenicity[J].Cell Stress Chaperones,2012,17(1):41-55.DOI:10.1007/s12192-011-0284-4.
[116]TAKAYA A,TOMOYASU T,MATSUI H,et al.The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages,leading to systemic infection[J].Infect Immun,2004,72(3):1364-1373.DOI:10.1128/IAI.72.3.1364-1373.2004.
[117]K?HLER S,EKAZA E,PAQUET J Y,et al.Induction of dnaK through its native heat shock promoter is necessary for intramacrophagic replication of Brucella suis[J].Infect Immun,2002,70(3):1631-1634.DOI:10.1128/IAI.70.3.1631-1634.2002.
[118]MALONEY K E,VALVANO M A.The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages[J].Infect Immun,2006,74(10):5477-5486.DOI:10.1128/IAI.00798-06.
[119]MOIGNEV V L,BELON C,GOULARD C,et al.MgtC as a host-induced factor and vaccine candidate against Mycobacterium abscessus infection[J].Infect Immun,2016,84(10):2895-2903.DOI:10.1128/IAI.00359-16.
[120]BELON C,GANNOUN-ZAKI L,LUTFALLA G,et al.Mycobacterium marinum MgtC plays a role in phagocytosis but is dispensable for intracellular multiplication[J].PLoS One,2014,9(12):e116052(1-23).DOI:10.1371/journal.pone.0116052.
[121]RETAMAL P,CASTILLO-RUIZ M,MORA G C.Characterization of MgtC,a virulence factor of Salmonella enterica serovar typhi[J].PLoS One,2009,4(5):e5551(1-6).DOI:10.1371/journal.pone.0005551.
[122]CAFIERO J H,LAMBERTI Y A,SURMANN K,et al.A Bordetella pertussis MgtC homolog plays a role in the intracellular survival[J].PLoS One,2018,13(8):e0203204.DOI:10.1371/journal.pone.0203204.
[123]HAGINS J M,LOCY R,SILO-SUH L.Isocitrate lyase supplies precursors for hydrogen cyanide production in a cystic fibrosis isolate of Pseudomonas aeruginosa[J].J Bacteriol,2009,191(20):6335-6339.DOI:10.1128/JB.00692-09.
[124]BERNUT A,BELON C,SOSCIA C,et al.Intracellular phase for an extracellular bacterial pathogen: MgtC shows the way[J].Microb Cell,2015,2(9):353-355.DOI:10.15698/mic2015.09.227.
[125]MC KINNEY J D,H?NER ZU BENTRUP K,MU?OZ-EL?AS E J,et al.Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase[J].Nature,2000,406(6797):735-738.DOI:10.1038/35021074.
[126]LINDSEY T L,HAGINS J M,SOKOL P A,et al.Virulence determinants from a cystic fibrosis isolate of Pseudomonas aeruginosa include isocitrate lyase[J].Microbiology (Reading),2008,154(6):1616-1627.DOI:10.1099/mic.0.2007/014506-0.
(责任编辑:黄仲一 英文审校:刘源岗)
