Elite, transformable haploid inducers in maize
2024-03-07BrntDlzrDwiLingDviSzwrszrIsorRoriguzGonzloMronsSivmniElumliFrninJohnsonSmsonNlplliRhlEggrErinBurhKrryMirJunWiXiujunZhngHupingGuiHuiingJinHunGuoKunYuYuoLiuBkyBritingrAnPotsJsonNiholsWnShiDviSkiQiungQuTimothy
Brnt Dlzr, Dwi Ling, Dvi Szwrszr, Isor Roriguz, Gonzlo Mrons,Sivmni Elumli, Frnin Johnson, Smson Nlplli, Rhl Eggr, Erin Burh, Krry Mir,Jun Wi, Xiujun Zhng, Huping Gui, Huiing Jin, Hun Guo, Kun Yu, Yuo Liu,Bky Britingr,An Pots,Json Nihols,Wn Shi,Dvi Ski,Qiung Qu,Timothy Kllihr,*
a Seeds Development, Syngenta Seeds, Janesville, WI 53546, USA
b Seeds Research, Syngenta Biotechnology China Co., Ltd., Beijing 102206, China
c Centro de Investigación de Cultivos Arica, Syngenta R&D, Arica 1000000, Chile
d Syngenta S.A., Proyecto la Soledad, Graneros 2880000, Chile
e Seeds Research, Syngenta Crop Protection LLC, RTP, NC 27709, USA
f Seeds Development, Syngenta Seeds, Slater, IA 50244, USA
Keywords:Zea mays L.Doubled haploids Transformation Genome editing QTL
ABSTRACT The introduction of alleles into commercial crop breeding pipelines is both time consuming and costly.Two technologies that are disrupting traditional breeding processes are doubled haploid (DH) breeding and genome editing(GE).Recently,these techniques were combined into a GE trait delivery system called HI-Edit (Haploid Inducer-Edit).In HI-Edit, the pollen of a haploid inducer line is reprogrammed to deliver GE traits to any variety, obviating recurrent selection.For HI-Edit to operate at scale, an efficient transformable HI line is needed,but most maize varieties are recalcitrant to transformation,and haploid inducers are especially difficult to transform given their aberrant reproductive behaviors.Leveraging marker assisted selection and a three-tiered testing scheme, we report the development of new Iodent and Stiff Stalk maize germplasm that are transformable, have high haploid induction rates, and exhibit a robust,genetically-dominant anthocyanin native trait that may be used for rapid haploid identification.We show that transformation of these elite ‘‘HI-Edit” lines is enhanced using the BABYBOOM and WUSCHEL morphogenetic factors.Finally,we evaluate the HI-Edit performance of one of the lines against both Stiff Stalk and non-Stiff Stalk testers.The strategy and results of this study should facilitate the development of commercially scalable HI-Edit systems in diverse crops.
1.Introduction
HI-Edit is a combination of two revolutionary crop breeding technologies: haploid induction (HI) and genome editing (GE) [1].In HI-Edit,a cross pollination between any line and a haploid inducer line carrying a GE transgene produces an edited haploid plant comprising only the non-inducer genome.The potential benefit of HIEdit is to rapidly deliver novel GE traits directly to commercial varieties using a single cross and the doubled haploid(DH)process,obviating generations of backcrossing and marker selection.Another research group has published a related method known as Inducer Mediated Genome Editing [IMGE] [2]).
The first report of HI-Edit[1]was based on NP222RS,which had low transformation frequency (TF), low haploid induction rate(HIR), and poor seed set.The genetic backgrounds capable of efficient transformation and haploid induction have never been combined into a single line.To breed an elite HI-Edit line, biparental crossing schemes were designed to combine the HIR of a modern haploid inducer with the transformability of elite stiff stalk and non-stiff stalk lines.
In this study we show how modern inducers may be bred via genotypic selection on quantitative trait loci (QTL) for haploid induction rate qhir1 [3] and qhir8 [4].The gene underlying qhir1 is matl,a loss of function of a patatin-like phospholipase A2α called MATRILINEAL [MATL][5–7], which confers a maternal HIR of 1% to 7%.The gene underlying qhir8 is DUF679 domain membrane protein 7 (DMP) [8].Genome editing of both genes has created novel inducer lines in diverse crops [9–11].Modern inducer lines may also have dominant anthocyanin alleles like R1 [12] to support visual haploid identification, and they lack anthocyanin pathway inhibitors [13].
Unlike QTL for HIR,QTL supporting efficient transformation are not penetrant across maize germplasm [14].Therefore, after marker-assisted selection on HI traits and multigenerational HIR testing,a screen was performed on the transformability of 80 putative elite HI-Edit lines.After selection of the most efficient, transformable haploid inducer lines, CRISPR/Cas12a genome editing constructs [15–17] were used to test the overall HI-Edit pipeline efficiency.
2.Materials and methods
2.1.Breeding and HIR selection
To stack TF,agronomic performance,and high HIR into one line,we crossed two transformable elite parents with RWKS/Z21S//RWKS, a backcross1(BC1)stage elite inducer with a 16% HIR (owing to qhir1 and qhir8), and strong color induction from R1-SCM2.This inducer was crossed as a male onto SYN-INBB23, an Iodent(non-stiff stalk) with a ~5% TF, and SYN-INBC34, a stiff-stalk with a ~45% TF.Twenty F1s from each population were self-pollinated,and a few F1from SYN-INBB23 were backcrossed by RWKS/Z21S//RWKS.Both Z21S and RWKS are derived from the foundational inducer Zarodyshevy Embryo Marker Saratovskiy ‘‘ZMS” (7% HIR)[18] through the intermediate RWS (8% HIR).
The breeding scheme is shown (Fig.S1).Selections were made on MATRILINEAL/qHIR1 [3,5–7], DMP /qHIR8 [8], R1-SCM2 [12],and a putative anthocyanin inhibitor locus at C1 [19] (Table S1).The haploid induction rate(HIR)is a ratio of haploid embryos over total; R1-SCM2 is less susceptible than other R1 alleles are to instances of germplasm-specific anthocyanin pathway inhibition[20].To avoid false positive haploids, rare instances of ears with light-purple embryos (indicative of color inhibition) were discarded.For seed set determination, pollinated ears were photographed post-harvest.Ears with bare sections at the tip or base were marked ‘‘poor nicks” as this indicates a large anthesissilking interval.In such cases, HIR was the selection criterion.When 80%+ of the ear was successfully pollinated, seed set and HIR were selection criteria.
2.2.Transformation testing and genome editing
Agrobacterium strain LBA4404(pAL4404,pVGW7)was used for transformation[16,21,22].Inoculation and co-cultivation of immature embryos were performed as described[21]on mannose selection [22].Transformation frequency (TF) was the proportion of Cas12a+events divided by the total embryos.Haploid editing rate(HER) was determined by outcrossing pollen from T2homozygous plants onto tester ears, recovering haploids via color sorting, and assessing the editing status using Taqman assays.Haploidy was confirmed by the absence of Cas12a and PMI Taqman markers.Candidate edited haploids were sequenced to confirm the editing status.
2.3.GWAS analysis
Associations between the genotypes of 480 single nucleotide polymorphisms (SNPs) and the transformation rate of 45 SYN-IN BC34 × RWKS//Z21S/RWKS F4lines were detected using a general linear model (GLM) in GAPIT 3.0 [23].The SNP Taqman markers were spaced roughly every 5 cM along the 10 maize chromosomes.The phenotypes were estimated using best linear unbiased predictor (BLUP) values for the TF of 45 lines.The GWAS was calculated with data sets representing the following two distributions of transformation rate: a) including lines with > 5% compared to 0%,and b) including lines with TF > 5% compared to < 5%.QQ plots(Fig.S2A, B) show that the phenotypes did not exhibit a normal distribution.The Manhattan plot in Fig S2C and S2D shows the GWAS results across the 10 chromosomes of maize.
3.Results
3.1.Breeding scheme and marker assisted selection
To stack TF,agronomic performance,and high HIR into one line,we crossed two transformable parents (SYN-INBB23 and SYNINBC34) with the RWKS/Z21S//RWKS inducer to produce F2and BC1seeds segregating roughly 3:1 for R1-SCM2 purple aleurone(Fig.S1).Purple seeds were germinated and genotyped for qhir1,qhir8, R1-SCM2, and C1 (SYN-INBC34 only).The genotypic ratios from 5219 SYN-INBB23 and 3251 SYN-INBC34 individuals (Tables S2,S3)led to selection of 117 and 194 self-pollinations.Due to segregation distortion against matl and qhir8 in both populations,plants heterozygous for R1, matl or qhir8 were among those selected for the next generation, which enabled a wider genetic diversity but necessitated F3genotyping.In the SYN-INBC34 family all F3s were selected to be heterozygous or homozygous for the BC1 inducer allele near C1,to avoid carrying a suspected color inhibitor allele from SYN-INBC34.The selection efficiency of fixing these HI traits in SYN-INBB23 was 14.5% for the BC1and 1.3% for the F2; in SYN-INBC23 F2, it was 0.92%.
3.2.Haploid induction rate testing
F3or BC1F2generation families were sown at Syngenta breeding stations: 120 SYN-INCC23 and 187 SYN-INBC34 families were planted in quadruplicate with intervening pairs of hybrid tester rows.Genotyping for qhir8, R1, and C1 in SYN-INBC34 identified over 1500 individuals, including controls, for HIR test crossing(Tables S4, S5) following screening for agronomic performance,anthesis-silking interval, and pollen production phenotypes.The HIR of lines fixed for all haploid inducer alleles ranged from 10%to 19%,similar to the RWKS/Z21S//RWKS parent.In contrast,inducer materials without matl and qhir8 had lower induction rates(Table S4).
3.3.Transformation testing and HIR validation
Seed from the best performing F3/BC1F2individuals were sown the next season.HIR testing of 81 SYN-INBB23 and 71 SYN-INBC34 F4/BC1F3lines was performed using pollen bulks onto two testers each.A subset of lines from each population were also tested for TF(Fig.S1),using embryos pooled from 3 to 10 self-pollinated ears[21].Results from this F4generation testing are shown in Fig.1A and Table S6 for SYN-INBB23; Fig.1B and Table S7 for SYNINBC34.Few F4s were transformable, but their high HIR reflected the selective pressure applied in the F2and F3.In the non-stiff stalk set, fewer ears nicked with the inducers, so seed set was not used as an F4selection metric.Three lines from this population had a 10% or higher TF: 19BD915147, 19BD915875 and 19BD915158.The first two also had>15%HIR.Retesting HIR in the F5generation of these and related families led to identification of 3 lines(Table S8) with high HIR or evidence of transformability.These were forwarded to F6TF testing with CRISPR-Cas12a vectors.The derivatives of 19BD915875 and 19BD915158 had < 10% HIR and were dropped.
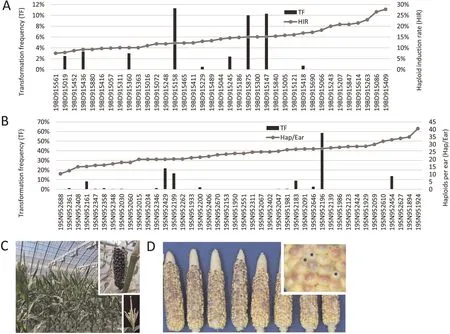
Fig.1.Elite HI-Edit line selection.(A)Transformation frequency(TF)and haploid induction rate(HIR)for F4 or BC1F3 lines from INBB23.(B)TF and haploids per ear for F4 lines from INBC34.(C) The F7 stage Iodent (21BD900056) crop habit, appearance, seed set after self-pollination (large inset) and tassel morphology (small inset).(D) Photo of representative set of ears from F5 out-crossing, showing full seed set and minimal embryo abortion.Inset, close-up showing of fertilized haploid or diploid kernels, and aborted kernels (black asterisks).
In the SYN-INBC34 population,the anthesis-silking interval was lower, so seed set was used as a selection metric.A wide TF range was observed (0 to 58.5%); 19SN952196 was the outlier at 58% TF with a 13% HIR and good seed set.Line 19SN952454 had a 13.7%TF, HIR > 15%, and good seed set.These lines averaged 27 and 33 haploids per ear, respectively, which is ten times higher than NP2222RS.F5seed lots from these lines were subjected to HIR and TF evaluation; the latter used a CRISPR-Cas12a construct to evaluate the HI-Edit rate across different varieties.Some embryos from 19SN952454 were provided BBM-assisted cotransformation vectors to test if that improved the TF.Three lines with high HIR but no TF data (19SN951958, 19SN952019, and 19SN952072) were also transformed in the F5stage with BBMassisted transformation.In isolated tests,BBM assisted transformation increased the TF of the parent inbred (Table S9).
3.4.Transformation with LbCas12a constructs to identify the top HIEdit lines
The next round of transformations used a Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR)-Cas12a cassette[16] with gRNAs targeting the Opaque2 (O2) gene [24] at the sequence CTGTATCTCGAGCGTCTGGCTGA, and gRNAs targeting Wx (Waxy1) [25] (GGGAAAGACCGAGGAGAAGATCT), Y1 [26](CTATCTTATCCTAAAGATGGTGG), and two predicted ubiquitin ligases, Zm00001d004139 (GGAGGGAAAAGGTGTCTGAGGC) and Zm00001d014920(GGAAGGAAAAGGTATCTGAAGG)(Fig.S3).Most lines were co-transformed sorghum WUSCHEL (cSbWUS-01) and Brassica napus BABYBOOM1 (cBnBBM1-02) cassettes to boost the TF [27], along with a CRE-LOX excision system to remove the WUS, CRE, and BBM1 cassettes after rooting.High HIR and seed set was also confirmed for all lines.The top F6derivative of SYNINBB23 (20BD917233) had a 15.8% HIR for 32.3 haploids per ear and a 1.5% transformation rate (Fig.S3) that was enhanced by BBM1+WUS co-transformation to 8.0%,or with the Brachypodium WUSCHEL alone to 7.0% (Fig.S3).
For the SYN-INBC34 families,the top line was less clear.The F5line 20ALL1134VG_MM was a strong inducer and had a 7% TF in the presence of BBM/WUS but exhibited leaf yellowing and other quality issues in field tests.Another transformable option was 20ALL1134VK_MM which had a 20.7% TF but a low 6% HIR in 2021 testing,despite the presence of matl and qhir8.As these failed performance criteria, we tested the HIR of selected F6lines from 19SN952454 and 19SN952196(Table S10),including ploidy confirmation of the higher HIR lines (Fig.S4).
Offspring lines from those bolded in Table S10 were tested for TF and HIR using a modified version of the first CRISPR construct where the gRNAs for O2, Y1, the ubiquitin ligases were replaced with a gRNA for Glossy2 [28] (GTCACAGATCACAAACTTCAAATG).We found that 21BD900077 had a 7% TF (Table S11) and 15%HIR, with strong seed set and 45 haploids/ear.In a parallel experiment,an F7-stage derivative(21BD900056)of the top Iodent inducer 20BD917233 produced 33 haploids per ear and a 4.2% TF(Table S11)without BBM or WUS enhancement.The HIR of the stiff stalk inducer was much higher than that of the Iodent inducer,but most of the events were sterile.From the HIR and HI-Edit rates(Table 1) of three events from the 21BD900056 Iodent inducer, it appears that this line can HI-Edit both stiff stalk and non-stiff stalk inbred lines, though the average haploid editing rate (HER) varies from event to event.The line has strong agronomic performance(Fig.1C)and seed set(Fig.1D).Table S12 summarizes the outcome of this work combining HI and transformability into an Iodent(21BD900056) and stiff stalk (21BD900077) chassis.
3.5.Transformation QTL discovery
In crop biotechnology, one challenge is the routine identification of transformable lines [14].To identify genetic loci associated with transformability, the SYN-INBC34 × RWKS//Z21S/RWKS F4materials transformed in Table S6 were genotyped by 480 polymorphic SNP markers spread evenly across the genome.GWAS analysis led to identification of a putative minor quantitative trait locus (QTL) for transformation ability on chromosome 3, between SM3158 and SM3814(Fig.S2;Table S13).Seven of the top 10 lines with TF>2%had the favorable genotype whereas 32 out of 36 lines showing a < 2% TF did not (Table S13).Comparing the set of lines with > 5% TF to those with 0%, SM4787 has a GWAS log10value of 1.7 and a P-value < 0.02, just above background (Fig.S2).Comparing lines with > 5% TF to those with < 5% TF, SM3814 has a GWAS log10value of 1.6,and SM4787 and SM3158 log10is 1.3,just above background.
To evaluate the relative importance of this QTL outside of this study’s biparental population, a panel of diverse lines were tested for TF and genotyped for 172 polymorphic qTF3.1 interval markers(Table S14).Ten out of 16 lines with TF>2%had the favorable haplotype for this QTL,including 8/9 lines that had a TF>10%,while all 7 showing<2%TF did not have it(Table S14).We cautiously posit that selection for plants having the beneficial allele at this QTL(qTF3.1) may enrich the resulting lines for transformability.qTF3.1 has not been identified in prior work.
4.Discussion
HI-Edit is an efficient method to deliver genome edits to commercial germplasm, but this new biotechnology platform requires the cross-breeding of rare maize traits: haploid induction and transformability.To explore if highly transformable elite haploid inducers could be developed through marker assisted selection and industrial-scale phenotyping operations, we developed two biparental populations between an elite inducer (> 15% HIR) and a transformable line from the stiff stalk (45% TF) and Iodent (5%TF) heterotic groups.After three generations of marker selection(F2, F3, and F4), three to five generations of haploid induction testing (F3through F7), and two to four generations of transformation testing(F4to F7),we identified one F7-stage line from each population that exceeded the target metrics for a commercial scale HIEdit platform, which were > 5% transformation rate, and >20 haploids/ear (combination of HIR and seed set).
These two inbred materials have a far superior combination of haploid induction performance and transformability than the first generation of HI-Edit germplasm.The same Cas12a vector used in the final testing of elite HI-Edit inducers,was transformed into the original parents,and the TF was comparable(Table S12).The stark difference in the editing efficiencies between the Waxy1 and Glossy2 gRNAs (Table 1) is common in the Cas12a system.It will be interesting to test new genomic targets and different gRNA design or expression methods in the new platform.
Fixation of haploid inducer and anthocyanin genes (qhir1 [3][matl],qhir8[4][dmp],R1-SCM2[12],and C1[19])via marker selection allowed setting of a high HIR baseline (> 7%) (Tables S4 and S5), whereas the loss of either qhir1 or qhir8 strongly reduced HIR.Importantly, the strategy of fixing R1-SCM2 and C1 in the inducer populations at an early stage allowed for a rapid and accurate selection on HIR due to the reliable dark purple marker expression it produced in the diploid embryos derived from the testcrosses.Notably, there was not much difference in between qhir1/qhir1;qhir8/+(~6.1%)and qhir1/qhir1;+/+sets(~5.9%),indicating that matl alone can give an HIR above 5% in maize (consistent with some prior reports [9] but in contrast to others [11])and that heterozygosity for qhir8 does not increase the induction rate of matl.Lines that were homozygous for matl had a lower TF compared to near-isogenic wild-type lines.
A variety of breeding schemes have been used to develop elite inducers haploid[29].One flaw in our approach of focusing on haploid induction QTL selection in the F2generation is that we narrowed the genetic diversity for transformability: as a result, only 10%of the SYN-INBB23 BC1F3/F4materials had a>5%TF,and those were all from F4rather than BC1F3populations which had 75%inducer genome.Thus,while making a BC to the inducer improved selection efficiency of progeny fixed for the HI genes, we reduced our chances of making a transformable inducer.We pursued thisscheme of prioritizing HIR traits because transformation testing is so resource intensive, and the transformation rate has lower heritability than HIR.The alternative strategy, to select first on transformation rate, then on HI traits, could be used to develop HI-Edit platforms in crops where the TF tests are scalable, and the transformation QTL are well-defined.A better way to approach the maize HI-Edit breeding could have been to BC to the transformable parent and use HIR marker assisted selection on purpleseeded BC1s and BC1F2s, with a goal of fixing matl and qhir8 in>200 BC1F3s.However,this approach may have yielded lower HIR.

Table 1 HI-Edit metrics from crosses between six Cas12a homozygous events.
GWAS analysis of the F4 stiff-stalk populations identified a candidate QTL for transformability on chromosome 3(Table S12),and the haplotype associated with this interval in the INBC34 parental line was found to be present in all but one maize germplasm with> 10% TF from the diverse panels tested at Syngenta and was absent in all but one maize germplasm with<5%TF.We also tested whether the BBM and WUS transformation-enhancer[27]morphogenetic factors can improve the transformability of the maize inducers, and found a significant improvement for lines that already had a moderate transformation rate (e.g., 19SN952821 and 19SN952822).However, these embryo regeneration factors were not able to convert non-transformable lines into transformable lines (e.g., for 19SN952871, 19SN952763, or 19SN9530981) (Table S8) in the small batches of experiments we performed.Protocol optimization for those specific lines and morphogenetic factor enhancers is needed to clarify if that recalcitrance may be overcome.
CRediT authorship contribution statement
Brent Delzer:Conceptualization, Investigation, Supervision.Dawei Liang:Conceptualization,Supervision,Data curation,Investigation.David Szwerdszarf:Investigation, Supervision.Isadora Rodriguez:Investigation, Supervision.Gonzalo Mardones:Investigation, Supervision.Sivamani Elumalai:Methodology, Supervision.Francine Johnson:Investigation.Samson Nalapalli:Investigation.Rachel Egger:Methodology, Supervision, Investigation.Erin Burch:Investigation.Kerry Meier:Methodology, Investigation.Juan Wei:Formal analysis.Xiujuan Zhang:Investigation.Huaping Gui:Investigation.Huaibing Jin:Investigation.Huan Guo:Investigation.Kun Yu:Investigation.Yubo Liu:Methodology,Investigation.Becky Breitinger:Data curation.Ana Poets:Formal analysis.Jason Nichols:Data curation.Wan Shi:Methodology.David Skibbe:Methodology.Qiudeng Que:Methodology.Timothy Kelliher:Conceptualization, Formal analysis, Methodology, Project administration.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The patent WO2022/212318 covering the information in this manuscript published in September 2023.
Acknowledgments
Thanks to Kristin Setliff and Anna Prairie for greenhouse and transformation support.Thanks to Tim Strebe for making the F1and F2generation crosses and leading the greenhouse team on the F2sampling effort that took place over the 2018 holiday break.Thanks to Dawn McNamara for seed support.Thanks to Donna Waite and Brad Waite for field support, including making crosses and shipping ears.Thanks to Rafaela Miranda Lunny Castro for sampling and to Ping Wu and Kayla Beam for preparing media.Thanks to Ian Jepson and Liang Shi for project support.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.10.016.
杂志排行
The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
