Transcription factor OsSPL10 interacts with OsJAmyb to regulate blast resistance in rice
2024-03-07ZofZhongLijingZhongXingZhuYiminJingYihongZhengToLnHitoCuib
Zof Zhong, Lijing Zhong, Xing Zhu, Yimin Jing, Yihong Zheng, To Ln*, Hito Cuib,,*
a College of Life Science, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
b Key Laboratory of Genetics, Breeding and Multiple Utilization of Crops, Ministry of Education, College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
c Fujian Provincial Key Laboratory of Crop Breeding by Design, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
d College of Plant Protection, Shandong Agricultural University, Tai’an 271018, Shandong, China
Keywords:Immunity Jasmonate Oryza sativa OsSPL10 Transcription factor
ABSTRACT Transcription factors (TFs) play essential roles in transcriptional reprogramming during activation of plant immune responses to pathogens.OsSPL10 (SQUAMOSA promoter binding protein-like10) is an important TF regulating trichome development and salt tolerance in rice.Here we report that knockout of OsSPL10 reduces whereas its overexpression enhances rice resistance to blast disease.OsSPL10 positively regulates chitin-induced immune responses including reactive oxygen species(ROS)burst and callose deposition.We show that OsSPL10 physically associates with OsJAmyb,an important TF involved in jasmonic acid (JA) signaling, and positively regulates its protein stability.We then prove that OsJAmyb positively regulates resistance to blast.Our results reveal a molecular module consisting of OsSPL10 and OsJAmyb that positively regulates blast resistance.
1.Introduction
Rice production is seriously affected by blast disease caused by the fungal pathogen Magnaporthe oryzae.This disease inflicts an annual yield loss of 10%–30%worldwide,posing a significant threat to global food security.Compared with application of pesticides,breeding disease-resistant cultivars with continuing genetic improvement in yield potential is the most economical and environmentally friendly approach to prevent blast and other diseases[1].To develop effective and durable resistance to blast, great efforts have been made to identify resistance (R) genes and quantitative trait loci (QTL) in rice, and to unravel immune signaling networks in response to infection by M.oryzae.
In the long-term arms race between pathogens and hosts,plants have evolved two-layered innate immune system consisting of pathogen-associated molecular pattern-triggered immunity(PTI) and effector-triggered immunity (ETI) [2].Both PTI and ETI immune signaling pathways converge in the nucleus and activate defensive transcriptional reprogramming[3].Transcription factors(TFs) are key regulators controlling gene expression.Several families of TFs, such as WRKY, MYB, ERF, and TGA, play essential roles in mediating immunity in plants [4].
SQUAMOSA promoter-binding protein (SBP) and SBP-like (SPL)proteins containing a plant-specific SBP domain are putative transcription factors [5].In rice, the SBP-box family contains 19 members [6], most of which were shown to be involved in regulating aspects of plant development, including tiller number, plant height, heading date, number of spikelets per panicle, number of grains per panicle, grain size, and grain shape [7,8].One of them has been intensively studied;OsSPL14,also named IPA1(ideal plant architecture1), is regulated by microRNA OsmiR156.Remarkably,elevated IPA1 protein levels enhance blast resistance[9].IPA1 promotes both yield and blast resistance through a phosphorylationdephosphorylation cycle at amino acid Ser163within its DNA binding domain.IPA1 is an excellent natural variation of OsSPL14 and has been widely used in rice breeding [10,11].Similarly, overexpression of OsSPL7,a close homologue of IPA1,enhances rice resistance against bacterial blight[12].The functions of other SPL genes in regulating rice immunity are still to be determined.
We recently reported that OsSPL10 plays essential role in trichome development,and negatively regulates salt tolerance in rice[13].In this study,we investigated the role of OsSPL10 in resistance to blast.We found that knockout of OsSPL10 reduced the level of resistance whereas overexpression enhanced resistance.OsSPL10 positively regulated immune responses, such as ROS production and callose deposition.Furthermore, we identified transcription factor OsJAmyb, an important positive regulator in JA signaling,as an interactor of OsSPL10.We show that OsSPL10 likely supports accumulation of OsJAmyb by promoting its stability.We also found that OsJAmyb positively regulates blast resistance.
2.Materials and methods
2.1.Plant and fungal materials
Cultivar (cv.) Zhonghua 11 (ZH11, Oryza sativa L.ssp.japonica)and Nipponbare (NIP, O.sativa L.ssp.japonica) were the wild type(WT)rice in this study.The Osspl10 knock-out and OsSPL10 overexpression lines (OsSPL10-OE) in ZH11 background, OsJAmyb mutants (Osjamyb#2, Osjamyb#5) and OsJAmyb overexpression lines(OsJAmyb-OE#3 and#4)in NIP background were previously described [13,14].Seedlings were grown in a growth chamber at 28°C under a 12 h light and 12 h darkness cycle.M.oryzae strains Guy11,Zhong1,and GFP-labeled Zhong1 were cultured on CM agar plates for 5 days and then on SRB medium for conidia production for 1 week at 28 °C in darkness.Conidia were induced by white light (20,000 lx) for 2 to 3 days after removing surface mycelium.
2.2.Pathogen inoculation
Three-week-old rice plants were sprayed with M.oryzae conidial suspensions in water plus 0.02% Tween-20 at a concentration of 3 × 105conidia mL-1.Punch-inoculation assays were made on four-week-old rice leaves using 10 μL of conidial solution (105conidia mL-1).Following both inoculation procedures the plants were placed in a dew growth chamber at 26°C for 24 h in darkness,and then grown at 26 °C with 12 h/12 h photoperiod and 90% relative humidity for 4–7 days for development of disease symptoms.The symptoms on the leaves were imaged and the relative fungal growth was determined using quantitative PCR to measure the relative amount of MoPot2 transposon DNA by normalizing to the rice reference gene OsUBQ (LOC_Os03g13170).The PCR primers are listed in Table S1.
The rice sheath inoculation assays with M.oryzae strain Zhong1 labeled by GFP were performed as previously described with a slight modification [15].Conidial suspensions (105mL-1) were injected into the inner leaf sheath using a syringe,and then the leaf sheaths were incubated at 28°C in darkness.Fungal growth in the leaf sheaths after 24 h and 48 h was determined using confocal microscopy (LSM880; Zeiss).
2.3.DAB staining and callose deposition
3,3′-Diamino-benzidine (DAB) (Sigma-Aldrich, St.Louis, MO,USA) was used for H2O2staining according to a previously described method [16].Rice tissue stained in 10 mL DAB solution[1 mg mL-1DAB, 10 mmol L-1MES, pH 3.8 with 0.2% (v/v)Tween-20] was vacuumed for 30 min to eliminate air from the plant tissues.Staining was continued for 8 h before de-staining with 95% ethanol in a boiling water bath until the tissues were transparent.
Observations of callose deposition in 7-day-old rice seedling leaves treated with 800 nmol L-1chitin(Hexa-N-acetylchitohexaose, Cayman Chemical, Ann Arbor, MI,USA) for 16 h followed a previously described procedure [17].The callose deposition was observed by confocal microscopy (340 to 380 nm).
2.4.RNA extraction and qRT-PCR analysis
Total RNAs for quantification of gene expression were extracted using TRIGene reagent according to the manufacturer’s instructions (GenStar, p118-05).One μg of RNA was reverse-transcribed into cDNA using a All-in-One First-Strand cDNA Synthesis Super-Mix for qPCR kit with gDNA Removal (TransGen, Q30518).qRTPCR was performed using Perfect Start Green qPCR SuperMix(TransGen, AQ601) with a Bio-Rad CFX96 real-time PCR detection system.Fungal biomass was examined by qPCR using specific primers for MoPot2 from M.oryzae and normalized to the reference gene OsUBQ.Relative transcript abundances were calculated using the 2-ΔΔCT method.The primers are listed in Table S1.
2.5.Protein interaction assays
For co-immunoprecipitation experiments, p35S:OsSPL10-YFP and p35S:OsJAmyb-HA constructs were co-transformed into rice protoplasts.Total proteins were extracted from the rice protoplasts after 12 h of incubation and subjected to immunoprecipitation according to a previously reported the protocol [18].The antibodies used in immunoblotting were anti-GFP antibody (TransGen,HT801) and anti-HA antibody (Roche, cat.no.11867423001).
For GST pull-down assays, the coding sequence of OsSPL10 cloned into pGEX-6p-1 vector (with an GST tag at the N terminus)and the OsJAmyb coding sequence were cloned into the pET28a-CYFP vector (with an YFP tag at the C terminus).The recombinant proteins were purified according to the manufacturer’s instructions and the GST pull-down assay was performed as previously described[19].The fusion proteins were detected by immunoblotting using anti-GFP antibody and Coomassie brilliant blue staining,respectively.
A yeast two-hybrid (Y2H) system (Clontech, USA, Cat.No.630489) was used for testing interaction between OsSPL10 and OsJAmyb.OsSPL10, OsSPL10-N, OsSPL10-SBP and OsSPL10-C were previously described [20].The coding region of OsJAmyb was cloned and inserted into the vector pGBKT7 between the NdeI and EcoRI restriction sites.Constructs were transformed into Saccharomyces cerevisiae strain AH109 according to the manufacturer’s instructions and coated onto SD/-Trp-Leu medium(Coolaber, PM2220).The yeast clones were then transferred to SD/-Trp-Leu-His-Ade medium (Coolaber, PM2110) to detect the protein interactions.
For split-luciferase complementation assays, leaves of N.benthamiana were infiltrated with Agrobacterium tumefaciens strain GV3101 harboring OsSPL10 fused to nLUC in combination with OsJAmyb fused to Cluc, or the negative control.Before infiltration,the bacteria were kept in infiltration buffer for 2–3 h and mixed with GV3101 strain harboring RNA interference suppressor P19.Two days after infiltration,the leaves were infiltrated with 1 mmol L-1luciferin and kept in darkness for 5 min to quench the fluorescence.LUC images were captured using a cooled CCD imaging apparatus.
2.6.Cell-free protein stability assay
Recombinant GST-OsJAmyb was expressed in Escherichia coli strain BL21(DE3)and purified with Glutathione Sepharose 4B agarose (Cytiva, 17075605).Total plant proteins were extracted with extraction buffer(25 mmol L-1Tris-HCl,pH7.5,10 mmol L-1NaCl,10 mmol L-1MgCl2, 10 mmol L-1ATP, 5 mmol L-1DTT) from leaves of 4-week-old plant.Protein concentration was determined by Bradford assay and 300 μL of total proteins were mixed with GST-OsJAmyb and then incubated at 30 °C for 0, 4, 8,12 min.The reaction was stopped by adding 5 × Laemmli/SDS sample buffer.
3.Results
3.1.OsSPL10 positively regulates resistance to blast
As OsSPL10 mRNA was rapidly induced following infection by the blast fungus (Fig.S1), we investigated whether OsSPL10 was involved in blast resistance.We spray-inoculated Osspl10 knockout mutant and OsSPL10-OE overexpression lines [13] with isolates Guy11 and Zhong1.Compared with the cv.Zhonghua 11(ZH11) WT, leaves of the Osspl10 mutant displayed larger disease lesions and had more fungal biomass, whereas leaves of OsSPL10-OE plants had less and smaller lesions with reduced fungal biomass(Figs.1A,B,S2A,B).Similarly punch-inoculated leaves showed that Osspl10 mutant plants were more susceptible and OsSPL10-OE plants were more resistant than the WT (Fig.1C, D).Examination of growth of strain Zhong1 tagged by GFP showed that the invading hyphae spread to the neighboring cells more rapidly in the Osspl10 mutant and more slowly in OsSPL10-OE plants than that in the WT (Fig.S3).We concluded that OsSPL10 positively regulates blast disease resistance in rice.
3.2.OsSPL10 positively regulates PTI response
The PTI response of cellular reactive oxygen species(ROS)plays a major role in resistance to blast.We firstly measured the rapid ROS burst in the leaves of WT, Osspl10 mutants and OsSPL10-OE following treatment with chitin, a well-known fungal pathogenassociated molecular pattern (PAMP).Compared with WT, the ROS burst was lower in the Osspl10 mutant but higher in OsSPL10-OE plants (Fig.1E).We then examined H2O2levels in Guy11-inoculated leaf cells using 3,3′-diamino-bezidine (DAB)staining.Similarly, the Osspl10 mutant produced much less H2O2than ZH11 at 48 hpi (h post inoculation), whereas OsSPL10-OE plants produced more (Fig.1F, G).Callose deposition at infection sites in plant cell walls is another typical PTI response that slows pathogen invasion and spread [21].Compared with ZH11, there was a nearly 50%decrease of callose deposition on Osspl10 mutant leaves and about a 40%increase in OsSPL10-OE leaves after 24 h of treatment with chitin(Fig.1H,I).The results indicate that OsSPL10 positively contribute to ROS induction and callose deposition.
3.3.OsSPL10 physically interacts with OsJAmyb
To study the molecular mechanisms underlying OsSPL10-regulated immunity, we attempted to identify its interactors by immunoprecipitation (IP) followed by liquid chromatography and tandem mass-spectrometry (LC-MS/MS).The immunocomplexes were purified from rice protoplasts transiently expressing OsSPL10-YFP or YFP (as control) with GFP-trap agarose and then were applied for protein identification by LC-MS/MS.We identified 112 proteins(Table S2)that co-purified with OsSPL10-YFP but not with YFP control.Among these potential interactors,OsJAmyb was selected for further verification as it had been shown to be involved in JA-regulated abiotic responses [22] and antiviral defense [14].Interaction between OsSPL10 and OsJAmyb was validated by Co-IP assays of transiently expressed proteins in rice protoplasts(Fig.2A)and by Split-LUC complementation assays in N.benthamiana leaves (Fig.2B).Furthermore, in an in vitro pull-down assay,OsJAmyb-YFP was co-purified with GST-OsSPL10 but not with the GST control (Fig.2C).Although interaction between OsJAmyb and the full length OsSPL10 was not observed in yeast-twohybrid (Y2H) assays (Fig.S4), interaction was detected between OsJAmyb and OsSPL10 C-terminal domain, but not with the Nterminal or SBP domains(Fig.S4).Taken together,the results indicate OsSPL10 directly interacts with OsJAmyb through its Cterminal domain.
3.4.OsSPL10 promotes protein stability of OsJAmyb
OsJAmyb mRNA was induced by M.oryazae infection (Fig.S5)[23].As there was no clear trend in expression of OsJAmyb among ZH11, Osspl10 mutant and OsSPL10-OE plants (Fig.S5), it seemed that OsSPL10 plays no significant role in regulating OsJAmyb expression.However,we noticed that in Co-IP experiments,protoplasts co-expressing OsJAmyb-HA and OsSPL10-YFP accumulated more OsJAmyb-HA protein (Fig.2A).To validate the observation,we expressed OsJAmyb-HA and OsSPL10-YFP individually or together in rice protoplasts.Indeed, co-expression of OsJAmyb-HA and OsSPL10-YFP greatly promoted OsJAmyb-HA protein accumulation (Fig.2D).A cell-free protein stability assay was performed to test whether OsSPL10 promotes protein stability of OsJAmyb.The same amount of GST-OsJAmyb was incubated in total protein extracts of each of ZH11, Osspl10 mutant or OsSPL10-OE plants at 30 °C for 4, 8 and 12 min.The immunoblots showed that the levels of GST-OsJAmyb declined faster in the Osspl10 mutant than in ZH11 (Fig.2E), but more slow in OsSPL10-OE lines(Fig.2E),indicating that OsSPL10 positively regulates the stability of OsJAmyb protein.
It was recently shown that OsJAmyb directly binds to the AGO18 promoter to activate AGO18 transcription which enhances RNA silencing and antiviral defense in rice [14].As OsSPL10 promotes OsJAmyb protein stability, we examined whether AGO18 expression is compromised in Osspl10 mutants.Compared with WT ZH11, the JA-induced AGO18 transcription was significantly reduced in the mutants(Fig.S6),thus correlated with the function of OsSPL10 in stabilizing OsJAmyb.
3.5.OsJAmyb positively regulates resistance to blast
To establish the biological significance of OsSPl10 stabilization of OsJAmyb in rice resistance to blast,we examined the resistance of Osjamyb knockout mutant and OsJAmyb overexpression lines(OsJAmyb-OE) to M.oryzae.At 5 d post spray-inoculation with strain Guy11, leaves of Osjamyb mutant plants showed more lesions and higher elevated biomass than WT Nipponbare(Fig.2F, G).On the contrary, leaves of OsJAmyb-OE plants displayed less lesions and lower fungal biomass than the WT(Fig.2F, G).Punch inoculation assays produced similar results(Fig.2H, I).Thus, like OsSPL10, OsJAmyb also positively regulates resistance to blast.
4.Discussion
Here, we document that transcription factor OsSPL10 is a positive regulator of resistance to blast in rice.OsSPL10 physically interacts with transcription factor OsJAmyb and increases its protein stability.We also confirmed that OsJAmyb positively contributed to resistance to blast.The results reveal a molecular module of OsSPL10 and OsJAmyb positively regulating blast resistance.
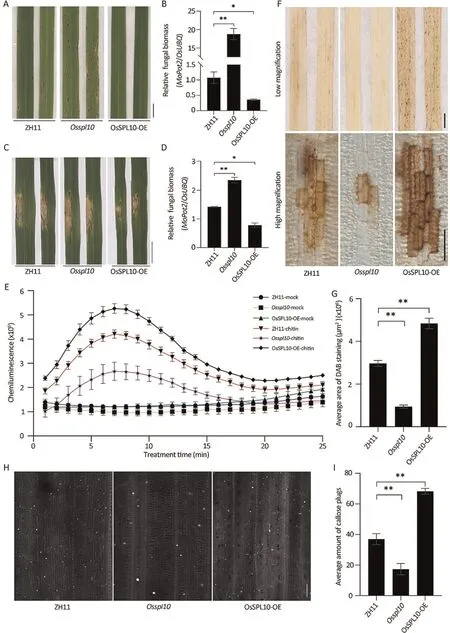
Fig.1.OsSPL10 positively regulates resistance to blast fungus.(A) Three-week-old seedlings of ZH11, Osspl10 mutant and OsSPL10-OE plants were sprayed inoculated with conidia of M.oryzae isolate Guy11.Disease symptoms on leaves were photographed 6 days post inoculation(dpi).Scale bar,1 cm.(B)Relative fungal biomass of M.oryzae in(A)measured using qPCR.Fungal biomass was determined by normalizing the expression of fungal MoPot2 gene against the rice OsUBQ gene.Values are means±SE(n=3).*,P<0.05;**,P<0.01 compared to WT ZH11 according to Student’s t-tests(two-tailed).(C)Five-week-old rice plants were subjected to punch inoculation with Guy11 conidia.Images were taken 7 dpi.Scale bar, 1 cm.(D) Quantification of lesion length in (C).Values are means ± SE (n = 6) *, P < 0.05; **, P < 0.01 compared to ZH11 according to Student’s t-tests(two-tailed).(E)Time course of ROS burst generated from ZH11,Osspl10 and OsSPL10-OE leaf sheaths treated with 800 nmol L-1 chitin.Data are means±SE(n=9).The data are from one of three independent experiments.(F)Detection of H2O2 levels in M.oryzae-infected leaves of ZH11,Osspl10 and OsSPL10-OE by DAB staining.The size of dark-brown area is corelated with the H2O2 level.Scale bars, 1 cm (top) or 500 μm (bottom).(G) Quantification of the area of DAB staining under high magnification of(F)using ImageJ software.Values are mean±SE(n=6).**,P<0.01 compared to ZH11 according to Student’s t-tests(two-tailed).(H)Chitin-induced callose deposition on ZH11, Osspl10 and OsSPL10-OE leaves.Leaves were treated with 800 nmol L-1 chitin and callose deposition was imaged after 16 h under UV light.Scale bar,50 μm.(I)Callose deposition in(H)quantified with ImageJ software.Values are means±SE(n=6).**,P<0.01 compared to ZH11 according to Student’s t-tests(two-tailed).The experiments were repeated three times similar results.
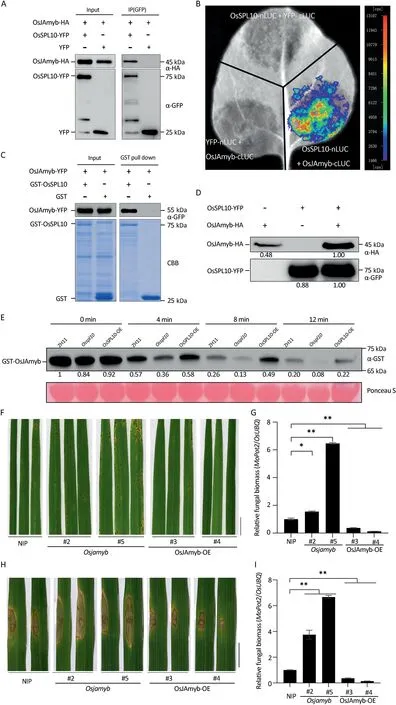
Fig.2.OsSPL10 promotes protein stability of OsJAmyb.(A)Co-immunoprecipitation(Co-IP)analysis of the interaction between OsSPL10-YFP and OsJAmyb-HA in transfected rice ZH11 protoplasts.Proteins in total extracts (input) and after IP with GFP-trap beads (IP(GFP)) were detected on immunoblots using anti-HA or anti-GFP antibodies.(B)Split-luciferase assay of interaction between OsSPL10-nLUC and OsJAmyb-cLUC in N.benthamiana.Combinations OsSPL10-nLUC+YFP-cLUC and YFP-nLUC+OsJAmyb-cLUC were included as negative controls.Luciferase complementation imaging was performed 2 d after agroinfiltration.(C) GST pull-down assays reveal that OsSPL10 directly interacts with OsJAmyb.The recombinant OsJAmyb-YFP,GST-OsSPL10 and GST(as negative control)proteins purified from Escherichia coli were subjected to GST pull-down analysis.The purified protein complexes were visualized by immunoblotting with anti-GFP antibodies or by Coomassie blue staining (CBB).(D) Immunoblot analysis with anti-HA or anti-GFP antibodies showing OsJAmyb-HA or OsSPL10-YFP protein accumulation in rice protoplasts expressing OsJAmyb-HA or OsSPL10-YFP individually or both.Values indicate relative band densities calculated using ImageJ software.(E)Immunoblot analysis of GST-OsJAmyb fusion protein in a cell-free degradation assay.The purified GST-OsJAmyb proteins from E.coli were incubated for the indicated times with total protein extracts from WT, Osspl10 mutant and OsSPL10-OE plants.Values are relative band densities of GST-OsJAmyb calculated using ImageJ software.Ponceau staining of the blot shows equal protein loading.The experiments were repeated three times with consistent results.(F)Blast symptoms of cultivar Nipponbare(NIP),Osjamyb knock-out mutants,and transgenic lines overexpressing OsJAmyb(OsJAmyb-OE)at 5 dpi with M.oryzae strain Guy11 using spray inoculation.Scale bars,1 cm.(G)Quantified relative fungal biomass in(F)by measuring fungal MoPot2 gene against host OsUBQ DNA level.Values are means±SE(n=3).*,P<0.05;**,P<0.01 compared to NIP according to Student’s t-tests(two-tailed).(H)Four-week-old rice plants subjected to punch inoculation with strain Guy11 conidia.Images were taken 7 dpi.Scale bar,1 cm.(I)Quantified relative fungal biomass in(H).Values are means±SE(n=3).**,P<0.01 compared to NIP according to Student’s t-tests (two-tailed).
In a previous report, we showed that OsSPL10 negatively contributed to salt tolerance in rice [13].In agreement with this,OsSPL10 was shown to negatively regulate drought tolerance by positively regulating TF OsNAC2[24],which is a negative regulator in rice response to drought stress [25].Consistent with a positive role of OsNAC2 in regulating ROS accumulation in rice under salt stress [26], overexpression of OsSPL10 accumulated more H2O2whereas Osspl10 knockout lines had less ROS production under drought stress conditions [24].It is well known that abiotic and biotic stress responses usually antagonize each other in plants.In line with this,we show here that OsSPL10 positively regulates rice resistance to blast fungus.Interestingly,consistent with its positive role in regulating ROS accumulation under drought stress,OsSPL10 positively regulated ROS production in defense against blast(Fig.1E, F, G).Thus, the low level of ROS in Osspl10 mutant plants enhances drought tolerance but compromises defense against blast.This at least partly explains the antagonism between abiotic and biotic stress responses regulated by OsSPL10.It was similarly shown that IPA1/OsSPL14 positively regulates resistance to blast while negatively regulating salt tolerance [27].
OsSPL10 is required for trichome development in rice [13,28].We found recently that OsSPL10 physically interacts with transcription factor OsWOX3B to regulate trichome formation [20].The OsSPL10-OsWOX3B interaction attenuates each other in binding to the promoter and in transcribing the target gene OsPLT2[20].Here, we show that OsSPL10 associates with OsJAmyb (Fig.2), an important transcription factor regulating JA signaling.It was shown that OsJAmyb expression was induced by M.oryzae infection and overexpression of OsJAmyb enhanced resistance to blast [23].Our genetic analysis of OsJAmyb knockout and overexpression lines further demonstrated that OsJAmyb positively regulates resistance to blast (Fig.2).Therefore, it is possible that OsSPL10 regulates blast resistance through OsJAmyb.Indeed, we found that OsSPL10 stabilizes protein stability of OsJAmyb(Fig.2E)and thus affects JAinduced expression of OsAGO18 (Fig.S6).Interestingly, the Nterminal region of OsSPL10 is responsible for association with OsWOX3B[20],whereas its C-terminal region interacts with OsJAmyb (Fig.S4).Thus, we speculate that through different regions,OsSPL10 can interact with multiple transcription factors and differently regulate their activity or protein stability.Therefore,we cannot exclude the possibility that OsSPL10 regulates blast response by directly or indirectly regulating defense gene expression.
Overexpression of OsSPL10 enhances rice resistance to blast,but also causes reduced salt and drought tolerance [13,24], and hence could be limited in application in rice breeding.It was shown that overexpression of OsSPL14 enhanced resistance to bacterial blight,but reduced yield [12].However,controlled expression of OsSPL14 with the pathogen-inducible promoter OsHEN1 in transgenic rice produced plants with both enhanced disease resistance and increased yield [12].It will be interesting to test whether similar strategy can be applied to OsSPL10 to control its expression to enhance blast resistance without compromising salt/drought tolerance.
CRediT authorship contribution statement
Zaofa Zhong:Investigation, Visualization, Writing – original draft.Lijing Zhong:Investigation.Xiang Zhu:Investigation.Yimin Jiang:Investigation.Yihong Zheng:Investigation.Tao Lan:Resources,Funding acquisition,Writing–review&editing.Haitao Cui:Conceptualization, Supervision, Visualization, Project administration, Resources, Funding acquisition, Writing – original draft,Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Prof.Yi Li(School of Life Sciences,Peking University)for providing seed of Osjamyb#2,Osjamyb#5,OsJAmyb-OE#3 and OsJAmyb-OE#4.Prof.Dingzhong Tang (State Key Laboratory of Ecological Control of Fujian-Taiwan Crop Pests, Fujian Agriculture and Forestry University) for providing the pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors; Prof.Zonghua Wang (College of Life Science,Fujian Agriculture and Forestry University)for providing the M.oryzae strain Guy11; and Prof.Xuewei Chen (Rice Research Institute, Sichuan Agricultural University) for providing the GFP labeled M.oryzae strain Zhong1.This work was supported by grants from Natural Science Foundation Key Program of Fujian Province (2023J02011), National Natural Science Foundation of China (31970281, 31671668), and a Sino-German Mobility Program funded jointly by National Natural Science Foundation of China and German Research Foundation (M-0275).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.10.015.
杂志排行
The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
