Flowering-time regulation by the circadian clock: From Arabidopsis to crops
2024-03-07MingkngYngWnjiLinYrouXuBiyuXiBiyinYuLingChnWiHung
Mingkng Yng, Wnji Lin, Yrou Xu, Biyu Xi, Biyin Yu, Ling Chn,c,d, Wi Hung,c,d,*
a State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Sciences, South China Agricultural University, Guangzhou 510642,Guangdong, China
b Henry Fok School of Biology & Agriculture, Shaoguan University, Shaoguan 512005, Guangdong, China
c Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, South China Agricultural University, Guangzhou 510642, Guangdong, China
d Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou 510642, Guangdong, China
e Guangdong Provincial Key Laboratory of Utilization and Conservation of Food and Medicinal Resources in Northern Region, Shaoguan University, Shaoguan 512005,Guangdong, China
Keywords:Circadian clock Photoperiod Short-day plants Long-day plants
ABSTRACT Precise timing of flowering in plants is critical for their growth and reproductive processes.One factor controlling flowering time is the cycle of light and darkness within a day, known as the photoperiod.Plants are classified into long-day,short-day,and day-neutral plants based on light requirements for floral initiation.Although the molecular mechanisms that govern this differentiation remain incompletely understood, studies have consistently shown that the circadian clock plays a central role in regulating photoperiod response across diverse plant species.However, there is a scarcity of reviews describing the regulatory network linking the circadian clock with photoperiodic flowering.This review summarizes that regulatory network, focusing on the distinct roles of clock genes in long-day and short-day plants.We also discuss the strategies of clock gene mutations contributing to geographic variation in longday and short-day crops.
1.Introduction
1.1.Photoperiodic flowering in plants
Flowering plants have developed complex mechanisms to align their vegetative and reproductive periods with seasonal variation[1].Photoperiod, the most dependable clue by which plants sense the time of the year for floral transition,was first described by Garner and Allard[2,3]in the 1920s.They found that artificially shortening day length led to an accelerated flowering response in‘‘Maryland Mammoth”, a tobacco strain distinguished by its plant height and biomass but whose commercial application is limited by the delay in flowering.Later, Hamner and Bonner [4] reported that the duration of the night is more critical for the photoperiodic response,as evidenced by the finding that a short pulse of red light at midnight inhibited flowering in tobacco and soybean.These experiments eventually led to the identification [5] of phytochrome (PHY), one of the photoreceptors involved in the nightbreak and photoperiodic response.It has long been known that flowering plants can be divided into three groups based on the response to day length: long-day (LD) plants, which flower earlier under LD conditions; short-day (SD) plants, which flower earlier under SD conditions;and day-neutral plants,which are insensitive to daylength variations in their flowering process.
Photoperiodic response mechanisms can be described by an external-coincidence model, the one best supported by current molecular studies (Fig.1) [6].In this model, the photoperiodic response is driven by the coincidence of the external light signal and the inherent rhythms [1,7].The model posits the presence of a light-triggered enzyme and a substrate regulated by the circadian clock.When exposed to light,the enzyme becomes activated,leading to its interaction with the substrate.The substrate undergoes diurnal oscillations, and induces the photoperiodic response only when its level passes a specific threshold,thereby limiting the process to a narrow time window.Alternatively, an internalcoincidence model [7] offers another explanation, proposing that the photoperiodic response is generated by the synchronization of two light-entrained regulators whose expression phases exhibit distinct sensitivities to photoperiod entrainment (Fig.1).
In plants responding to photoperiod, flowering signal culminates in the control of several critical flowering regulators.The most important one is FLOWERING LOCUS T (FT), the florigen gene responsible for inducing changes in gene expression that reprogram the shoot apical meristem(SAM)to trigger flower formation rather than leaf growth [8].FT is synthesized in leaves under inductive photoperiods and subsequently transported to the SAM[9].Once at the SAM, FT collaborates with FLOWERING LOCUS D(FD)to directly activate the expression of the floral meristem identity gene APETALA 1 (AP1), and indirectly stimulate the expression of SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), a MADS-box transcription factor that links floral induction [10,11].SOC1 interacts with AGAMOUS-LIKE 24 (AGL24) at the SAM, promoting the transcription of another meristem identity gene LEAFY (LFY) [12].Up-regulation of floral identity genes eventually leads to the initiation of flower development.

Fig.1.Models of photoperiodic response induction.The external-coincidence model proposes a hypothetical substrate whose expression is regulated by the circadian clock and slightly entrained by light.A photoperiodic response is triggered when the amount of substrate surpasses a required threshold,coinciding with the transformation of its corresponding enzyme from an inactive state(Ei)to an active state(Ea)upon exposure to light.In the internal-coincidence model,two hypothetical regulators are regulated by the circadian clock,but exhibit differing sensitivities to the entrainment of photoperiod.A photoperiodic response is triggered when the overlap of the regulators exceeds a critical threshold.
1.2.Circadian clock in plants
In the external- and internal-coincidence models, the crucial factors in induced photoperiodic response are regulated by the circadian clock,highlighting that a self-sustained circadian rhythm is necessary for day-length measurement.The plant circadian clock is an endogenous molecular oscillator that confers control of the circadian rhythm of plants across a roughly 24-h period [13].The clock regulates physiological processes including photosynthesis,growth, flowering, and responses to biotic and abiotic stresses, to ensure efficient resource utilization at optimal times during the day [13].The input pathway is a regulatory mechanism that facilitates the synchronization of the circadian clock with external cues such as light and temperature.Light signals that orchestrate this entrainment rely on a series of photoreceptors:the red-light receptors PHYs and blue-light receptors CRYPTOCHROMES(CRY),ZETLUPE(ZTL), FLOWERING KELCH FACTOR 1 (FKF1), and LOV KELCH PROTEIN2 (LKP2) [14].These photoreceptors regulate both the phase and amplitude of the circadian clock by directly interacting with clock components or engaging downstream signal transduction pathways.
The core oscillator in plants is composed of several transcription activators and repressors that establish a complex network of intertwined feedback loops.The morning-expressed transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) repress the expression of PSEUDORESPONSE REGULATOR (PRR) family genes [15–17].In turn, PRR9,PRR7, PRR5, and PRR1 (also known as TIMING OF CAB EXPRESSION1, TOC1), which are expressed sequentially from morning to afternoon,repress the expression of CCA1 and LHY,thereby closing the feedback loops [18,19].CCA1/LHY suppress the expression of the components of evening complex (EC), EARLY FLOWERING 3(ELF3), ELF4, and LUX ARRHYTHMO (LUX) [20–22].EC, in turn, suppresses multiple clock genes, including PRR9, PRR7, TOC1, and GIGANTEA (GI), and indirectly promotes the expression of CCA1 and LHY, thus completing additional feedback loops within the intricate network [23].REVEILLE 8 (RVE8, also known as LHYCCA1-LIKE 5 or LCL5) acts as a transcriptional activator of PRR5,TOC1, and ELF4 to strengthen circadian rhythms [24,25].GI, a plant-specific protein, induces the expression of CCA1/LHY by an unidentified mechanism and is essential for circadian timekeeping[26].
2.Mechanism of clock-gene control of flowering time in Arabidopsis
Most of the molecular knowledge about the floweringregulatory mechanism of the circadian clock stems from genetic and molecular studies conducted in Arabidopsis thaliana.In this LD plant,a transcriptional activator CONSTANS(CO),is at the heart of the molecular network that induces FT expression in a photoperiod-dependent manner [27].Consistent with the external-coincidence model, FT is activated in the LD afternoon when the stabilization of CO protein in the light phase coincides with the peak expression of CO mRNA [1].The circadian clock is intricately involved in the regulation of both processes.The clock component GI participates in forming the GI-FKF1 complex,which is activated by light,leading to an increase of CO expression under LD conditions[28].Simultaneously,the circadian clock restricts the activity of CO in the afternoon via regulatory mechanisms at both the transcript and protein levels [29].Beyond the GI-CO-FT cascade,clock genes exert their influence on flowering by modulating the other flowering regulators SOC1,SVP,TEMPRANILLO 1(TEM1),TEM2, and PIF4, forming an intricate regulatory network (Fig.2).
CCA1 is an MYB-like transcription factor whose overexpression causes circadian rhythm disruption and delays flowering [30].CCA1 prolongs the flowering time of Arabidopsis by directly repressing the expression of the flowering activators GI, SOC1,and FT.During the daytime, CCA1 binds to the CCA1-binding site(CBS) in the promoters of SOC1 and GI, thereby decreasing their expression [21].CCA1 binds directly to the CBS and evening element (EE) in the FT promoter to impede its transcription [31].LHY encodes a flowering repressor closely related to CCA1, and its gain-of-function mutant delays flowering [32].LHY and CCA1 have partially redundant roles in flowering control, as the cca1 lhy double mutant showed an earlier flowering phenotype than the individual single mutants [33].The mechanism by which LHY influences flowering regulation is unclear, but it is suggested [31]that LHY exerts its effects by modulating circadian rhythms.Under continuous-light conditions, LHY/CCA1 promotes flowering by inhibiting the SHORT VEGETATIVE PHASE (SVP) protein through a mechanism independent of the conventional photoperiodic pathway[34].RVE8,another MYB-like gene that regulates the circadian rhythm antagonistically to LHY/CCA1,is also implicated in controlling circadian rhythm and flowering time [35].In contrast to the profound impact of CCA1 and LHY on flowering, the effects of RVE8 are relatively moderate, as evidenced by the slightly earlier flowering of rve8 mutant plants and the delayed flowering of RVE8-overexpressing plants [35].
PRR9,PRR7,PRR5,and TOC1 positively regulate flowering time in Arabidopsis[36].The double and triple mutants of PRR9,PRR7,and PRR5 genes display an extreme late-flowering phenotype despite the relatively moderate flowering-time changes in PPR single mutants and overexpression plants [36–38].All four PRR genes physically interact with CO, increasing the stability of CO during the daytime as PRR9, PRR7, PRR5, and TOC1 are expressed sequentially at 2–3 h intervals from dawn to dusk [38].CO is targeted by the E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) for degradation in darkness.However,it is protected in the light phase when the activity of COP1 is suppressed by the photoreceptors PHYA, CRY1, and CRY2 [39,40].The mutant of COP1 is epistatic to toc1 prr5 prr7 prr9 in CO accumulation, indicating that PRRs contribute to reducing COP1 activity in addition to photoreceptors[38].PRR9, PRR7, and PRR5 directly repress the expression of CYCLING DOF FACTOR (CDF) family genes, including CDF1, CDF2,CDF3, and CDF5, which are transcriptional repressors of CO[36,41,42].
Mutations in ELF3,ELF4,and LUX genes result in an acceleration of flowering time compared to the wild type, indicating that they function as flowering repressors [43–45].LUX is responsible for DNA binding activity in the EC, while ELF3 functions as a protein scaffold to facilitate the interaction between ELF4 and LUX [46].ELF4 functions to increase the stability of EC, particularly under elevated temperatures [47].The regulatory process governing flowering time involves direct binding of the EC to the GI promoter,inhibiting its transcriptional activity[48].This mechanism includes the direct interaction of ELF3 and LUX with the components of the histone-modifying complex,HISTONE DEACETYLASE 9(HDA9)and WD40 repeat-containing protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15(HOS15), which are linked to histone deacetylation at the GI promoter [49,50].EC medicates heatinduced flowering by directly repressing the transcription of PHYTOCHROME INTERACTING FACTOR4 (PIF4), a transcriptional activator of FT, in response to temperature changes [46,51].The activity of EC is reduced at high temperatures owing to the liquid–liquid phase separation of ELF3,which serves as a direct thermosensor, subsequently leading to an increase in PIF4 expression[47,52].At post-transcriptional levels,ELF3 bridges the interaction between COP1 and GI and facilitates the COP1-mediated degradation of GI [53].However, COP1 can also target ELF3 for its ubiquitination and degradation [53].In a study [54] under natural longday conditions, ELF3 interacted with CO to promote its degradation, suggesting its involvement in the COP1-CO protein complex.ELF4 directly interacts with and recruits GI to nuclear bodies,thereby sequestering GI from its association with the CO promoter[55].

Fig.2.The flowering regulation network of clock genes in Arabidopsis.The left panel shows the molecular mechanism controlling flowering by clock genes.Clock genes are positioned roughly according to their expression time throughout the day, with yellow and gray shading indicating the light and dark phases, respectively.The rounded rectangles indicate proteins,with floral activators in red and floral repressors in blue.Arrows indicate transcriptional activation;perpendicular bars indicate transcriptional repression; dashed lines indicate antagonism.Placement of proteins together indicates direct interaction.Ub indicates ubiquitin-dependent degradation.The right panel shows the hierarchy inferred from genetic analysis.
GI was one of the first genes isolated from flowering mutants in Arabidopsis during the 1960s[56].Genetic analysis showed that gi flowers much later than the wild type under LD conditions, displaying a phenotype similar to those of co and ft[57].GI promotes flowering by increasing the expression of CO and FT, independent of its effect on the circadian rhythm [58].A typical example of the external-coincidence model is provided by the FKF1-GI complex and its induction of CO expression.FKF1 is an F-box protein involved in the SKP1-Cullin-Rbx1-F-box protein (SCF) E3 ligase complex, with expression peak in the afternoon [59,60].FKF1 interacts with the GI through the light-activated LOV (Light, Oxygen, or Voltage) domain in a blue light-dependent manner [28].Under LD conditions,the formation of the FKF1-GI complex is promoted by the light phase and the synchrony of FKF1 and GI expression in the afternoon.FKF1-GI complex targets CDF1 and CDF2 proteins for ubiquitin-dependent degradation via the interaction between GI and CDF proteins,resulting in an increase in CO mRNA expression [28,61,62].In contrast, under SD conditions, the interaction between FKF1 and GI is hindered by the asynchrony between the light period and the expression phase of GI and FKF1 [28].GI directly activates FT mRNA expression by binding to the promoter of FT and forms complexes with several FT repressors, including SVP, TEMPRANILLO 1 (TEM1), and TEM2 [63].
3.Interaction between clock genes in control of flowering time in Arabidopsis
Genetic studies revealed that the functions of most clock genes in flowering regulation are independent (Fig.2, right panel).Among the MYB-like transcription factor, flowering time is advanced in cca1 lhy plants and slightly delayed in rve4 rve6 rve8 plants(a phenotype opposite to that of rve8).However,the flowering time of the quintuple mutant resembles that of the wild type,suggesting that LHY-clade and RVE8-clade genes act antagonistically on flowering [24].Among the core feedback loops, genetic analysis of cca1 lhy toc1 and its parents showed [64] that cca1 lhy has a more predominant role in flowering control than toc1,consistent with the finding that GI expression is advanced in cca1 lhy toc1 and cca1 lhy, but not toc1.The slight change in flowering time observed in toc1 might be attributed to the compensatory functions of other PRR genes.Another study[36]conducted by crossing cca1 lhy with prr7 prr5 showed that the quadruple mutant flowered at a time intermediate between that of its parents,suggesting that PRR genes act parallel with and antagonistically to CCA1/LHY.Both CCA1-ox and elf3 display an elongated hypocotyl but opposite flowering-time phenotypes.Genetic study of CCA1-ox elf3 showed that CCA1-ox and elf3 exert additive effects on flowering, but ELF3 plays a more dominant role in controlling the expression of flowering promoting genes, including SOC1, GI, CO, and FT [21].
GI acts epistatically to many clock genes.The flowering time of cca1 lhy gi is highly similar to that in gi,but not cca1 lhy,indicating that the influence of CCA1 and LHY on flowering is dependent mainly on GI activity [58].ELF3 and ELF4 regulate GI activity via both EC-dependent and -independent pathways.Consistent with this regulation, gi is genetically epistatic to elf3 and elf4, as evidenced by genetic studies showing that elf3 gi [65] and elf4 gi[66] double mutants exhibit flowering times similar to those of the gi single mutant.But another study [67] suggested an alternative perspective: under LD conditions, GI is epistatic to ELF3,whereas,under SD conditions,ELF3 acts as an epistatic factor over GI.In distinction to the formation of feedback loops in circadian rhythm regulation, clock genes control flowering time through a linear hierarchy in which clock genes engage in GI-CO-FT pathway.In either case, as depicted in the left panel of Fig.2, the multilayered regulatory network may explain the rarity of observing epistatic effects among clock genes, except for GI.
4.Distinct roles of clock genes in flowering-time regulation in LD and SD plants
Conserved clock components, including those belonging to the CCA1/LHY, PRR, EC, and GI clades, are present across the range from green algae to higher plants [68].Although the families of clock genes have expanded during plant evolution, numerous homologs in angiosperms have maintained consistent functions in photoperiodic flowering [69].However, clock genes exert control over flowering time via diverse mechanisms in different plants,and the activities of clock genes in flowering regulation are opposite in LD and SD plants,even within phylogenetically close species(Table 1).
4.1.CCA1/LHY homologs
In the LD plant Medicago truncatula, loss of function of MtLHY results in early flowering, suggesting that MtLHY is a flowering repressor that is equivalent to its homologs in Arabidopsis [70].In contrast, in the SD plants soybean(Glycine max) and rice (Oryza sativa),the flowering activities of LHYs are opposite to their homologs in LD plants.Loss of function of GmLHY1a/1b/2a/2b (also known as LHY-CCA1-LIKEs, LCLs) postpone flowering in soybean[71,72].The mutation of OsLHY (also known as OsCCA1 or Nmediated heading date1,Nhd1)generally causes a delay in heading date [73–75].However, an early-heading phenotype is also observed in oslhy under SD conditions, suggesting that OsLHY may play a bifunctional role in heating regulation depending on photoperiod [73].Although GmLHYs and OsLHY promote flowering,their mechanisms are different.GmLHYs bind to the promoter and inhibit the expression of E1,the crucial transcriptional repressor of florigen GmFT2a and GmFT5a [72].OsLHY directly controls the expression pattern of OsGI by a mechanism similar to those in Arabidopsis [73].OsLHY directly activates the expression of the florigen Hd3a while repressing the expression of floral repressors OsPRR37 and DTH8[74,75].In addition to flowering,the functions of CCA1/LHY are associated with agronomic traits including nodulation [70], nitrogen assimilation [74], tiller growth [76],and drought tolerance [77].In particular, the activities of the CCA1/LHY homologs for the regulation of nitrogen assimilation are positive in Medicago [70] and negative in rice [74] and Arabidopsis [78], indicating that those functions do not differ in LD and SD plants as does photoperiodic flowering.

Table 1 Clock homologs that alter flowering time and their association with agronomic traits.
4.2.PRR homologs
Loss of function of PRR homologs usually causes late flowering in LD plants but early flowering in SD plants.In the LD plant barley(Hordeum vulgare), Photoperiod-H1 (HvPpd-H1 or HvPRR37)induces flowering under LD conditions by positively regulating HvFT1, while HvELF3, HvLUX1, and HvPHYC repress HvPpd-H1 expression during the night [79–82].In another LD plant, sugar beet(Beta vulgaris),BOLTING TIME CONTROL 1(BvBTC1 or BvPRR7)also promoted flowering, acting like its homolog in Arabidopsis and barley [83].BvBTC1 acts with BvBBX19 to repress BvFT1 and activate BvFT2 under LD conditions [84].It is conceivable that BvBTC1 physically interacted with BvBBX19, as an interaction between AtPRRs and AtBBX19 proteins has been shown [85] in Arabidopsis.In contrast, in the SD plant rice, both OsPRR37 (also known as Hd2) and OsPRR73 act as flowering suppressors by repressing the expression of Early heading date 1 (Ehd1, a direct activator of Hd3a) and Hd3a [86–88].Overexpression of OsTOC1 delays heading date, indicating that it is a flowering suppressor,but the underlying mechanism remains unknown [89].In another SD plant, soybean, GmPRR3a/Tof11 and GmPRR3b/Tof12 act as flowering repressors by directly down-regulating GmLHYs genes to modulate E1 expression [72].
4.3.EC homologs
The functions of EC components have been extensively characterized in plant species because loss-of-function mutants strongly affect flowering phenotype.Floral repression has been identified as one of the functions of EC components in LD plants.In barley,mutation of HvELF3(also known as EAM8)and HvLUX1(also known as EAM10)leads to early flowering by up-regulating the expression of Ppd-H1, HvCO1, and HvFT1 [81,82,90].Similarly, in peas (Pisum sativum),mutations of HR/PsELF3,DNE/PsELF4,and SN/PsLUX result in the elevation of FT family gene expression, leading to an earlyflowering phenotype [91–93].In Medicago, mutation of MtLUX leads to the up-regulation of MtFTa1 expression and early flowering,but the mechanism is unclear[94].In SD plants,in distinction from flowering inhibition in LD plants, the homologs of the EC components are floral activators, and their mutations usually result in extremely late flowering.The oself3-1 oself3-2 double mutant and oslux mutant completely failed to flower in rice [95].Although the effect of OsELF4 is weaker than those of its partners,it still shows the ability to induce flowering in rice[96].OsEC binds to and inhibits the transcription of flowering repressors such as
OsGI, OsPRR37 and Grain number, plant height and heading date 7(Ghd7) [95,96].In soybean, the gmlux1 gmlux2-1 double mutant exhibits extremely late flowering under short days, resulting in a huge amount of biomass; for this reason, the name ‘‘Guangzhou Mammoth”is given in homage to the Maryland Mammoth tobacco[97].The EC components J gene/GmELF3 and GmLUX promote flowering by directly repressing the expression of E1 [97,98].In maize (Zea mays), ZmELF3.1, ZmELF3.2, ZmLUX1, and ZmLUX2 form a complex that promotes flowering, with ZmELF3.1 and ZmLUX1 exerting the largest effects.ZmEC directly inhibits the expression of the flowering suppressor genes ZmCCT9, ZmCCT10,ZmCOL3, ZmPRR37a, and ZmPRR73 [99].
4.4.GI homologs
In the LD plant pea,LATE1/PsGI promotes flowering by inducing the expression of FT-like genes [100], whereas in SD soybean and maize, GmGI and ZmGI1 act as flowering repressors by downregulating the expression of FT-like genes[101,102].The molecular mechanism by which OsGI represses flowering provides an example of flowering regulation in SD plants.In rice,OsGI has been identified [103] as a target of PHOTOPERIODIC SENSITIVITY 5 (SE5), a heme oxygenase involved in phytochrome–chromophore biosynthesis.Overexpression of OsGI results in delayed flowering under both SD and LD conditions [104].However, mutation of OsGI results in both early and delayed flowering depending on the photoperiod and temperature conditions [73,96,105].Consequently,OsGI is also recognized as a dual-function floral regulator in rice.Like Arabidopsis GI, OsGI physically interacts with OsFKF1 and OsCDF1, promoting the expression of Heading date 1 (Hd1), a rice homolog of Arabidopsis CO [104,106].Although the GI-CO/Hd1-FT/Hd3a pathway is conserved, Hd1 acts as a repressor of Hd3a expression in rice under LD conditions[107].This divergence offers an explanation for the differing photoperiodic responses in rice and Arabidopsis [104].OsGI functions in flowering regulation by modulating a Ghd7-Ehd1-Hd3a pathway, a mechanism absent in Arabidopsis but conserved in SD monocots [108].OsGI antagonizes the function of OsPHYs in interacting with Ghd7, leading to the facilitation of Ghd7 protein degradation [109,110].
5.Conservation of protein activities of clock components in plants
Although clock components display distinct activities for photoperiodic flowering in LD and SD plants,heterologous transformation of clock genes has been shown to rescue the phenotypic defects of corresponding mutants, even across opposite types of photoperiodic response plants.OsPRR37 and OsTOC1, clock genes of the SD plant rice,complemented atprr7-11 and attoc1-2 mutants and restored their normal hypocotyl length,circadian rhythm,and flowering time in the LD plant Arabidopsis [111].The maize clock gene ZmGI1 rescued the flowering time delay in atgi-101[102],and overexpression of ZmCCA1a and ZmCCA1b in Arabidopsis leads to a delay in flowering,similar to the role of AtCCA1[112,113].The soybean clock gene GmLUX2 perfectly complemented the atlux-6 mutant in regulating flowering time [114].
Clock components are well-conserved in protein activities,including binding DNA motifs, interacting partners, and transcriptional activities.It has been shown that LHY homologs bind to EE and CBS motifs [73,74,115], while PRR homologs bind to the Gbox motif [72,116,117] in Arabidopsis, rice, and soybean.LUX homologs bind to LUX-binding sites(LBS)in Arabidopsis,Medicago,rice and maize [47,96,99].Formation of a CCA1/LHY dimer and EC has been identified in diverse species [46,76,77,96,97,99,118].These observations suggest that the divergence of photoperiodic response between LD and SD plants may be attributed to the evolution of downstream flowering-regulatory networks rather than the protein activities of the core clock components.
6.Modification of clock genes to improve geographical adaptability in LD and SD crops
An endogenous period that coincides with the external light–dark cycle confers survival advantages for plants.The biomass and photosynthesis of wild-type Arabidopsis were maximized in 24-h cycles, whereas toc1-1 (circadian period ≈20.7 h) and ztl-1(circadian period ≈27.1 h) showed a photosynthetic advantage in 20-h and 28-h cycles,respectively[119].In day-neutral tomato,the delayed phase and the extended period of the circadian rhythm are associated with adaptation to high latitudes, caused by artificial selection of the mutations of EMPFINDLICHER IM DUNKELROTEN LICHT 1 (EID1) and NIGHT LIGHT-INDUCIBLE AND CLOCKREGULATED GENE 2 (LNK2) genes during domestication [120].This association between circadian period and latitude is also observed in Arabidopsis, soybean, and monkeyflower (Mimulus guttatus),suggesting that a slower clock may take advantage of longer day length in summer [121,122].
Despite the loss of function leading to disruptions in circadian rhythms and insensitivity to photoperiod,many clock components have been identified as targets of crop domestication, with their weak/nonfunctional alleles contributing to the enhancement of agronomic traits(Table.1)[123].A possible explanation is that certain survival disadvantages of clock mutants can be circumvented in crops by their cultivation in fields under human management,which provides them with reduced exposure to unfavorable conditions and diminished competition from other species.This hypothesis is supported by the observation that nonfunctional alleles of clock genes are consistently identified in cultivated varieties,while functional alleles are prevalent among wild-type varieties[72,79,83,91,98,124].Moreover, artificial selection is more likely to select crop varieties with clock gene mutations that exert lessadverse effects on agronomic traits (Table 1).
Clock damage modifies flowering time by reducing sensitivity to photoperiod and temperature, thereby permitting adaptation to climatic conditions that differ from the natural habitat of the wild type [83].LD and SD crops with clock gene defects seem to adopt different strategies to improve their geographic adaptation(Fig.3; Table 1).In SD crops, modifications of clock genes tend to increase latitudinal adaptability.The diminished function of floral activators (such as OsCCA1 and J) commonly facilitates the expansion of crops from higher to lower latitudes [75,98].In contrast,gain-of-function mutations of floral activators (such as OsELF3-1 and ZmELF3.1) or loss-of-function mutations of floral repressors(such as OsPPR37, GmPPR3a, and GmPPR3b) promotes the spread of crops to higher-latitude regions [72,86,99,124].Because of the rotation of the earth on its tilted axis, the summer day length is shorter at lower than at higher latitudes,leading to early flowering of SD crops.Selective breeding for mutations in floral activators prevents premature flowering, extending the vegetative growth phase and ultimately increasing yield [98].
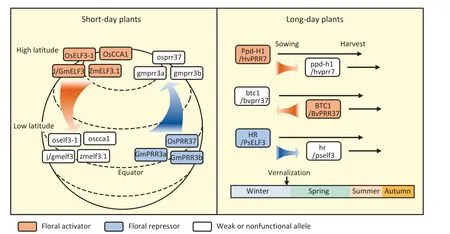
Fig.3.The strategy of clock-mediated geographic expansion in LD and SD plants.The defect of clock genes in SD plants contributes to latitudinal adaptation,and that in LD plants contribute to seasonal adaptation.The rounded rectangles indicate crop varieties carrying corresponding clock genes,with floral activators in red,floral repressors in blue,and weak/nonfunctional alleles in gray.The red and blue arrows indicate habit changes caused by the mutations of clock genes and black arrows roughly indicate the life cycle of the varieties.Os, Oryza sativa; Gm, Glycine max; Zm, Zea mays; Hv, Hordeum vulgare L.; Bv, Beta vulgaris; PS, Pisum sativum.
In LD in contrast to SD crops, mutations of clock genes tend to increase their adaptation to multiple seasons and modify their life cycles (Fig.3; Table 1).Mutations in HR/PsELF3 and HvPpd-H1 determine the spring-sown habit in cultivated varieties of peas and barley, respectively, in contrast to the winter-sown habit of wild-type varieties [79,91].The winter habits of biennial beets are controlled by a nonfunctional allele of the BvBTC1 gene [83].Although BvBTC1 and HvPpd-H1 are floral activators that are homologous to Arabidopsis PRR7, their differing effects on the spring–winter habit transition may be associated with the difference in growth habit between wild-type beet and barley.The transition from winter-sown to spring-sown habit usually accompanies the reduction of the vernalization requirement,implying potential interactions of clock genes with the vernalization pathway.In contrast to FLOWERING LOCUS C(FLC) in Arabidopsis, the core components of the vernalization pathway in barley include VERNALIZATION1 (VRN1), a homolog of AP1, and VRN3, a homolog of FT [125].The overlap of core components of floral initiation and vernalization may explain how clock genes influence both processes in LD crops.Although the mRNA expression of both PRR7 and ELF3 in Arabidopsis is subject to temperature influence[126], the precise interactions between the circadian clock and the vernalization pathway remain to be elucidated.
7.Discussion
Clock genes control photoperiodic flowering via species-specific mechanisms.The flowering activities of clock genes and the strategies employed for crop domestication by clock genes are contrasting between LD and SD plants, and these distinctions are even more varied than those observed between eudicots and monocots[127](Table 1).This finding suggests that clock genes could potentially serve as novel genetic markers to differentiate between LD and SD plants.The finding that clock-gene homologs exhibit conserved functions in heterologous transformations between LD and SD plants suggests that distinct photoperiod responses may be mediated primarily by the varying floral targets of these clock genes.Among these factors,variation in the activities of transcription factors targeting florigen emerges as critical: LD plants use transcriptional activators such as CO in Arabidopsis as master florigen regulators, whereas SD plants rely on transcriptional repressors like Hd1 in rice and E1 in soybean.As for day-neutral plants,the precise role of core clock genes in flowering control remains undetermined, but promotion by the slow clock of adaptation to high latitudes is known in day-neutral tomato [120].The mechanisms by which circadian rhythm promotes geographical adaptability in other LD and SD plants remain to be established.The convergent evolution leading to various types of photoperiodic response plants and the flowering network mediated by clock genes, particularly in crop species, awaits further study.
The multi-layered regulatory network connecting the circadian clock and flowering offers abundant targets for crop domestication.Gene mutations of various types in clock genes,including promoter insertions, codon substitutions, frameshift mutations, and premature stops, have been observed in crop domestication[72,75,79,83,86,91,98,99,124].Among the circadian clock components, the homologs of PPR7 and ELF3 have emerged as the most frequently identified quantitative-trait loci (QTL) for photoperiod response and geographic expansion (Fig.3).These genes exhibit high expression levels around the light–dark transition and are of ancient origin, possibly accounting for their strong functionality in photoperiod response [68].Based on current understanding, a promising strategy for SD crop improvement involves targeting PPR7 homologs to facilitate their introduction in high-latitude regions and targeting ELF3 homologs for low-latitude regions.In LD crops,modifying either PPR7 or ELF3 homologs has the potential to induce a shift in growth behavior from annual to biennial as an adaptation to varying climatic conditions.But it is imperative to evaluate the potential impact on agronomic traits before the manipulation of clock genes—mutations of PRRs and EC components in rice lead to a reduction in salt tolerance (Table 1).Given this caution, the exploitation of clock-gene targeting holds great potential as a robust approach to manipulating flowering time in the pursuit of higher crop yield and quality.
CRediT authorship contribution statement
Mingkang Yang:Formal analysis, Visualization, Writing–original draft.Wenjie Lin:Formal analysis,Visualization,Writing– original draft.Yarou Xu:Conceptualization, Writing – review &editing.Biyu Xie:Conceptualization, Writing – review & editing.Baiyin Yu:Conceptualization, Writing – review & editing.Liang Chen:Conceptualization, Writing – review & editing.Wei Huang:Funding acquisition, Supervision, Conceptualization, Writing –review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Laboratory of Lingnan Modern Agriculture Project (NZ2021001), State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKICUSAa202007), and Natural Science Foundation of Guangdong Province(2022A1515011027, 2021A1515012148), and the Double Firstclass Discipline Promotion Project (2023B10564004).
杂志排行
The Crop Journal的其它文章
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
- A telomere-to-telomere genome assembly of Zhonghuang 13,a widely-grown soybean variety from the original center of Glycine max
