Sugarcane transcription factor ScWRKY4 negatively regulates resistance to pathogen infection through the JA signaling pathway
2024-03-07DongjioWngWeiWngShoujinZngLiqinQinYnlnLingPeixiLinYchunSuYouxiongQue
Dongjio Wng, Wei Wng, Shoujin Zng, Liqin Qin, Ynln Ling, Peixi Lin, Ychun Su,*,Youxiong Que,b,*
a Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs, Key Laboratory of Genetics, Breeding and Multiple Utilization of Crops,Ministry of Education, College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian, China
b National Key Laboratory for Tropical Crop Breeding, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Sanya 572024, Hainan,China
Keywords:Disease resistance Expression profile Transcriptome analysis WRKY transcription factors
ABSTRACT WRKY transcription factors,transcriptional regulators unique to plants,play an important role in defense response to pathogen infection.However,the resistance mechanisms of WRKY genes in sugarcane remain unclear.In the present study,gene ontology(GO)enrichment analysis revealed that WRKY gene family in sugarcane was extensively involved in the response to biotic stress and in defense response.We identified gene ScWRKY4, a class IIc member of the WRKY gene family, in sugarcane cultivar ROC22.This gene was induced by salicylic acid (SA) and methyl jasmonate (MeJA) stress.Interestingly, expression of ScWRKY4 was down-regulated in smut-resistant sugarcane cultivars but up-regulated in smutsusceptible sugarcane cultivars infected with Sporisorium scitamineum.Moreover, stable overexpression of the ScWRKY4 gene in Nicotiana benthamiana enhanced susceptibility to Fusarium solani var.coeruleum and caused down-regulated expression of immune marker-related genes.Transcriptome analysis indicated suppressed expression of most JAZ genes in the signal transduction pathway.ScWRKY4 interacted with ScJAZ13 to repress its expression.We thus hypothesized that the ScWRKY4 gene was involved in the regulatory network of plant disease resistance,most likely through the JA signaling pathway.The present study depicting the molecular involvement of ScWRKY4 in sugarcane disease resistance lays a foundation for future investigation.
1.Introduction
Sugarcane(Saccharum spp.)is a cash crop grown mainly in subtropical and tropical regions and plays an important role in the agricultural economy of China[1].Like other crops,biotic and abiotic stresses, such as drought, low temperature and pathogenic fungi, are the main factors restricting sugarcane production [2].Sugarcane smut, which seriously endangers the healthy and sustainable development of the sugarcane industry, is a worldwide fungal disease caused by Sporisorium scitamineum [3].
When plants encounter stress during growth and development,they implement a complex defense regulatory network in which transcription factors perform critical functions[4].WRKY transcription factors, one of the largest families of transcription factors in plants, are widely involved in plant responses to biotic, abiotic and hormonal stresses [5], especially in the determination of disease resistance[6].They have a 60 amino acid long DNA binding domain,which is characterized by a highly conserved WRKYGQK core motif at the N-terminal and a CX4–5CX22–23HXH zinc finger motif at the C-terminal [7].WRKY family members can be divided into three groups according to the number of WRKY domains and type of zinc finger motif;Group I has two WRKY domains whereas Groups II and III have one WRKY [8].The zinc finger structure in Groups I and II members is C2H2(CX4–5CX22–23HX1H), where X can be any amino acid; in Group III it is C2HC (CX7CX23HXC) [8].Group II can be further classified into subgroups IIa, IIb, IIc, IId and IIe based on homology [9].
Plants have evolved a complex mechanism to respond to biotic stresses[10].The regulatory roles of WRKY transcription factors in response to biotic stresses have been studied extensively[11],with regulation having both positive and negative effects [12].For example, transient silencing of VqWRKY31 reduced resistance to powdery mildew in Vitis vinifera [13] and silencing of JrWRKY21 in Juglans regia significantly reduced the resistance of walnuts to Colletotrichum gloeosporioides.On the other hand, the resistance was significantly enhanced in walnuts overexpressing JrWRKY21[14].Knock out mutations of OsWRKY53 in Oryza sativa led to thickened sclerenchyma cell walls,thereby conferring higher resistance to rice bacterial blight[15].In Triticum aestivum, TaWRKY19 negatively regulated stripe rust resistance by enhanced production of reactive oxygen species (ROS) and transcriptional repression of TaNOX10 [16].WRKY transcription factors can be involved in phytohormone-mediated signaling pathways by acting as its downstream target genes or regulating phytohormone anabolism to modulate plant responses to biotic stresses.For example,AtWRKY70, an important member of the salicylic acid (SA)- and jasmonic acid (JA)-regulated defense signaling pathways, was induced by SA and repressed by JA, thereby activating SAinducible genes and repressing JA-responsive genes that were involved in defense responses[17].Previous research also demonstrated that overexpression of AtWRKY70 increased resistance in Arabidopsis to Pseudomonas syringae pv tomato [17] whereas GhWRKY70 negatively regulated tolerance to Verticillium dahliae in Gossypium hirsutum by up-regulating expression of SA-related genes and repressing expression of JA-related genes [18].The ScWRKY gene in sugarcane (GenBank accession number GQ246458) belonging to class IIc was cloned and its expression was induced by the stress of S.scitamineum and SA [19].In addition, expression of sugarcane class IIc gene ScWRKY3 (GenBank accession number MK034706) was up-regulated in smutsusceptible variety ROC22, but remained unchanged in smutresistant variety Yacheng05–179, but its expression was inhibited by both SA and methyl jasmonate(MeJA)treatments[20].RNA-seq analysis of 154 members of the SsWRKY gene family in Saccharum spontaneum revealed that SsWRKYs displayed different temporal and spatial expression patterns at various developmental stages;52 SsWRKY genes were expressed in all tissues examined, four were not expressed in any tissue, and 21 were likely involved in photosynthesis[21].Javed et al.described 53 ShWRKY genes in Saccharum spp.hybrid R570; four of them, ShWRKY13-2/39-1/49-3/125-3, were significantly up-regulated in sugarcane cultivar LCP85–384 resistant to leaf scald [22].From all the above examples, the regulatory mechanisms of disease response associated with WRKY genes were widely reported in across species,but only a few reports addressed disease resistance functions of WRKY genes in sugarcane.
We reported previously that expression of ScWRKY4, a class IIc gene could be induced under SA, MeJA and smut pathogen stress[23].Of most interest to us was that expression of ScWRKY4 was down-regulated in smut-resistant sugarcane cultivars but upregulated in smut-susceptible cultivars infected with S.scitamineum.In the present study, we also found both transient and stable overexpression of ScWRKY4 in Nicotiana benthamiana enhanced the susceptibility of tobacco plants to Fusarium solani var.coeruleum and caused down-regulated expression of immune marker genes.Strikingly, after inoculation with the pathogen,expression of most JAZ-related genes was repressed in transgenic tobacco plants stably overexpressing ScWRKY4.Further experiments indicated that ScWRKY4 protein interacted with ScJAZ13 inhibiting its expression.Our study explored the regulatory network/mechanism of disease resistance involving the ScWRKY4 gene and provided a theoretical basis for future studies on other members of the WRKY gene family in sugarcane.
2.Materials and methods
2.1.Plant materials, culture conditions and pathogen inoculation
The two smut-resistant sugarcane cultivars (YZ01–1413 and YT96–86) and two smut-susceptible sugarcane cultivars(YZ03–103 and FN39)provided by the Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs (Fuzhou) were inoculated with S.scitamineum according to the methods of Que et al.[24].Three buds from each cultivar were collected at 0, 1, 3 and 7 d post inoculation (dpi), and stored at -80 °C after freezing in liquid nitrogen.
Gateway primers (Table S1) were designed to construct and convert the ScWRKY4/ScJAZ13 gene into the overexpression vector pEarleyGate 203.The empty pEarleyGate 203 (35S::00) and the fusion vector pEarleyGate 203-ScWRKY4/ScJAZ13 (35S::ScWRKY4/ScJAZ13) were transiently overexpressed in N.benthamiana by the Agrobacterium-mediated method.After 1 d, two leaves were taken from each plant and used for real-time quantitative PCR(RT-qPCR)and 3,3′-diaminobenzidine (DAB) histochemical analysis [25].Symptoms (phenotype and DAB) in N.benthamiana leaves were observed and relative transcription levels of tobacco immunerelated marker genes were calculated 1 d after inoculation of fungal pathogen F.solani var.coeruleum into the over expressing N.benthamiana leaves [25].
Agrobacterium tumefaciens carrying pEarleyGate 203-ScWRKY4 was stably transformed into N.benthamiana by the leaf-disc method [26].T3generation plants overexpressing ScWRKY4 gene were screened on MS (Murashige and Skoog) medium supplemented with glufosinate.DNA was extracted from leaves of 5–8-leaf plants and diluted to 25 ng L-1.The pEarleyGate 203-ScWRKY4 plasmid was used as a positive control and wild type(WT)N.benthamiana as a negative control.Genomic DNA of transgenic plants was used as a template for PCR amplification and electrophoresis[27].RNA was extracted from the leaves of the transgenic plants and expression of ScWRKY4 gene was measured using WT plants as the control [28].Transgenic plants overexpressing ScWRKY4 and the WT were inoculated with the F.solani var.coeruleum and leaf tissues were analyzed by DAB and trypan blue staining [29].Samples were taken at 0 and 2 dpi for subsequent detection of immune marker-related genes, determination of physiological indicators, and transcriptome analysis.
2.2.RNA extraction and RT-qPCR analysis
The total RNA was extracted by the TRIzol method [30].RNA was reverse transcribed into first strand cDNA from plant tissue according to instructions with the Hifair III 1st Strand cDNA Synthesis SuperMix kit (gDNA digester plus) and used as a template for quantitative detection of target gene expression levels.The RNA and cDNA were tested for quality by 1.0% agarose gel electrophoresis.Primer Premier 5 software was used to design specific quantitative PCR primers (primer pair: ScWRKY4-Q-F/R) for ScWRKY4 using glyceraldehyde-3-phosphate dehydrogenase(GAPDH) (GenBank Accession Number CA254672) as the internal reference gene(primer pair:GAPDH-Q-F/R)(Table S1)[31].Expression of 10 immune-related tobacco marker genes,including hypersensitive response (HR) marker genes NbHSR201, NbHSR203 and NbHSR515, ethylene (ET) synthesis-dependent genes NbACO-like and NbACO,SA signaling pathway-related genes NbPR2 and NbPR3,JA signaling pathway-related genes NbLOX1 and NbDEF1,and ROSrelated genes NbCAT1 and NbGST1(Table S1),was assessed[28].An ABI 7500 Real-time PCR System(USA)was used for RT-qPCR detection,with the quantitative reaction system was set up according to instructions for using the SYBR Green PCR Master Mix Kit.There were three technical replicates for each sample,and negative controls were with sterile water.Relative gene expression was calculated using 2-ΔΔCT [32], and the significance levels of experimental data were analyzed using DPS 9.50 software and histograms were plotted using Origin 2022 software.
2.3.Gene ontology (GO) enrichment analysis of ShWRKY gene family members
GO annotation of our previously identified ShWRKY gene family members [33] based on the eggNOG-mapper v2 database (http://eggnog-mapper.embl.de/) using default parameters [34].Data were analyzed by GO enrichment using Tbtools software [35]and visualized by the online software Chiplot (https://www.chiplot.online/).
2.4.Bioinformatics analysis of ScWRKY4
Structural domain prediction of the ScWRKY4 protein was conducted using the SMART website (https://smart.embl.de/).Protein sequence alignment was performed using MUSCLE v3.7[36],and a phylogenetic evolutionary tree of ShWRKY proteins including ScWRKY4 protein was constructed using IQ-TREE in PhyloSuite software, with the replication value set to 1000 times [37].The evolutionary tree was prepared with the help of the EvolView online website (https://evolgenius.info//evolview-v2) [38].Conserved motif information for ScWRKY4 gene was obtained by analysis of the MEME online website (http://meme-suite.org/index.html)[39].Multiple sequence alignment of the ScWRKY4 gene with other ShWRKY genes was performed by DNAMAN software to obtain exon–intron structure information of the ScWRKY4 gene from the S.spp.hybrid R570 genome[40].GFF3 file and gene structure of the ScWRKY4 homologues were visualized using TBtools software [35].Two thousand bp promoter sequences upstream of homologous genes were retrieved and cis-acting regulatory elements were predicted using the PlantCARE online website(https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [41].
2.5.Hormone content and enzyme activity determination
The contents of endogenous hormones SA and JA were determined in leaves of WT and transgenic tobacco plants at 0 and 2 dpi, respectively, according to instructions with the plant SA ELISA and JA ELISA kits (Enzyme-linked Biotechnology, Shanghai,China).The enzymatic activities of catalase (CAT) and glutathione S transferases (GST) were also measured, according to the CAT ELISA and GST ELISA kits (Enzyme-linked Biotechnology).
2.6.Transcriptome data analysis
RNA samples from WT plants inoculated with F.solani var.coeruleum at 0 d and 2 d were named WT-CK and WT-T, respectively.RNA samples from transgenic tobacco plants overexpressing ScWRKY4 inoculated with F.solani var.coeruleum at 0 and 2 d were named ScWRKY4-CK and ScWRKY4-T,respectively.Three biological replicates were set up for each of the above trials,a total of 12 samples (WT-CK1, WT-CK2, WT-CK3, WT-T1, WT-T2, WT-T3,ScWRKY4-CK1, ScWRKY4-CK2, ScWRKY4-CK3, ScWRKY4-T1,ScWRKY4-T2 and ScWRKY4-T3).RNA-Seq was entrusted to Genedenovo.The raw reads were quality controlled using fastp to filter low quality data[42].A comparative analysis based on the N.benthamiana (https://sefapps02.qut.edu.au/benWeb/subpages/downloads.php) database was carried out and annotated using HISAT2 software [43].Bioinformatic analysis was performed using Omicsmart (http://www.omicsmart.com), a dynamic real-time interactive online platform for data analysis.
2.7.Yeast two-hybrid analysis
Recombinant plasmids pGBKT7-ScWRKY4 and pGADT7-ScJAZs(JAZ6,8,9,10,11,13 and 14)constructed earlier by our group were used in a yeast two-hybrid system.The positive control pGADT7-T + pGBKT7-p53, negative control pGADT7-T + pGBKT7-lam, and a combination of plasmids pGBKT7-ScWRKY4 + pGADT7-ScJAZs were each co-transformed into Y2HGold yeast strain according to instructions of Y2HGold Chemically Competent Cell.After transformation, the Y2HGold strain containing the above plasmid combinations was spread on media plates deficient in tryptophanleucine (SD/-Trp-Leu), and cultured in a 28 °C incubator for 2–3 d.Single colonies were selected and cultured in SD/-Trp-Leu liquid medium for about 18 h,and then diluted into four gradients(10-1,10-2, 10-3and 10-4) with sterile water, with 10 μL to spotted on SD/-Trp-Leu and tryptophan-leucine-histidine-adenine (SD/-Trp-Leu-His-Ade) deficient medium plates, respectively.After incubation at 28 °C for 2–3 d, the growth of the yeast colonies was recorded by photography.Interaction of proteins in the yeast was judged according to growth of the yeast colonies.
2.8.Bimolecular fluorescent complimentary (BiFC)
The Gateway method was used to transform the ScWRKY4 and ScJAZ13 genes into pEarleyGate 201 and pEarleyGate 202 vectors,respectively.The BiFC recombinant vectors pEarleyGate 201-ScWRKY4, pEarleyGate 202-ScJAZ13, empty-loaded pEarleyGate 201-YN and pEarleyGate 202-YC were transformed into Agrobacterium GV3101 strains, respectively, and injected into 5–8 leaf N.benthamiana leaves.After 3 d at room temperature, about 1 cm2leaf sections were examined under a Leica TCS SP8 laser confocal microscope(Leica Microsystems(Shanghai)Trading Co.,Ltd.,Mannheim, Germany).Yellow fluorescent protein (YFP) was observed with a 10× lens and YFP filter (561 nm excitation wavelength).
2.9.Detection of protein inhibition using green fluorescent protein(GFP)
The ScJAZ13 gene was transformed into the pCAMBIA2300 vector with GFP tag,and the fusion vector pCAMBIA2300-ScJAZ13 was transformed into A.tumefaciens strain GV3101.Agrobacterium containing the target fragment and NbH2B (histone H2B)-RFP [44]were subsequently collected and diluted with MS blank medium and mixed in equal proportions to OD=0.8.One mL bacterial solution(containing 200 μmol L-1acetosyringone)was injected with a sterile syringe into 5–8 leaf WT plants and transgenic plants overexpressing ScWRKY4, respectively.After incubation at 28 °C in a 16 h light/8 h darkness photoperiod for 2 d, the leaves were collected and GFP was observed under a Leica TCS SP8 laser confocal microscope using a 10× lens and 488 nm green fluorescence excitation wavelength.The above experimental materials were subjected to western blotting (WB).Anti-GFP (#HT801-01,TransGen) and anti-Myc (#HT101-01, TransGen) were used and Coomassie brilliant blue (CBB) staining was performed.
2.10.Co-Immunoprecipitation
Recombinant vectors pEarleyGate 202-ScWRKY4-FLAG and pCAMBIA2300-ScJAZ13-GFP were each transformed into A.tumefaciens GV3101,and co-injected into 5–8 leaf N.benthamiana leaves.After 48 h, the injected tobacco leaves were removed and total tobacco protein was extracted.The experimental method followed Ling et al.[45].The antibodies used in this experiment were anti-GFP (#HT801-01, TransGen) and anti-FLAG (#AE092, ABclonal).
3.Results
3.1.Characteristics of ScWRKY4 in sugarcane
In a previous study we identified 60 ShWRKY genes from the genome of S.spp.hybrid R570[33].These ShWRKY genes were constitutively expressed in sugarcane tissues and their expression was induced by the smut pathogen [33].GO annotation analysis was performed to further explore the function of the ShWRKY genes.The ShWRKY genes were annotated in biological process (38%),molecular function(36%),and cellular component(26%)categories(Fig.S1A; Table S2).In biological process, ShWRKY genes were mainly enriched in biotic stress response and defense response categories (Fig.S1B; Table S2).For molecular function, 26 ShWRKY genes were enriched in transcriptional regulatory activity and DNA-binding transcription factor activity (Fig.S1C; Table S2).In the cellular component, most of the ShWRKY genes were enriched in the nucleus (Fig.S1D; Table S2).
Our previous findings indicated that ScWRKY4 was downregulated in smut-resistant sugarcane cultivar Yacheng 05–179[23].In the smut transcriptome data, expression of ScWRKY4 was significantly down-regulated in the same cultivar [33].The ORF(137–877 bp) of this gene was 741 bp in length, encoding 246 amino acids, and contained a typical WRKY structural domain(Fig.1A).A phylogenetic tree of ScWRKY4 protein constructed using ShWRKY proteins as reference showed that ScWRKY4 protein belonged to subgroup IIc(Fig.1B).Multiple sequence comparison indicated that ScWRKY4 had the highest similarity of 99.19%with ShWRKY36 (Table S3), suggesting functional similarity.Ten conserved motifs in ScWRKY4 protein were observed using MEME software.The ScWRKY4 protein contained only Motifs 1,2,3,and 6(Fig.1C).ScWRKY4 had four exons and three introns(Fig.1D).Prediction of promoter elements in the 2000 bp upstream region of ScWRKY4 suggested several cis-acting regulatory elements associated with stress, growth, and hormone response (Fig.1E;Table S4);among them the stress-related cis-acting regulatory element LTR is involved in low temperature response.Growth and development-related cis-acting regulatory elements, such as the ARE, is essential for anaerobic induction, and the RY-element is involved in seed-specific regulation.There were also four hormone response-related cis-acting regulatory elements, namely MeJAresponsive element TGACG-motif, auxin-responsive element TGA-element, ABA-responsive element ABRE, and gibberellinresponsive element P-box (Fig.1E; Table S4).It is thus hypothesized that ScWRKY4 is involved in growth and development and in defense against biotic stresses in sugarcane.
3.2.Expression profile of the ScWRKY4 gene
Previous study showed that, the expression level of ScWRKY4 reached a peak at 12 h under both SA and MeJA treatments, with levels of 2.69-fold and 2.38-fold that of the controls, respectively(Fig.2A).Fig.2A demonstrated the expression levels of ScWRKY4 in four different sugarcane genotypes infected with the smut pathogen.The ScWRKY4 gene was down-regulated in resistant cultivars YZ01–1413 and YT96–86, and up-regulated in susceptible cultivars YZ03–103 and FN39 at 7 dpi.We speculated that ScWRKY4 negatively regulated the defense of sugarcane against the smut fungus.
ScWRKY4 was successfully detected in tobacco leaves at 1 d after transient transformation into N.benthamiana leaves(Fig.2B).DAB staining revealed darker browning in tobacco leaves transiently overexpressing the ScWRKY4 gene (35S::ScWRKY4) compared to the control group (35S::00) (Fig.2C).Expression of seven HR marker genes was significantly upregulated except for NbHSR201 (Fig.2D).At 7 d post inoculation with F.solani var.coeruleum, 35S::ScWRKY4 tobacco leaves began to wilt,and the degree was more widespread than for the 35S::00 control(Fig.2E).Then,the expression of immune marker genes in the leaves of 35S::00 and 35S::ScWRKY4 plants was examined at 2 dpi.Compared with 35S::00 plants the expression of seven immune-related genes were down-regulated in 35S::ScWRKY4 tobacco leaves at 2 dpi; expression of the NbPR-1a/c gene was unchanged (Fig.2F).We hypothesized that ScWRKY4 negatively regulated resistance to pathogens.
3.3.Stable overexpression of ScWRKY4 negatively regulates resistance to pathogen infection
We genetically transformed ScWRKY4 into N.benthamiana and cultured it to the T3generation to further examine the effect of ScWRKY4 overexpression(OE)on disease response(Fig.S2A).A target band consistent with the size of the overexpression vector plasmid was identified in all transgenic plants, indicating that the ScWRKY4 gene was present in the genome of N.benthamiana(Fig.S2B).
After inoculation with F.solani var.coeruleum, the leaves of OE plants at 7 dpi showed significant wilting and yellowing with obvious disease symptoms compared to the WT (Fig.3A).DAB (H2O2accumulation) and trypan blue staining (cell death) of OE leaves were darker than those of the WT.Besides,the SA and JA contents of WT and transgenic lines plants were measured at 0 and 2 dpi.There was no significant difference in SA content in the WT and OE lines, but JA content in the OE line was significantly higher(1.28-fold) than the control, higher than the control at 0 dpi.At 2 dpi, the contents of SA and JA in OE plants were significantly lower than that in WT plants, at 0.93-fold and 0.89-fold, respectively (Fig.3B).Enzyme activity assays revealed that the CAT and GST activities of the OE plants were significantly higher than the control before inoculation (2.44- and 1.16-fold) and both decreased to the control level at 2 dpi (Fig.3C).Expression of immune marker-related genes NbHSR201 and NbHSR515 at 0 and 2 dpi, SA signalling pathway-related genes NbPR2 and NbPR3, JA signaling pathway-related genes NbLOX1 and NbDEF1, and the ROS-related gene NbCAT1 were all lower in OE plants compared to the WT (Fig.3D–G).The above results demonstrated that overexpression of ScWRKY4 in N.benthamiana enhanced susceptibility to F.solani var.coeruleum.
3.4.Differentially expressed genes (DEGs) involved in the disease resistance regulatory network of the ScWRKY4 gene
To identify DEGs involved in disease response the WT and ScWRKY4-OE, WT-CK, WT-T, ScWRKY4-CK and ScWRKY4-T plants at 0 d and 2 d inoculated with F.solani var.coeruleum were sequenced.Comparisons were made of WT-CK vs.WT-T and treatment group ScWRKY4-CK vs.ScWRKY4-T.Among clean reads of all samples, Q20 and Q30 were all greater than 91%, and GC content was higher than 42%,indicating good sequencing quality for subsequent sample analysis (Table S5).The regions aligned to the genome were divided into exon regions, intron regions and intergenic regions,that most of the reads could be aligned to exon regions (Fig.S3A).The Pearson correlation coefficient between replicate samples within a group were all close to one (Fig.S3B),indicating good inter-sample repeatability allowing betweengroup difference analysis.
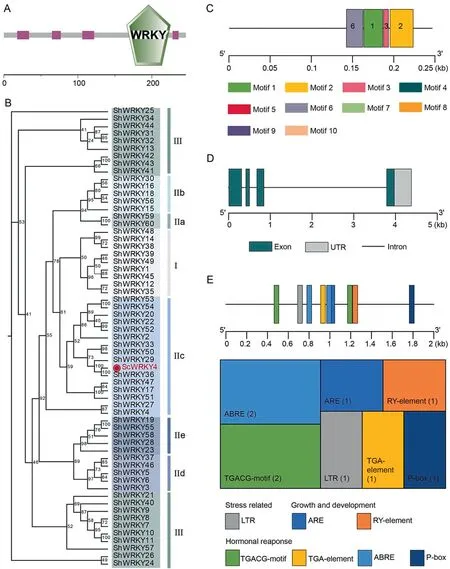
Fig.1.Characterization of ScWRKY4 in sugarcane.(A)Conserved domains of the ScWRKY4 protein.(B)A phylogenetic tree of the ScWRKY4 protein(shown in red font).(C)Conserved motifs in ScWRKY4 protein.(D) Structure of the ScWRKY4 gene.(E) Distribution and functional prediction of cis-acting regulatory elements in the ScWRKY4 promoter.The numbers in parentheses represent the number of cis-acting regulatory elements.
The number of up-regulated DEGs in WT was significantly lower than that in OE plants at 0 d (Fig.S4A).The up-regulated DEGs number increased in WT and decreased in OE plants at 2 dpi(Fig.S4B).A total of 4944 up-regulated DEGs and 3903 downregulated DEGs were identified in WT-CK vs.WT-T, whereas 8043 up-regulated DEGs and 8,013 down-regulated DEGs were identified in ScWRKY4-CK vs.ScWRKY4-T (Fig.S4C).The downregulated DEGs in ScWRKY4-CK vs.ScWRKY4-T was 2.05-fold higher than in WT-CK vs.WT-T (Fig.S4C).Among the DEGs, 5488 were unique to WT-CK vs.WT-T and 12,697 were unique to ScWRKY4-CK vs.ScWRKY4-T (Fig.S4D).Subsequent, GO and KEGG analyses found that 10.23% of the specific DEGs in WT-CK vs.WT-T were enriched in response to hormone and 10.62% of the specific DEGs were enriched in the response to endogenous stimulus item(Fig.4A;Table S6).In ScWRKY4-CK vs.ScWRKY4-T,32.34%specific DEGs were enriched in the response to stimulus item,18.73% in the response to stress item, and 6.51% in the response to external biotic stimulus item (Fig.4B; Table S7).Surprisingly,DEGs specific to both WT-CK vs.WT-T and ScWRKY4-CK vs.ScWRKY4-T were significantly enriched in the plant hormone signal transduction, MAPK signaling and plant-pathogen interaction pathways (Fig.4C, D).By contrast, the specific DEGs in WT-CK vs.WT-T were significantly enriched in the plant hormone signal transduction pathway (Fig.4C), whereas the specific DEGs in ScWRKY4-CK vs.ScWRKY4-T were significantly enriched in the plant-pathogen interaction pathway (Fig.4D).
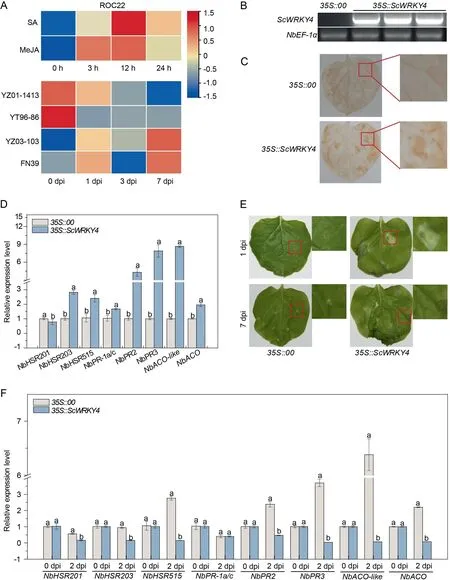
Fig.2.Function of the ScWRKY4 gene.(A)Relative expression of ScWRKY4 in sugarcane under SA,MeJA and smut pathogen stresses.The sugarcane material treated with SA and MeJA was ROC22.Sugarcane cultivars YZ01-1413 and YT96-86 were resistant to smut; YZ03-103 and FN39 were susceptible to smut.(B) Semi-quantitative PCR amplification of ScWRKY4 in N.benthamiana leaves.35S::00: empty vector pEarleyGate 203; 35S::ScWRKY4: pEarleyGate 203-ScWRKY4.(C) DAB staining to analyze H2O2 accumulation in Agrobacterium strains carrying 35S::ScWRKY4 after one day of injection into the leaves of N.benthamiana.(D) RT-qPCR expression level of eight immunityassociated marker genes in the 35S::ScWRKY4 transiently expressing leaves after infiltration for 1 day.(E)Phenotypes of N.benthamiana leaves after inoculation with F.solani var.coeruleum.(F)The transcripts of immunity-associated marker genes in the N.benthamiana leaves after inoculation with F.solani var.coeruleum at 0 d and 2 d.Data were normalized to the NbEF-1α expression level.All data points are means±standard errors(n=3).Different letters above the columns indicate significant differences(Duncan’s new multiple range test at P = 0.05).
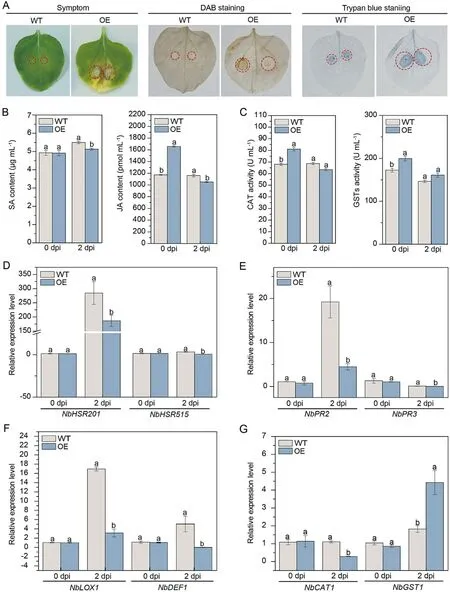
Fig.3.Disease resistance of ScWRKY4 transgenic N.benthamiana plants inoculated with F.solani var.coeruleum.(A) Phenotype at 7 dpi.Wilting is shown within the red circles.(B) SA and JA contents in WT and OE plants at 0 dpi and 2 dpi.(C) CAT and GST enzyme activities in WT and OE plants at 0 dpi and 2 dpi.(D–G) Expression of immunity-associated marker genes.Expression of HR marker genes (D), SA and JA signaling pathways (E, F), and ROS defense-response related genes (G).WT: wild type N.benthamiana;OE:transgenic N.benthamiana plants overexpressing ScWRKY4 gene.Data were normalized to the NbEF-1α expression level.All data points are means±standard errors (n = 3).Different letters above the columns indicate significant differences (Duncan’s new multiple range test at P = 0.05).
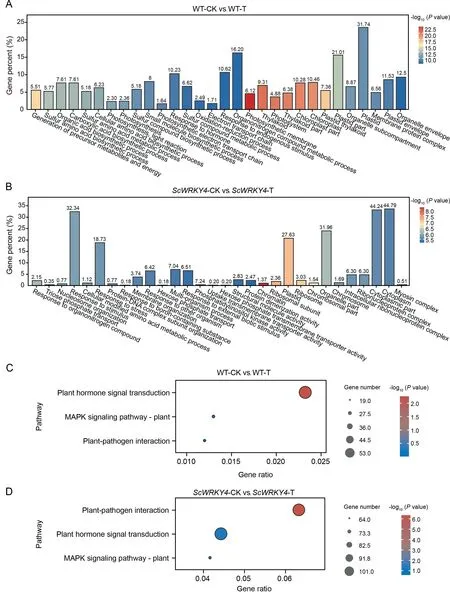
Fig.4.GO and KEGG enrichment of differentially expressed genes.Bar graphs of DEGs in(A)the control group and(B)the treatment group.(A)and(B)each show the first 30 significantly enriched GO terms.Bubble plots of DEGs enriched in disease resistance-related pathways in(C)the control group and(D)in the treatment group.Control group:WT-CK vs.WT-T; treatment group: ScWRKY4-CK vs.ScWRKY4-T.

Fig.5.Pathways related to disease resistance and the expression levels of key genes.(A–C) Expression patterns of specific DEGs in (A) plant hormone signal transduction pathway;(B) plant-pathogen interaction pathway;and (C)MAPK signaling pathway in the control and the treatment groups, respectively.Control group: WT-CK vs.WT-T;treatment group: ScWRKY4-CK vs.ScWRKY4-T.
3.5.ScWRKY4 mediated the regulation of disease resistance-related pathway genes in response to infection
To investigate the regulatory mechanism of ScWRKY4 gene in disease response, we analyzed the expression of key DEGs in the disease resistance-related pathway identified above.The specific DEGs TIFY10A (Nbv5tr6373992, Nbv5tr6373993, Nbv5tr6204331 and Nbv5tr6233370), TIFY10B (Nbv5tr6348185, Nbv5tr6349145,Nbv5tr6349147, Nbv5tr6374864 and Nbv5tr6405095), and TIFY11B(Nbv5tr6349144 and Nbv5tr6349145) in WT-CK vs.WT-T were up-regulated (Fig.5A; Table S8).However, expression of these DEGs was reduced in ScWRKY4-CK vs.ScWRKY4-T (Fig.5A;Table S8).In the plant-pathogen interaction pathway, CNGCs and CDPK were involved in HR (Fig.5B).CNGCs-related DEGs CNGC1(Nbv5tr6233756 and Nbv5tr6399602) were up-regulated, and CDPK-related DEGs CPK2 (Nbv5tr6321646) and CPK5(Nbv5tr6397003) were down-regulated (Fig.5B; Table S8).Most of the DEGs in the MAPK signaling pathway were inhibited in WT-CK vs.WT-T(Fig.5C).FLS2-related DEG XA21(Nbv5tr6263859)and VIP1-related DEGs VIP1 (Nbv5tr6405992) and BZIP18(Nbv5tr6405991) involved in defense response and late defense response to pathogens were up-regulated(Fig.5C;Table S8).Notably, most DEGs in the ScWRKY4-CK vs.ScWRKY4-T were downregulated,including pathogen attack-related DEGs Nbv5tr6231425,Nbv5tr6339859 and Nbv5tr6343569 were significantly downregulated, and participated in H2O2production (Fig.5C; Table S8).
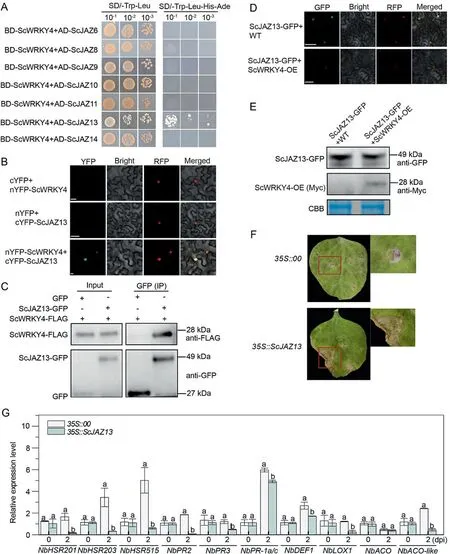
Fig.6.ScWRKY4 interacted with ScJAZ13 and inhibited its expression.(A) ScWRKY4 interacted with seven ScJAZ proteins in yeast.BD,pGBKT7 vector;AD, pGADT7 vector.(B)BiFC assay detecting interaction between ScWRKY4 and ScJAZ13 in N.benthamiana epidermal cells.NbH2B(histone H2B)-RFP was used to identify the nucleus in(B)and(D).(C)Co-IP assay detecting interaction of ScWRKY4 with ScJAZ13.(D)ScWRKY4 inhibited expression of ScJAZ13 in N.benthamiana epidermal cells.Images were obtained with a confocal microscope at 2 dpi.Scale bars,25 μm.(E)WB assay detecting ScJAZ13 protein in WT and ScWRKY4-OE plants.Anti GFP antibody application and Coomassie brilliant blue (CBB) staining showed uniform protein loading.WT, wild type N.benthamiana; ScWRKY4-OE, transgenic plants overexpressing ScWRKY4 N.benthamiana.(F)Phenotypes of tobacco leaves at 7 d post-inoculation with F.solani var.coeruleum.35S::00, empty vector pEarleyGate 203; 35S::ScJAZ13, pEarleyGate 203-ScJAZ13.(G)Expression of immune marker genes in N.benthamiana leaves at 0 d and 2 d post-inoculation with F.solani var.coeruleum.Data were normalized to the NbEF-1α expression level.All data points are means ± standard error (n = 3).Different letters above the columns indicate significant differences (Duncan’s new multiple range test at P = 0.05).
3.6.ScWRKY4 interacted with ScJAZ13 and inhibited its expression
Transcriptome analysis revealed that JAZ genes were mostly upregulated in WT-CK vs.WT-T and their expression was repressed in ScWRKY4-CK vs.ScWRKY4-T.Yeast two-hybrid assays to test interactions between ScWRKY4 and the seven previously cloned JAZ proteins indicated that all plasmid combinations grew normally on SD/-Trp-Leu medium, implying that all plasmids were successfully transformed into the yeast strain Y2HGold (Fig.6A) but only the BD-ScWRKY4 + AD-ScJAZ13 combination grew normally on SD/-Trp-Leu-His-Ade medium (Fig.6A).Interaction between ScWRKY4 and ScJAZ13 was verified by BiFC assays.When ScWRKY4 was fused to nYFP (nYFP-ScWRKY4), and ScJAZ13 was fused to cYFP (cYFP-ScJAZ13), fluorescent complexes were formed and detected in the nuclei of N.benthamiana leaf cells(Fig.6B).This demonstrated interaction between ScWRKY4 and ScJAZ13 and that the specific protein complex was produced in the nucleus.
The CDS of ScWRKY4 was fused to sequences encoding FLAG to form ScWRKY4-FLAG, whereas the CDS of ScJAZ13 was fused to GFP to form ScJAZ13-GFP.Co-IP assay revealed the interaction between ScWRKY4-FLAG and ScJAZ13-GFP in tobacco leaf cells(Fig.6C).
ScJAZ13-GFP was transiently overexpressed in the leaves of WT and OE plants.Fluorescence of ScJAZ13-GFP after 2 d was significantly more intense in the leaves of WT plants than in the leaves of OE plants (Fig.6D).A WB experiment performed with the same experimental materials confirmed that the protein level of ScJAZ13-GFP in WT was higher than that in OE plants (Fig.6E).At 7 d post-inoculation with F.solani var.coeruleum,tobacco plants transiently overexpressing ScJAZ13 showed more leaf wilting than that the controls (Fig.6F) and down-regulated expression of most immune marker genes(Fig.6G).Thus,ScWRKY4 may repress the of ScJAZ13 protein and negatively regulate resistance to pathogen infection.
4.Discussion
WRKY is one of the largest families of transcription factors in plants [8].Their functions have been reported in many plant species [11] but there are few reports on their functions in sugarcane[19,21–23,46].Here, we identified ScWRKY4 as a class IIc WRKY family member, induced by SA and MeJA stresses (Fig.2A).ScWRKY4 expression was up-regulated in smut-susceptible sugarcane cultivars YZ03–103 and FN39 and down-regulated in smutresistant cultivars YZ01–1413 and YT96–86 after inoculation with S.scitamineum(Fig.2A).It was thus speculated that ScWRKY4 negatively regulated resistance to infection.
Previous studies have shown that WRKY genes respond to infection caused by pathogenic fungi [47,48].ROS levels in plants change following infection [49].GhWRKY27a inhibited expression of the CAT and GST genes and negatively regulated resistance in tobacco to Rhizoctonia solani [50].HR is the defense response of resistant plants to pathogens [51].Overexpression of CaWRKY40 in tobacco altered the expression of HR-related genes NtHSR201,NtHSR203 and NtHSR515 and pathogenicity-related genes, thereby regulating tobacco response to R.solanacearum stress [52].In this study, the pre-inoculation CAT and GST enzyme activities in OE plants were significantly higher than those of the WT, but decreased to the control level after inoculation(Fig.3C).Expression of HR marker genes NbHSR201 and NbHSR515,and the ROS-related genes NbCAT1, was significantly decreased (Fig.3D, G).It is speculated that N.benthamiana plants overexpressing ScWRKY4 may reduce tolerance to oxidative stress after inoculation with F.solani var.coeruleum.Reasonably, inhibition of HR was associated with decreased CAT and GST activities and down-regulated expression levels of ROS and HR-related genes.
The widely studied SA and JA signaling pathways play important regulatory roles in plant responses to infection [53,54].WRKY25 was identified as a negative regulator of SA-mediated response to Pseudomonas syringae in Arabidopsis WRKY25 T-DNA insertion mutants and transgenic plants overexpressing [55].In rice, JA played an important role in OsWRKY30-mediated defense to fungal pathogens [53].Here, after 2 d of inoculation with F.solani var.coeruleum, the SA and JA contents in OE plants were lower than that in WT plants (Fig.3B), and genes related to the SA and JA signaling pathways(NbPR2,NbPR3,NbLOX1 and NbDEF1)were also down-regulated(Fig.3E,F).This indicated that ScWRKY4 is a negative regulatory transcription factor that suppresses the SA and JA signaling pathways by inhibiting expression of NbPR2,NbPR3, NbLOX1 and NbDEF1, and at the same time repressing expression of HR and ROS-related genes, NbHSR201, NbHSR515 and NbCAT1, thereby weakening the resistance of N.benthamiana to F.solani var.coeruleum.
Clearly, WRKY genes are involved in various plant defense signaling pathways [53,56].Overexpression of rice OsWRKY03 enhanced the resistance of transgenic plants to bacterial blight and induced expression of several pathogenesis-related genes in transgenic plants [57].Further studies revealed that OsWRKY03,located upstream of OsNPR1, acted as a transcriptional activator of SA-related or JA-related defense signaling pathways [57].AtWRKY57 competed with AtWRKY33 to interact with VQ proteins SIB1 and SIB2, and competitively regulated expression of key repressors JAZ1 and JAZ5 of the JA signaling pathway,thereby partially blocking JA signaling and attenuating the effect of WRKY33 on Botrytis cinerea resistance [58].OsWRKY13 enhanced rice defense responses against R.solani and Sarocladium oryzae, possibly affecting the TIFY9-mediated MAPK cascade signaling pathway[59].In the present study, transcriptome data from plants inoculated with F.solani var.coeruleum showed expression of most JAZ genes was significantly suppressed in N.benthamiana plants overexpressing the ScWRKY4(Fig.5A;Table S8).We also confirmed that ScWRKY4 interacted with ScJAZ13 and repressed its expression(Fig.6A–E).Therefore,we deduced that ScWRKY4 negatively regulated resistance to pathogens by repressing the expression of ScJAZ13.Fig.7 presents a model of the disease resistance regulatory mechanism of ScWRKY4.

Fig.7.A model for disease resistance regulation of the transcription factor ScWRKY4.Stable overexpression of ScWRKY4 in N.benthamiana causes downregulated expression of JAZ-related genes.ScWRKY4 interacts with ScJAZ13 to repress its expression.The hypothesis is that ScWRKY4 enhances susceptibility to diseases and is a negatively regulated transcription factor.
5.Conclusions
Stable overexpression of ScWRKY4 negatively regulated the resistance of transgenic plants to pathogen infection and caused down-regulated expression of JA-related genes.During the transcription process, most of the JAZ genes were repressively expressed in ScWRKY4 transgenic plants.In addition, ScJAZ13 interacted with ScWRKY4 and was repressed by ScWRKY4.We thus proposed that ScWRKY4 may enhance susceptibility to pathogens by repressing expression of ScJAZ13.This work is expected to lay the foundation for in-depth analysis of the biological function and mechanism of sugarcane WRKY transcription factors.
CRediT authorship contribution statement
Dongjiao Wang:Writing–original draft,Data curation,Formal analysis.Wei Wang:Data curation, Formal analysis, Resources.Shoujian Zang:Data curation, Formal analysis, Resources.Liqian Qin:Data curation, Resources.Yanlan Liang:Data curation,Resources.Peixia Lin:Data curation, Resources.Yachun Su:Writing–review&editing.Youxiong Que:Writing–review&editing,Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2022YFD2301100 and 2019YFD1000503), the Natural Science Foundation of Fujian Province (2021J01137), the Special Fund for Science and Technology Innovation of Fujian Agriculture and Forestry University(CXZX2020081A), and the China Agriculture Research System(CARS-17).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.12.002.
杂志排行
The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
