Simultaneously quantifying hundreds of acylcarnitines in multiple biological matrices within ten minutes using ultrahigh-performance liquid-chromatography and tandem mass spectrometry
2024-03-21JingxinZhngQinshengChenLinglongZhngBiruShiMenYuQingxiHungHuiruTng
Jingxin Zhng , Qinsheng Chen , Linglong Zhng , Biru Shi , Men Yu ,Qingxi Hung ,**, Huiru Tng ,*
a State Key Laboratory of Genetic Engineering, School of Life Sciences, Human Phenome Institute, Zhangjiang Fudan International Innovation Center,Metabonomics and Systems Biology Laboratory at Shanghai International Centre for Molecular Phenomics, Zhongshan Hospital, Fudan University,Shanghai, 200032, China
b Wuhan Laboratory for Shanghai Metabolome Institute (SMI) Ltd., Wuhan, 430000, China
Keywords:Acylcarnitine UPLC-MS/MS Quantitative structure-retention relationship Molecular phenotype
A B S T R A C T Acylcarnitines are metabolic intermediates of fatty acids and branched-chain amino acids having vital biofunctions and pathophysiological significances.Here, we developed a high-throughput method for quantifying hundreds of acylcarnitines in one run using ultrahigh performance liquid chromatography and tandem mass spectrometry (UPLC-MS/MS).This enabled simultaneous quantification of 1136 acylcarnitines (C0-C26) within 10-min with good sensitivity (limit of detection <0.7 fmol), linearity (correlation coefficient >0.992), accuracy (relative error <20%), precision (coefficient of variation (CV),CV <15%),stability(CV <15%),and inter-technician consistency(CV <20%,n=6).We also established a quantitative structure-retention relationship(goodness of fit >0.998)for predicting retention time(tR)of acylcarnitines with no standards and built a database of their multiple reaction monitoring parameters(tR,ion-pairs,and collision energy).Furthermore,we quantified 514 acylcarnitines in human plasma and urine, mouse kidney, liver, heart, lung, and muscle.This provides a rapid method for quantifying acylcarnitines in multiple biological matrices.
1.Introduction
Acylcarnitines are esters of carnitine (3-hydroxy-4-trimethylaminobutyric acid) derived from fatty-acids and branched-chain amino acids in mitochondria, peroxisome, and endoplasmic reticulum [1-10].Although several thousand acylcarnitine species were predicted to be possibly present in the human samples [9], only a few hundred acylcarnitines were collectively reported with their acyl-chains containing 2-28 carbons(C2-C28)[11,12].Based on the acyl-chain length,they can be categorized as short-chain (C2-5), medium-chain (C6-12), longchain (C13-20), and very-long chain (>C20) acylcarnitines [8-10](Fig.1).The acyl-chains may also contain hydroxyl,carboxylic,and side-chain methyl groups so that these subgroups can be termed as hydroxylated, carboxylated, and branched-chain acylcarnitines,respectively [9,10].More recently, some acylcarnitines conjugated with numerous hydrophilic compounds including amino acids,glucuronic acid,and sulfoacid(Fig.1)were discovered though only in urine so far which were probably derived from phase II biotransformation [12].
These acylcarnitines play many vital roles including metabolic homeostasis of lipids and amino acids, fatty acid transportation,and host-microbial symbiosis [3-10].Numerous acylcarnitines have been reported as biomarkers for type-2 diabetes, cardiovascular diseases, Alzheimer's disease, non-alcoholic fatty liver disease, tumors, and inborn errors of metabolism [13-21].However,the functions of branched-chain, hydroxylated, carboxylated, and conjugated acylcarnitines are far from fully understood.Conceivably, simultaneous quantification of all these acylcarnitines is the prerequisite to effectively investigate their diverse functions thus reveal acylcarnitine-related molecular phenotypes of biological systems.Unfortunately, such analysis is nontrivial with huge diversity for the structural and physiochemical properties of acylcarnitines.Concurrent high-coverage and high-throughput quantification of acylcarnitines is still needed especially for large cohort population studies.
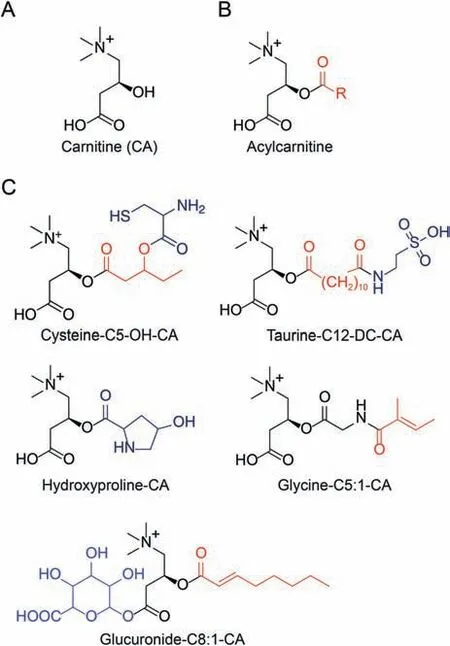
Fig.1.Structure of carnitines detected in biological samples.(A) Carnitine (CA).(B)Typical acylcarnitines whose acyl-group R (C2-C26) may contain straight chains and branched-chains, double bonds, hydroxyl (OH), and carboxyl (DC) groups.(C) Some typical conjugated acylcarnitines.
Ultrahigh performance liquid chromatography coupled with mass spectrometry (UPLC-MS) has shown great potentials for simultaneous detection and quantification of acylcarnitines[11,12,22-24], whilst flow injection MS methods [25] cannot distinguish isomers.For example, 733 acylcarnitines in the pooled samples from rat liver tissue, human plasma, and urine were reportedly detected with UPLC-coupled high-resolution mass spectrometry(HRMS)although the method required more than 10 runs (with 19 min each) [11].A recent HRMS study also reported coverage of 586 acylcarnitines and 125 conjugated acylcarnitines from human urine[12]with 108 individual runs(22 min each)but without quantitative data.For simultaneous quantifications,HRMS is limited by its scanning-speed and dynamic range hence accuracy for low abundant acylcarnitines such as those having polyunsaturated, hydroxylated, and carboxylated acyl-chains [24].UPLC-MS/MS in scheduled multiple reaction monitoring (MRM)mode should be the best choice.However, the reported UPLC-MS/MS methods either had thermal instability problems or limited coverage, throughput or isomer differentiability [26-30].Even with the combination of reversed- and normal-phase liquid chromatography (LC), such LC-MS/MS method could only quantify 117 acylcarnitines in biofluids with a 26-min run[29].All the reported LC-MS/MS methods covered only a few hydroxylated and carboxylated acylcarnitines with no conjugated acylcarnitines [9].
One of the bottlenecks for high-coverage LC-MS/MS quantification of acylcarnitines is the lack of standards to reliably obtain the retention time(tR)of all possible acylcarnitines for scheduled MRM analysis.For hundreds of acylcarnitines reportedly detected, only dozens of them have commercial standards.Under such circumstances, reliable modelling for quantitative structure-retention relationship (QSRR) becomes helpful to predict their tR[31,32] for setting up the MRM-needed time-windows and to assist identification.Previous studies have already shown potential linear relationship between tRand some structural descriptors for acylcarnitines [11,30,33].Unfortunately, these required massive calculation resources and gave no information for the tRdependence on functional groups.Understandably, the retention behaviors of acylcarnitines collectively depend on structural characteristics of their acyl-chains including the number of carbons, double-bonds, side-chains, hydroxyl and carboxyl groups,and their positions.So far, however, the best such study has considered only the number of carbon atoms and double bonds in acyl-chains with several linear models constructed for different classes of acylcarnitines[34].A unified QSRR model for demanded structural features has not been reported so far, to the best of our knowledge.
Here, we developed an MRM-based sensitive UPLC-MS/MS method for simultaneous quantification of all identifiable acylcarnitines in biofluids and tissues with a single 10-min run.We also built a QSRR model for predicting acylcarnitine tRvalues with their seven structural features and constructed a database of MRM parameters (tR, MRM ion-pairs, and collision energy) for 1136 acylcarnitines detected in literatures and with UPLC-based quadrupole time-of-flight MS (UPLC-QTOFMS) in this study.We further quantified acylcarnitines in two typical human biofluids (plasma and urine) and five typical tissues of animal model to reveal their molecular phenotypic characteristics.
2.Materials and methods
2.1.Chemicals
HPLC grade methanol (MeOH), isopropyl alcohol (IPA), and dichloromethane were obtained from Merck(Darmstadt,Germany)whereas acetonitrile (ACN), formic acid, and acetic acid were purchased from Sigma-Aldrich (St.Louis, MO, USA).Ultrapure water(H2O)was obtained from a Milli-Q system(Millipore,Billerica,MA,USA).49 acylcarnitine standards and 26 deuterated ones as internal standards (IS) were purchased from commercial sources with detailed information listed in Table S1.
2.2.Collection of biological samples
Human plasma and urine were obtained from Chinese adult volunteers recruited for the Human Phenome Project which was approved by the Ethic Committee of Fudan University (Approval No.:FE21087)with informed consent from all participants.Human lung cancer cell lines (A549, 16HBE) were obtained from China Center for Type Culture Collection.The New Zealand rabbit liver and five tissue samples of C57BL/6 mice were collected according to the National Guidelines for Experimental Animal Welfare(Ministry of Science and Technology of China, 2006).All biological samples were snap-frozen with liquid-nitrogen and stored at -80°C until further analysis.
2.3.Preparation of stock and working solutions
Each acylcarnitine standard was dissolved in MeOH or CH2Cl2:MeOH (3:2, V/V) to prepared individual stock solution(1-15 mM).These solutions were mixed, dried with nitrogen-gas,and reconstituted into aqueous MeOH (90%, V/V) to obtain the stock solution of acylcarnitine mixture.This mixture solution was then sequentially diluted to obtain 13 solutions for constructing calibration curves (Levels 1-13, Tables S2-S4).The mixture of 26 deuterated acylcarnitines in 90% (V/V) aqueous MeOH was used as IS.
2.4.Preparation of biological samples for analysis of acylcarnitines
For UPLC-QTOFMS analysis, acylcarnitines were extracted from multiple biological matrices including rabbit liver tissue, human plasma,urine,and cells as described in the Supplementary data.For quantification of acylcarnitines in biofluids and tissues, analytes were extracted with an optimized procedure.Briefly,20 μL of each human plasma or urine was mixed with 500 μL pre-cooled IPA containing 0.5%(V/V)acetic acid,20 μL mixed IS solution,and 30 μL water, and sonicated in an ice-bath for 5 min.After centrifugation(4°C,16,000 g)for 10 min,520 μL of supernatant was obtained.The above procedure was repeated once more and resultant 2 supernatants were combined and dried with nitrogen-gas.Each tissue sample (10 mg) was added with 400 μL pre-cooled IPA containing 0.5% (V/V) acetic acid, 20 μL mixed IS, and 30 μL water, and then homogenized with a tissuelyser (20 Hz) for 3 min to obtain supernatant after 10-min centrifugation (4°C, 16,000 g).This extraction was repeated two more times and three resultant supernatants were pooled and dried.These dried extracts were individually reconstituted into 90% (V/V) MeOH (40 μL) for UPLCMS/MS analysis.
2.5.UPLC-MS analysis
A ZenoTOF 7600 and TripleTOF 5600 plus (SCIEX, Chromos,Singapore) coupled to a Shimazu UPLC system were used in the information-dependent acquisition (IDA) mode (Supplementary data) to detect acylcarnitines and their fragment ions in multiple biological samples.A QTRAP 6500 Plus (SCIEX) coupled to two Shimadzu Nexera UPLC systems (Kyoto, Japan) was used for simultaneous quantification of acylcarnitines with scheduled MRM.Data were acquired and processed with Analyst and OS (v1.7,SCIEX).After comprehensive optimization of various C18columns and chromatographic conditions, the best performer Agilent ZORBAX Eclipse Plus C18column (2.1 mm × 100 mm, 1.8 μm; Santa Clara,CA,USA)was chosen at 40°C with 0.5 μL of sample injection.H2O and ACN(both containing 0.1%(V/V)formic acid)were used as mobile phase A and B,respectively,with elution gradients listed in Table S5.The dual-column-switching mode (Fig.S1) with Multiplexing (MPX) software was used to double throughput.By using acylcarnitine standards, mass spectrometric parameters were optimized including ionspray voltage, curtain gas, ion source temperature, collision energy, and declustering potential(Supplementary data).Collision energy values were further adjusted to ensure simultaneous quantification of all acylcarnitines in a single run for all biological matrices (Table S6).
2.6.Method validation
Method validation was conducted for sensitivity, linearity, accuracy, precision, stability, and inter-technician consistency.Linearity and sensitivity were assessed using acylcarnitine standards with variable concentration and IS.The limit of detection(LOD)was obtained as the amount of analytes on column at the signal-tonoise ratio (S/N) of 3 whereas the low limit of quantification(LLOQ) was obtained from the lowest amount in calibration curve according to the U.S.Food and Drug Administration guideline[35].Accuracy, precision, and stability were evaluated with plasma,urine,and liver samples at three different concentration levels(L3,L5,and L7).Accuracy was assessed with relative error(RE)at high,medium, and low levels whilst precision was assessed with the coefficient of variation (CV) for the intra- and inter-day measurements.Stability was assessed with extracted samples stored at -4°C for 24 h and at -80°C for 72 h.Inter-technician consistency was evaluated using the same batch of plasma samples(n = 6) with the independent results from three different technicians.
2.7.QSRR and MRM-parameter database
The QSRR model was built with multivariate polynomial regression approach [36,37] using experimental data from 45 known acylcarnitines (Table S1) containing saturated (18), unsaturated (12), hydroxylated (9), and dicarboxylic (6) acyl-chains.LM functions in R software package (v4.1.0) was used to optimize models with tRdependence on various structural features for acyl chains with stepwise regression and full subset regression.These features included the number of carbon atoms(c),branched-chain methyl groups (b), carbon-carbon double bonds (d), hydroxyl groups(h),carboxyl groups(j),position of branched-chain(p),and double bonds(q).10-fold cross-validation was conducted to obtain the optimized model with minimum prediction error and favorable adjusted-goodness of fit (R2a, Eq.(1)).Based on the experimental and calculated tRresults, mean absolute error (MAE, Eq.(2)) and Pearson correlation coefficient (r2) were used to assess the overall performance of model.

3.Results and discussion
3.1.Detection of acylcarnitines in multiple biological matrices with UPLC-QTOFMS
Although thousands of acylcarnitines have been suggested in the Human Metabolome Database,so far,only dozens of them have experimental mass spectrometric data due to limited standards available.To obtain such data,we employed UPLC-QTOFMS in fullscan and IDA modes to detect all possible acylcarnitines present in multiple biological matrices including biofluids, cells, and tissues.Chromatographic parameters were systematically optimized including column, temperature, elution gradients, flow-rate, and injection volume to achieve maximum separation (data not shown).Since carnitine moiety has a quaternary ammonium cation,(CH3)3N+R,with permanent charge, acylcarnitines are expected to have some unique mass spectrometric features.In the negative ion mode, all acylcarnitines gave parent ions as [M+HCOO--H]-,fragment ions for carnitine moiety (m/z 188.0934 and 144.1042)and [FA-H]-from fatty-acid chains (Fig.S2).In the positive ion mode,in contrast,much more intense signals including parent ions[M]+due to the carnitine quaternary ammonium cations,[M-59.0735]+due to neutral loss of N(CH3)3, and three fragment ions from carnitine moiety (m/z 144.1019, 85.0284, and 60.0808)were observable for all acylcarnitines(Figs.2A and S2)as reported[38-40].All conjugated acylcarnitines gave parent ions [M]+,[M-conjugator-59.0735]+,and three fragment ions from carnitine moiety (Figs.2B and S2) as reported [12,38].Therefore, acylcarnitines and conjugated acylcarnitines were both readily identifiable collectively with their aforementioned five ions, respectively.Acylcarnitines gave the most intense fragment ion m/z 85.0284,thus MRM ion-pair [M]+/85.0284 was useable for quantifying acylcarnitines(Fig.S3).In contrast,[M-conjugator-59.0735]+was a unique intense fragment ion for conjugated acylcarnitines(Fig.2B),thus[M]+/[M-conjugator-59.0735]+was employable for their quantification.Typical ions were also recorded for 26 deuterated acylcarnitines, which were commercially available and used as internal standards for quantification.
By concurrently extracting the above ions, we identified 471 acylcarnitines including 395 acylcarnitines and 76 conjugated ones from above biological matrices (rabbit liver tissue, human plasma,urine, and cells) and their pooled samples (Fig.S4).After taking structural isomers having different retention time into consideration,the coverage of acylcarnitines having different retention time was expanded to 1136 species (Table S7) including 1124 possible species derived from these collectively reported in literatures[11,12,29,41,42].Unambiguous identification was achieved for acylcarnitine isomers having commercial standards (Tables S7 and S8)such as butyryl-carnitine(C4:0)and isobutyry-carnitine(2-M-C3:0),succinyl-carnitine (C4-DC), and methylmalonyl-carnitine (2-M-C3-DC) as well as adipoyl-carnitine (C6-DC) and 3-methylglutarylcarnitine (3-M-C5-DC).For acylcarnitines with saturated fivecarbon chains (C12H23NO4), for instance, four isomers were detected with clearly different retention times.By using the commercially available standards,three of them were unambiguously assigned as valerylcarnitine (C5:0), 2-methylbutyryl-carnitine (2-M-C4:0), and isovalerylcarnitine (3-M-C4:0), respectively; pivaloylcarntine (2-2M-C3:0)became the only possibility for the remaining isomer and was further confirmed using its standard.Furthermore,the isomers without commercial standards were only considered as present here when detected with clearly different retention times(Table S8).They can be unambiguously identified with extensive syntheses of their standards, which is beyond the scope of this study.Nevertheless,we identified these isomers as analyte-a,analyte-b,etc.here to ensure their separate identities.For instance, five isomers were detected for acylcarnitines with saturated six-carbon chains giving same precursor and fragment ions in both Q-TOFMS and MS/MS.Hexanoyl-carnitine was unambiguously assigned (C6:0) with its standard and the other four were tentatively assigned to be C6:0-a,C6:0-b, C6:0-c, and C6:0-d (Table S8) respectively.It is conceivable that the identification of isomers can also be assisted to some extent by reliable prediction of their tR.
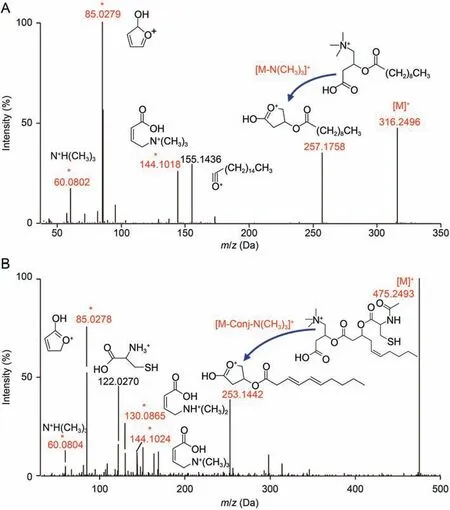
Fig.2.Fragment assignments for (A) decanoylcarnitine (C10:0) and (B) N-acetylcysteine-conjugated-hydroxydecenoylcarnitine (N-acetylcys-C10:1-OH-CA) in the positive ion mode.* Fragment ions for carnitine moiety.Conj: conjugator.
3.2.QSRR and MRM parameter database
The parameters for tRand precursor-product ion pairs for MRM transitions (MRM ion-pairs) are necessary for simultaneous quantification of all acylcarnitines in the scheduled MRM mode using UPLC-MS/MS [43].However, only less than 50 acylcarnitines have standards available currently.Nevertheless, our tRresults of these available acylcarnitines clearly showed some dependence on their structural features(Fig.S5A)as indicated previously[44,45].These structural features included the number of carbons, branchedchains, double-bonds, hydroxyl, carboxylic groups, and positions in their acyl-chains.With the same number of acyl-chain carbons,the presence of these groups resulted in earlier elution.Amongst four isomers of acylcarnitines with five acyl-chain carbons, for example, C5:0 had the greatest tRfollowed with 3-M-C4:0, 2-MC4:0, and then 2-2M-C3:0.
By using 45 acylcarnitine standards (Table S1) excluding carnitine and acetylcarnitine for their close-to-void elution, a unified QSRR model was built for tRvalues (i.e., the model-calculated retention times (tRC)) obtained from our chromatographic method(including column and elution schemes)as a function of acyl-chain structural characteristics (Eq.(3)) with the fitted k0-k10data obtained (i.e., k0= -0.252, k1= 8.54 × 10-4, k2= -4.82 × 10-2,k3= 1.06, k4= -0.487, k5= -0.172, k6= -1.65, k7= -1.55,k8=0.152,k9=-1.59×10-2,and k10=7.20×10-2).In the model,variables c, b, d, h, and j denote the number of acyl-chain carbon,branched-chain methyl group, carbon-carbon double-bond, hydroxyl, and carboxylic groups, respectively, whilst p and q denote the positions of the branched-chains and double-bonds.
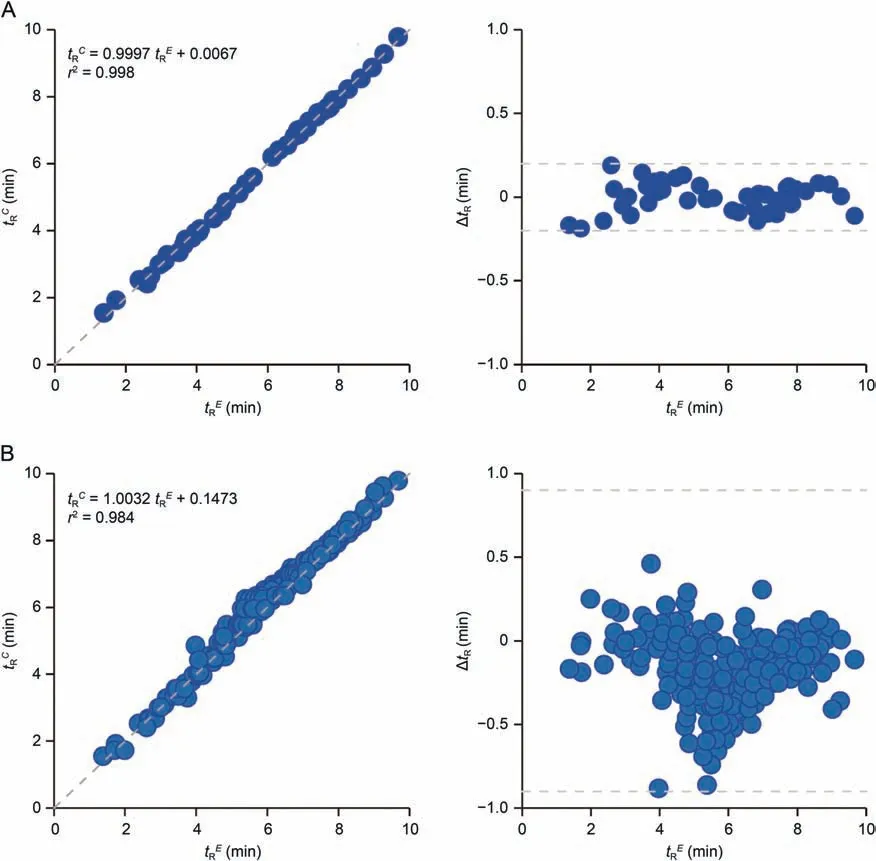
Fig.3.Comparison of the model-calculated retention times (tRC) and experimental results (tRE) for (A) 45 acylcarnitine standards and (B) acylcarnitines detected in biological samples.
Such a unified model could predict tRvalues for all different classes of acylcarnitines with good performance (R2a>0.998) for chain length of C3-C26,which were only describable with multiple linear models previously [11,30,34].The tRCand experimental results (tRE) showed excellent linear correlation (r2, 0.998) and accuracy (ΔtR<0.20 min and MAE <0.07 min) for 45 acylcarnitine standards (Fig.3A).Such tRdifferences between the experimental and predicted values were 0.46 and 0.01 min for 2-2M-C3:0 and C4-DC,respectively,when using the present chromatographic method.
The model also showed good tRCand tREcorrelation(r2~0.984)and prediction accuracy(MAE <0.21 min and ΔtR<0.9 min)for all 211 acylcarnitines detected here from the pooled sample having variable acyl-chain carbons, branch-chains, double-bonds, or hydroxyl or dicarboxylic groups (Fig.3B).Even with concurrent presence of these structure features,the model showed reasonable tRCand tREcorrelation (r2~0.956) and accuracy (MAE <0.25 min,ΔtR<0.9 min) for 97% acylcarnitines identified from the pooled sample(Fig.S5B).By setting the tRrange of 2 min for MRM window,even the acylcarnitine isomers without commercial standards can be reliably covered in UPLC-MS/MS quantification.Taking all this together, we built a database of MRM parameters (tR, MRM ionpairs, and collision energy) to cover 1136 acylcarnitine species with 398 formulae including acylcarnitines, conjugated ones and their isomers (Table S7).Among them, hydroxylated, unsaturated,dicarboxylated,conjugated,and saturated acylcarnitines accounted for 31.6%, 24.9%,18.5%,11.5%, and 7.9%, respectively; acylcarnitines with dicarboxylated chains and hydroxyl group accounted for 5.5%.
3.3.Development of quantitative method for acylcarnitines using UPLC-MS/MS
Based on the above investigations, we established an efficient method for simultaneous quantification of all possible carnitines within 10 min using UPLC-MS/MS in MRM mode.With all standards available commercially, the method showed good separation powers for acylcarnitine isomers with acyl-chains containing C0-C26(Fig.4),which was exemplified with resolution of 1.14-1.24 for acylcarnitine isomers with C5:0 (peaks 16-18).All deuterated standards with N-CD3(peaks 50-53 and 55-75) had similar retention times to the corresponding acylcarnitines(Fig.4).This is in contrast to methyl-D3-malonylcarnitine(peak 54)and suggests the N-CD3-labeled acylcarnitines as good IS for quantifications.
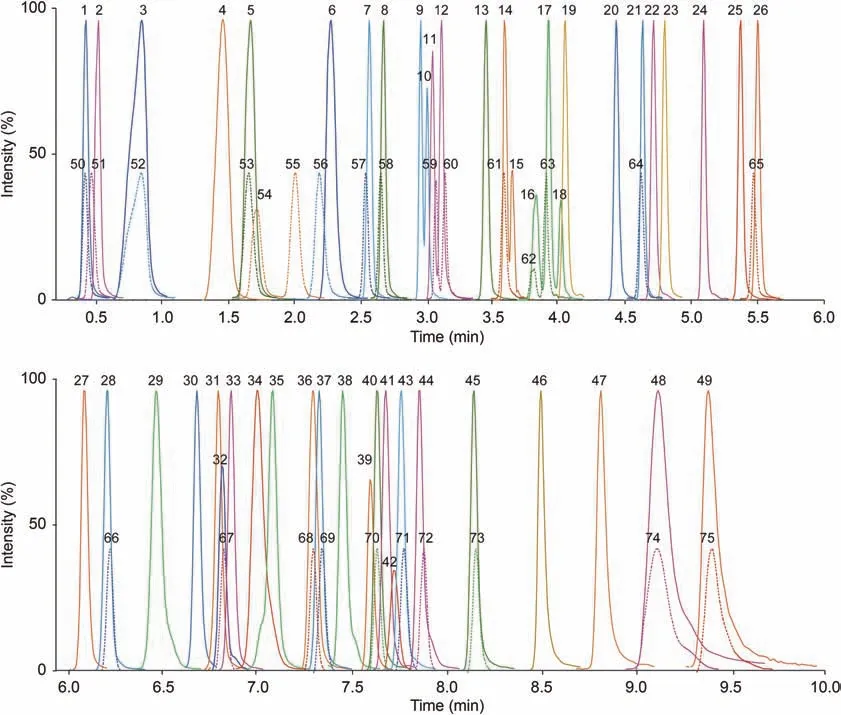
Fig.4.Ultrahigh performance liquid chromatography and tandem mass spectrometry (UPLC-MS/MS) chromatogram (up: 0-6.0 min; bottom: 6.0-10.0 min) for 49 acylcarnitine standards (solid line) and 26 deuterated internal standards (dotted line) with keys listed in Table S1.
Fragment ion m/z 85.0284 reached maximum with collision energy above 30 consistently for short chain(C4:0),medium chain(C10:0), long chain (C16:0), and very-long chain (C22:0) acylcarnitines hence being employable as a quantitative ion(Fig.S3).Other MS parameters were further optimized using all acylcarnitine standards available(Fig.S6),which were used with the deuterated standards to generate calibration curves.For these without authentic standards, such MS parameters for analytes in the same subclass with the closest retention time were used and the results were considered as relative quantification.For instance, MS parameters for C10:0 were used for quantification of C9:0, whose standard was not available.
The method was further validated in terms of linearity, sensitivity, accuracy, precision, stability, and inter-technician consistency.For all acylcarnitines having authentic standards, excellent linearity (r2>0.992) was observed with concentration over 3-4 orders of magnitude and the LOD below 0.7 fmol on column(Table S9), with the exception for only carnitine (<3.5 fmol).The method had good accuracy (RE <20%), intra- and inter-day precision (CV <15%), as well as stability (CV <15%) at 4°C (24 h)and-80°C(72 h)for all spiked standards in plasma,liver,and urine samples (Figs.S7-S9).Acceptable inter-technician consistency(CV <20%) was further confirmed from independent experiments conducted by three different technicians (Fig.S10).With dualcolumn switching (MPX mode), finally, the method was capable of quantifying 1136 carnitines within 10 min;both throughput and coverage were better than the existing methods (Table S10)[11,12,26-30].
3.4.Quantification of acylcarnitines in multiple biological matrices to characterize their molecular phenotypes
By using this method, we managed to quantify 514 acylcarnitines collectively in two typical human biofluids(plasma and urine)and five mouse tissues (kidney, liver, heart, lung, and muscle) to characterize their metabolome-based molecular phenotypes(Fig.5 and Table S8).The results showed carnitine itself as the most abundant in all samples.However, clear acylcarnitine phenotypic differences were observable between biofluids and tissues, between two biofluids and between different tissues from the same animal(Fig.5 and Table S8).Human urine samples contained more acylcarnitine species (421) than human plasma (149) and mouse tissues including kidney (211), liver (195), heart (189), lung (188),and muscle (169), especially conjugated acylcarnitines and these with hydroxyl and carboxylic groups (Fig.5 and Table S8).Noticeably, some carnitine species were only detected in urine including 3-dehydrocarnitine, pivaloylcarnitine, and isomers of C6:0, C8:0,C10:0,C11:0,C12:0,C7:1,C9:1,C12:1,C8:2,C10:2,C11:2,C12:2,and C14:2.Although some of these urinary acylcarnitines (e.g., Phe-C4:1, Phe-C4:0, and odd-carbon chain acylcarnitines) were probably from diets and gut microbiota[46],their sources and functions clearly warrant further investigations.

Fig.5.Concentration (c) in multiple biological matrices for acylcarnitine species and different types of acylcarnitines with saturated (sAC), unsaturated (uAC), hydroxylated (ACOH), carboxylated (AC-DC) chains, and conjugated acylcarnitine (CAC) with bold line (sum) and thin lines indicating total concentration of each class and each species.
Additionally,acylcarnitines had a huge concentration range from 0.31 nM(butenoylcarnitine)to 58.73 μM(carnitine)in human plasma and even greater range in other samples (Fig.5 and Table S8).The abundances of acylcarnitines also differed greatly in two human biofluids(urine and plasma).For example, the concentration of isobutyrylcanitine (2-M-C3:0) in urine was 164 times higher than in plasma.In contrast, butyrylcarnitine (C4:0) and valerylcarnitines were not detected in urine but abundant in human plasma and all mouse tissues.The acylcarnitine compositions further showed obvious diversities for these mice tissues in terms of different acylcarnitines with the levelsranging from 0.14 pmol/mg(C9:0 and C11:0)to 1.66 μmol/mg (carnitine).Kidney had more acylcarnitine species especially conjugated ones with unsaturated and hydroxylated acylchains than the other tissues.In heart and lung tissues,the levels of carnitine (1.3-1.6 μmol/mg) and γ-butyrobetaine (21-24 nmol/mg)were about twice as high as in the other three tissues, whilst acetylcarnitine levels in heart,lung,and muscle tissues(0.16-0.29 μmol/mg) were three times higher than in the other two tissues(0.04-0.06 μmol/mg).These acylcarnitines are broadly attributable to oxidation of fatty acids and branched-chain amino acids in mitochondria and peroxisomes, although the functions of all these detected acylcarnitines warrant further investigations.
Numerous acylcarnitine isomers were concurrently detected in this study although they could be derived from completely different metabolic pathways.Amongst isomers for acylcarnitines with five acyl-carbons, for example, 2-methylbutyrylcarnitine and isovalerylcarnitine are from isoleucine (Ile) and leucine (Leu) metabolism,respectively,whereas pivaloylcarnitine and valerylcarnitine are adducts of exogenous metabolites pivalic acid and valeric acid,respectively, originated from antibodies [47], microbes [48], and plants [49].Interestingly, both 2-methylbutyrylcarnitine and isovalerylcarnitine from two essential amino acids (Ile and Leu) had low levels in all mouse tissues.Hydroxyisobutyrylcarnitine and 3-hydroxybutyrylcarnitine are two isomers but derived from valine and β-hydroxybutyrate, respectively [4,50].The former had high concentration in heart, lung, and muscle, whereas the latter had high concentration in heart, lung, kidney, and liver (Fig.S11).Apparently, new strategies are required to ambiguously assign these acylcarnitines detected but with no standards.
Moreover, obvious level differences were observable for acylcarnitines in human blood plasma and serum from the same blood sample (Fig.S12).The concentrations of most acylcarnitines were lower in serum than in plasma,including about 80%acylcarnitines with short chains and very long chains as well as 60%-70% acylcarnitines with medium chains and long chains.Therefore,plasma samples are recommended for metabolomic studies of acylcarnitines.
4.Conclusions
We developed a high-throughput UPLC-MS method and detected several hundred acylcarnitines and conjugated acylcarnitines in tested multiple biological matrices with their typical fragment ions recorded.Based on the experimental data from our method, we built a good quality model of QSSR for predicting the tRvalues of acylcarnitines with no standards.We also established MRM parameters (tR, MRM ion pairs, and collision energy) to enable simultaneous quantification of 1136 acylcarnitines using a 10-min UPLC-MS/MS method with good linearity, accuracy, precision, stability, and inter-technician consistency.To the best of our knowledge, this method has the higher coverage and throughput than reported methods.By using this method,moreover,we managed to quantify more than 500 acylcarnitine species in two human biofluids and five mouse tissues offering data for molecular phenotyping these biological matrices.This provides some essential basic biochemical data for these biological matrices and a highthroughput method to quantify acylcarnitines for large cohort studies, which are already ongoing in our lab for various diseases.
CRediT author statement
Jingxian Zhang:Conceptualization, Methodology, Validation,Formal analysis,Investigation,Data curation,Visualization,Writing- Original draft preparation;Qinsheng Chen:Validation, Formal analysis, Visualization, Writing - Original draft preparation;Lianglong Zhang:Validation,Formal analysis;Biru Shi:Validation;Men Yu:Conceptualization, Validation;Qingxia Huang:Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Visualization, Writing - Original draft preparation;Huiru Tang:Resources, Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation,Visualization, Writing - Reviewing and Editing, Supervision,Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We acknowledge financial supports from the National Key R&D Program of China(Grant Nos.:2022YFC3400700,2022YFA0806400,and 2020YFE0201600),Shanghai Municipal Science and Technology Major Project(Grant No.:2017SHZDZX01),and the National Natural Science Foundation of China(Grant No.:31821002).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.10.004.
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Platycodin D inhibits angiogenic vascular mimicry in NSCLC by regulating the eIF4E-mediated RNA methylome
- Identification of different degrees of processed ginger using GC-IMS combined with machine learning
- A proteomic landscape of pharmacologic perturbations for functional relevance
- Licorice-saponin A3 is a broad-spectrum inhibitor for COVID-19 by targeting viral spike and anti-inflammation
- Distinct molecular targets of ProEGCG from EGCG and superior inhibition of angiogenesis signaling pathways for treatment of endometriosis
- Hydralazine represses Fpn ubiquitination to rescue injured neurons via competitive binding to UBA52
