The ABA synthesis enzyme allele OsNCED2T promotes dryland adaptation in upland rice
2024-03-07LiyuHungYchongBoShiwenQinMinNingQinynLiQingmoLiShiliZhngGungfuHungJingZhngWenshengWngBinyingFuFengyiHu
Liyu Hung*, Ychong Bo Shiwen Qin Min Ning Qinyn Li Qingmo Li Shili ZhngGungfu Hung Jing Zhng Wensheng Wng, Binying Fu, Fengyi Hu*
a Key Laboratory of Biology and Germplasm Innovation of Perennial Rice from Ministry of Agriculture and Rural Affairs, School of Agriculture, Yunnan University, Kunming 650091, Yunnan, China
b National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Keywords:Upland rice Dryland adaptation ABA Root development Drought tolerance
ABSTRACT Upland rice shows dryland adaptation in the form of a deeper and denser root system and greater drought resistance than its counterpart,irrigated rice.Our previous study revealed a difference in the frequency of the OsNCED2 gene between upland and irrigated populations.A nonsynonymous mutation (C to T, from irrigated to upland rice) may have led to functional variation fixed by artificial selection, but the exact biological function in dryland adaptation is unclear.In this study, transgenic and association analysis indicated that the domesticated fixed mutation caused functional variation in OsNCED2,increasing ABA levels, root development, and drought tolerance in upland rice under dryland conditions.OsNCED2-overexpressing rice showed increased reactive oxygen species-scavenging abilities and transcription levels of many genes functioning in stress response and development that may regulate root development and drought tolerance.OsNCED2T-NILs showed a denser root system and drought resistance, promoting the yield of rice under dryland conditions.OsNCED2T may confer dryland adaptation in upland rice and may find use in breeding dryland-adapted, water-saving rice.
1.Introduction
Natural and artificial selection has driven the evolution of rice ecotypes such as upland and irrigated rice, leading to phenotypic adaptation to their respective environments and simultaneous genetic differentiation among ecotypes [1,2].Upland rice provides a model system for studying plant adaptation to aerobic and drought conditions[3,4].Although the yield of upland rice is lower than that of irrigated rice, it serves as a staple food for nearly 100 million people[5].Accordingly,upland rice cultivars are often cultivated under sloping field conditions to optimize crop yields,owing to their more efficient use of water and their adaptation to aerobic and drought-prone low-fertility soils[3].Compared with their irrigated counterparts, upland rice plants show greater height, lower tillering potential, and longer and denser roots[6,7].Although it is unclear whether upland or irrigated ecotypes emerged first, the genetic trade-offs between drought resistance and productivity likely constrained the evolution of upland rice[8].Ecotype-differentiated genes potentially contribute to the aerobic adaptation of upland rice to drylands [8], and balancing selection signatures observed in the genome explains the differentiation between the two ecotypes [3].
Abscisic acid (ABA) is a stress-responsive phytohormone that regulates multiple pathways to adapt to unfavorable environmental conditions[9,10].First,ABA-mediated root tropic response and stomatal closure under water stress are responses of plants that ensure survival under water-limited conditions[11].Furthermore,ABA signaling functions in the regulation of root development and inhibits aboveground growth, which increases root/shoot ratio to promote soil moisture deficit [12,13].Moreover, ABA influences plant physiological status through the use of antioxidant enzymes and osmotic adjustment substances to adapt to unfavorable environmental conditions[9,10,14].In plants,abiotic stress can induce the production of cellular reactive oxygen species (ROS), which have dual functions depending on the amount of ROS involved:at a low level, ROS trigger defenses and developmental responses at early stages; at a high level, ROS attack the cell membrane to breakdown the defense barrier and thus destroy the cell [15–17].
ABA concentration has been reported to contribute to the aerobic adaptation of upland rice [18].9-cis-epoxycarotenoid dioxygenase (NCED), oxidatively cleaves both 9′-cis-neoxanthin and 9′-cis-violaxanthin to produce xanthoxin, which plays vital roles in ABA biosynthesis [19,20].The NCED encoding gene(LOC_Os12g24800), named OsNCED2, which may function in the ABA synthesis pathway, was identified as a domestication gene with a fixed SNP(C to T)that may contribute to the dryland adaptation of upland rice[18].However,how OsNCED2 regulates stress responses has not been determined and the function of the SNP mutation requires further clarification via transgenic verification.
Plants modify their development and physiological status to adapt to the environment and thus protect themselves from detrimental conditions by triggering a variety of signaling pathways,and ABA likely interacts with other hormones,such as auxins,gibberellins (GAs) and brassinosteroids (BRs), to regulate these processes [21–23].ABA has also been reported to interact with other pathways in a tissue-dependent manner [12].For example, phosphatase enzymes, especially PP2C, have been shown to function as negative regulators of ABA signaling and the MAPK pathway,which regulate stomatal closure in leaves in response to drought conditions [24,25].In plants, the MAPK pathway also functions in signal transduction in response to drought,ROS,pathogen defense,wounding, and low temperature [26,27].Because rice contains seven paralogous genes in the whole genome and might play different roles in rice development and response to the environment,the functions of OsNCED2 in different tissues at different developmental stages needs to gain insight into the genes and pathways that contribute to aerobic adaptation regulated by OsNCED2.
For the base transition of the domesticated fixed SNP of OsNCED2 from C to T, which results in an amino acid change from valine to isoleucine, there is currently no direct evidence of functional variation.CRISPR/Cas9 has emerged as a promising genetic perturbation system, enabling breakthroughs in fundamental and translational research in agriculture [28].A cytidine deaminase such as APOBEC1 can convert cytidines into uridines within a five-nucleotide window specified by the sgRNA [29], which can realize fixed SNP from C to T in OsNCED2 to give functional variation evidence in chief.
Dryland adaptation traits are mainly selected through grain yield under stress conditions in crop plants[3,30].To demonstrate whether the OsNCED2T(T-type OsNCED2) locus contributes to root development and drought resistance in rice production under dryland conditions, yield traits of near-isogenic lines (NILs) with OsNCED2Tfrom upland rice and plants with edited fixed SNP should be evaluated to elucidate its application potential.
Here, we generated OsNCED2Tand OsNCED2Coverexpression lines and mutants to study their function in ABA mediated dryland adaptation.We revealed that OsNCED2Tplay roles in promoting ABA accumulation, root development and drought resistance.Using RNA-seq,we identified a set of OsNCED2Tdownstream genes involved in MAPK and BR signaling pathways and in root development-related genes.We provide in planta evidence that the SNP(C/T)results in an increase in ABA concentration,root system and drought resistance.In addition, we showed that the import of OsNCED2Tinto irrigated rice enhanced grain yield under dryland conditions and therefore could be used in the breeding of water-saving and drought-resistant rice.
2.Materials and methods
2.1.Plant materials, growth conditions, and phenotyping
Oryza sativa L.cvs.Koshihikari and IRAT104 were used as recipient rice lines for genetic transformation and NIL construction.Three NILs containing the OsNCED2Tlocus (named OsNCED2TNIL1~3) were obtained by crossing the upland rice cultivar IRAT104 with the irrigated rice cultivar Koshihikari followed by backcrossing to Koshihikari and DNA marker-assisted selection(MAS) from BC4F9lines with a cleaved amplified polymorphism sequence (CAPs) marker (Table S1).
For root phenotype measurement at the seedling stage(30 days after germination, DAG), plants were grown in Yoshida nutrient solution [31] in a growth chamber with 14 h of daylight at 28 °C and 10 h of darkness at 25 °C.Root phenotyping was performed with Epson Perfection V850 Pro Photo Scanner (Epson Portland Inc., Hillsboro,Oregon,)that determined total length,surface area,volume and tip number of roots.Six plants of each genotype were used for phenotyping.
Drought tolerance was evaluated in a pot experiment with 200 g of sterilized field soil.Nine plants per pot and three replicates of each line were used,grown under the same controlled conditions described above.In the stress treatment,drought stress was initiated by withholding water at weekly time intervals when the soil water content reached 10% at 5 days after the drought treatment as measured with a soil moisture meter (TZS-W, Zhejiang Top Instrument Co.Ltd., Hangzhou, Zhejiang, China).
Phenotyping was performed in the field under paddy and dryland conditions throughout the growth stage at the breeding center of Yunnan University, Xishuangbanna, Yunnan Province,China.For paddy conditions, seeds were germinated in a seedbed and the seedlings were transplanted to a paddy field,where water was ponded on the soil surface throughout the growth and developmental period.For aerobic or upland conditions, direct seeding was performed by dibbling the seeds into dry soil.During the growth stage, no irrigation was applied, and the plants were dependent on rainfall.The available water content (AWC) in the soil reached 30%–40%, a level defined as moderate drought via measurements performed at weekly time intervals.Another experiment involving severe drought stress treatment was performed under dryland conditions using a rainout shelter that controlled the soil AWC at 15%–20%.The seedling cultivation and transplanting methods used were the same as those used for the paddy field,and drought treatment was applied at 10 days after transplanting.For each line, three replicates were planted, with each replicate consisting of 12 plants in two rows (six plants in each row), with a row spacing of 20 cm and a plant spacing of 20 cm.For each line,approximately six plants from each replicate were randomly selected and phenotyped for the grain yield, which included the number of panicles per plant,seed-setting rate,1000-grain weight and yield per plant.
2.2.Phylogenetic analysis of the NCEDs and identification of ciselement across the promoter of OsNCED2
Protein sequences from the completely sequenced genomes were downloaded from UniProt (https://www.uniprot.org/).Clustalw 2.0 (https://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align the protein sequences of the NCEDs with default parameters.The construction of phylogenetic trees was conducted through MEGA 5.0 [32] via the neighbour-joining (NJ) method.Bootstraps with 1000 replicates for Poisson correction model were performed to assess node support.
The cis-element in 1.5 kb upstream of OsNCED2′s start codon was analyzed via Plant CARE online software(https://bioinformatics.psb.ugent.be/webtools/plantcare)[33]with default parameters.
2.3.Vector construction and genetic transformation
The coding region of OsNCED2 was amplified from genomic DNA of IRAT104 and Koshihikari by PCR using primers containing Pst I and Spe I restriction sites (Table S1).The resulting OsNCED2 fragments were inserted into the Pst I and Spe I sites of pCUbi1390 plasmid [34] to generate UbiPro::OsNCED2Tand UbiPro::OsNCED2C.Then, 3.4 kb fragments from the OsNCED2 promoter of IRAT104 were amplified from rice genomic DNA by PCR and inserted into the pMDC162 vector [35] using Gateway technology (Invitrogen,Thermo Fisher Scientific, Shanghai, China) to generate OsNCED2-Pro::GUS, which aims to test promoter activity of OsNCED2 by the β-glucuronidase (GUS) reporter gene.CRISPR/Cas9 gene editing[36] was used to produce OsNCED2 mutants.The target sequence(GTGTAGATTACGGTGAGCCG) before a protospacer-adjacent motif(PAM)in the coding region of OsNCED2 was designed and selected using targetDesign (http://skl.scau.edu.cn/targetdesign/).The single-guide RNA (sgRNA) containing the fused target sequence,crRNA and trans-activating crRNA was inserted into a modified CRISPR/Cas9 vector as previously described [36].23-bp targeting sequences across the fixed SNP (including the PAM) (Table S1)were selected within the target gene OsNCED2Cof Koshihikari,and their targeting specificity was confirmed with a BLAST(https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the rice genome.The designed targeting sequences were synthesized and annealed to form oligo adaptors.The pCSGAPO1 vector [29] was digested with Bsa I and purified using a DNA purification kit(TIANGEN Biotech (Beijing) Co., Ltd., Beijing, China).A ligation reaction mixture(10 μL) containing 10 ng of the digested pCSGAPO1 vector and 0.05 mmol L-1oligoadaptor was prepared and directly transformed into competent E.coli cells to produce CRISPR/Cas9 editing plasmids.
All the vectors were introduced into Agrobacterium tumefaciens strain EHA105.Then, the overexpression cassettes (UbiPro::OsNCED2Tand UbiPro::OsNCED2C), GUS reporter gene expression cassette (OsNCED2Pro::GUS), and CRISPR/Cas9 editing cassette were transferred into Koshihikari by Agrobacterium-mediated transformation following a standard protocol [37].IRAT104 was used to generate osnced2 knockout mutants.More than 20 independent transformation lines were obtained for each transgenic vector.
2.4.RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from rice tissues with TRIzol reagent(Invitrogen, USA).cDNA synthesis was performed with EasyScript First-Strand cDNA Synthesis SuperMix(TransGene,Beijing,China).qRT-PCR analysis was performed using TaKaRa SYBR Premix Ex Taq.The relative expression level of target gene was calculated with the 2-ΔCTor 2-ΔΔCTmethod [38], depending on whether there was a WT control available.The primers (Table S1) used for qRT-PCR were subsequently tested according to unimodal dissociation curve analysis for the absence of nonspecific amplification.The rice ubiquitin gene was used as the reference.All analyses were performed with three biological replicates.
2.5.Subcellular localization of GFP-OsNCED2 fusion proteins
Subcellular localization prediction of OsNCED2 protein was performed by an online software with default parameters [39].The open reading frames (ORFs) of OsNCED2Tcloned from IRAT104 were inserted into pMDC43 as C-terminal fusions to the green fluorescent protein(GFP)reporter gene driven by the CaMV 35S promoter [35].For transient expression, GFP-OsNCED2 fusion vector constructs were transformed into the leaves of 3-week-old tobacco(Nicotiana benthamiana) plants by A.tumefaciens infiltration [40].The green fluorescence resulting from GFP-OsNCED2Texpressing leaves and isolated protoplasts at 60 h after infiltration was observed with a confocal laser scanning microscope (LSM700,Zeiss, Jena, Germany).The 35S::GFP construct was used as a control.
2.6.Histochemical staining assay
Sample tissues from OsNCED2Pro::GUS transgenic plants were submerged in GUS staining buffer (containing 2 mmol L-15-bromo-4-chloro-3-indolyl glucuronide, 0.1 mol L-1sodium phosphate buffer [pH 7.0], 0.1% [v/v] Triton X-100, 1 mmol L-1potassium ferricyanide, 1 mmol L-1potassium ferrocyanide, and 10 mmol L-1EDTA),vacuum infiltrated for 10 min,and then incubated overnight at 37°C.The staining buffer was removed,and the samples were cleared with 95%(v/v)ethanol and observed under a stereoscope [41].
2.7.Measurement of ABA concentration
For ABA content assays, shoots (containing the coleoptile and first leaves) and roots of plants at seedling stage were harvested.For each sample, 0.2 g of fresh tissue was homogenized in liquid nitrogen,weighed,and extracted for 24 h with cold methanol containing 5% formic acid and 6 ng of 2H6-ABA (internal standard).Endogenous ABA was purified and measured as previously described [42] with some changes in detection conditions [43].
2.8.Physiological trait response to drought stress
Fresh weight loss monitoring was performed at indicated [44]time points (hourly) to measure the water loss rate (WLR) in detached leaves.Samples were collected as described above for the pot experiment under well-watered normal conditions and 1 day after drought treatment.Relative water content (RWC, in %)was calculated as [(FM - DM)/(TM - DM)] × 100, where FM,DM, and TM are respectively the fresh, dry, and turgid (water saturated) masses of the weighed leaves.Relative electrolyte leakage(REL) or solute leakage from rice leaves was evaluated following Arora et al.[45].The REL induced by each treatment was calculated from electrical conductivity as follows: [(Lt - Lc)/(100 - Lc)]× 100), where Lt and Lc are the percent conductivities of treated and control samples, respectively.Fresh leaves (0.5 g) of seedlings under normal and stress conditions were harvested and used to measure malondialdehyde (MDA) content and superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) activity levels as previously described [46,47].The second leaves from plants subjected to drought stress for 1 d were subjected to scanning electron microscopy (SEM) following You et al.[44] with minor modifications.Fresh leaf samples were prefixed for 3 h in 3% glutaraldehyde–sodium phosphate buffer (0.1 mol L-1) at room temperature and rinsed three times with 0.1 mol L-1sodium phosphate buffer.Postfixation was performed with 2%OsO4at 4°C.The samples were dehydrated through an ethanol series, infiltrated with an isoamyl acetate series, and coated with metal particles for SEM observation of guard cells in upper and lower epidermises.A SEM instrument (Hitachi S750, Japan) was used to acquire images, and the numbers of guard cells in 30 randomly selected visual fields were recorded.
2.9.RNA-seq analysis
Total RNA was extracted from seedlings of OsNCED2T-OE3 and WT rice plants using TRIzol reagent (Invitrogen, USA).For each sequencing library,100 mg of RNA from each replicate was mixed and three replicates were conducted.The libraries were sequenced using an Illumina HiSeq 2000 sequencer.Low-quality nucleotides(< Q20) were trimmed from the raw sequences obtained for each sample,and paired-end reads with at least one end with length<30 nt were removed using an in-house Perl script.The retained highquality reads were mapped to the Nipponbare reference genome MSU 7.0 (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0)using Bowtie [48].Differential expression analysis of two samples(two biological replicates per sample) was performed with the DESeq2 R package (1.20.0) [49].The resulting P values were adjusted using Benjamini and Hochberg’s approach [50] for controlling false discovery rate.Genes with an adjusted P value<0.001 and a fold change > 2 according to DESeq2 were considered to be differentially expressed[49].Gene Ontology(GO)and Kyoto Encyclopedia of Genes and Genomes(KEGG)pathway enrichment analyses of the rice DEGs were performed with EXPath 2.0[51,52]and visualized with Pathview [53].The raw sequence data have been deposited in the Genome Sequence Archive [54] at the National Genomics Data Center [55], China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences(GSA: CRA008878) and are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
3.Results
3.1.Characterization and expression pattern of OsNCED2
The full-length OsNCED2 gene sequence consists of a single exon of 1731 bp and encodes a 576-amino-acid polypeptide annotated as a 9-cis-epoxycarotenoid dioxygenase.A phylogenetic analysis(Fig.S1) suggested that all NCED paralogs originated from two ancestral genes in rice, and OsNCED2 clustered with other paralogous but not orthologous genes, suggesting that OsNCED2 might have arisen from gene duplication in the rice genome.OsNCED2 contains several signal-responsive cis-elements in its promoter region(1.5 kb upstream of the start codon),including light,anaerobic condition, GA, MeJA and ABA response elements (Table S2),suggesting that the expression of OsNCED2 could be regulated by environmental and hormone signaling.A subcellular localization prediction indicated that OsNCED2 might be localized in chloroplasts, as confirmed by our subcellular localization experiment(Fig.S2), consistent with its function as a 9-cis-epoxycarotenoid dioxygenase that catalyses the synthesis of ABA in chloroplasts[56].
QRT-PCR and histochemical staining indicated the occurrence of OsNCED2 promoter activity in OsNCED2Pro::GUS reporter transgenic plants.OsNCED2 was expressed mainly in mature leaves,stems, and young roots and only weakly in young panicles and seedlings(Fig.1A).Although differences in the OsNCED2 promoter were found between upland and irrigated rice, no differences in expression were detected between IRAT104 and Koshihikari(Fig.1B).GUS histochemical staining of the OsNCED2Pro::GUS reporter transgenic plants also revealed consistent expression in related tissues (Fig.1C).Notably, OsNCED2 was expressed mainly in new or young roots and not in mature roots (Fig.1D).
3.2.OsNCED2 is involved in ABA accumulation,root development,and drought tolerance in rice
Two independent homozygous novel alleles in T1marker-free plants were obtained for the OsNCED2Tknockout mutants(Fig.S3A).One allele had an insertion (+1 bp) in OsNCED2T, and the other had a deletion(-5 bp)(Fig.S3A).These new mutant alleles were named osnced2t-1 and osnced2t-2.To confirm its potential function in ABA synthesis and root development, we determined the ABA content and root phenotype of the mutants and WT plants.The osnced2t-1 and osnced2t-2 plants showed lower ABA content and fewer lateral roots than wild type (WT) plants (Fig.2A, B;Table 1).Overexpression of OsNCED2Tin Koshihikari promoted ABA accumulation and root development,resulting in a denser root system with a greater number of crown and lateral roots(Figs.2C,D,S3B;Table 1).Knockout of OsNCED2Treduced the number of lateral roots,as reflected in root surface area,volume,tip number,and dry weight (Fig.S3C; Table S3).In contrast, overexpression of OsNCED2Tpromoted root development (Fig.S3C; Table S3).These results indicate that OsNCED2 can mediate rice root development by regulating ABA contents.In comparison with nontransgenic WT plants,knockout and overexpression of OsNCED2Trespectively reduced and increased drought tolerance (Fig.2E–H).At 3 d after drought stress, OsNCED2Toverexpression lines showed fewer curled leaf blades than the WT.After 5 d of drought treatment,all the rice leaf blades were curled, and the plants were subsequently rewatered.The OsNCED2Toverexpression lines showed higher seedling survival rates than the WT after 7 d of rewatering(Fig.2E–H).In contrast, the osnced2tmutants were more sensitive to drought stress, with lower survival rates than the WT (Fig.2E–H).
3.3.Physiological manifestations mediated by OsNced2T promoted plant drought resistance
Leaf surfaces of the OsNCED2T-OE1 and OE3 plants showed higher stomatal closure rates than those of WT plants (Fig.3A).To investigate the physiological responses mediated by OsNCED2T,several indices of drought-induced effects on OE and WT leaves at the seedling stage under both normal and drought conditions were measured.OE leaves showed lower WLR than WT leaves (Fig.3B).RWC in OsNCED2T-OE1 and OE3 plants after 1 h of exposure to drought stress was higher than that in WT plants(Fig.3C).OE seedlings experienced less REL and showed lower MDA concentrations after drought treatment than WT seedlings, revealing that overexpression of OsNCED2Tcould protect plants from extensive cell membrane injury (Fig.3D, E).OE lines showed increased ability to regulate redox homeostasis even under normal conditions(Fig.3E–H).Moreover, under drought stress conditions, the CAT,POD and SOD activities in the OsNCED2T-OE1 and OE3 plants were significantly higher than those in the WT plants (Fig.3F–H), indicating that the OsNCED2T-OE plants exhibited active detoxification via the regulation of ROS scavenging in response to drought [57].Thus, the DT of OsNCED2T-OE plants was greater than that of the WT plants, indicating that overexpression of OsNCED2 increased DT via physiological regulation.
3.4.A C-to-T SNP contributed to functional differentiation of OsNCED2 between irrigated and upland rice
OsNCED2T-OE plants showed higher ABA content than OsNCED2C-OE plants (Figs.4A, S3B), supporting the notion that the mutation leads to functional variation in proteins.To determine whether the fixed SNP, from C to T, distinguishing irrigated rice from upland rice was responsible for the functional variation in OsNCED2, the newly developed CRISPR/Cas9 gene editing technology[29]was used to edit the C-SNP of OsNCED2 in Koshihikari.Eight positive independent lines identified as heterozygous at this site were identified from 32 transgenic T0lines via both the CAPs marker and Sanger sequencing.The ABA content and root system of both T-type and C-type plants of the T1generation were evaluated.The ABA content in both leaves and roots of the T-type plants was significantly higher than that in the C-type plants (Fig.4B).T-type plants had denser lateral roots than C-type plants, which had greater total surface area,volume,tip number,and dry weight of roots (Fig.4C–E).The grain yield of the T-type plants was significantly higher than that of the C-type plants, owing mainly to increased productive panicles and seed-setting rate (Fig.4F).These results confirm that the C-to-T SNP distinguishing irrigated rice from upland rice results in functional variation in OsNCED2,indicating that this fixed SNP site functions in the adaptation and artificial selection of upland rice.
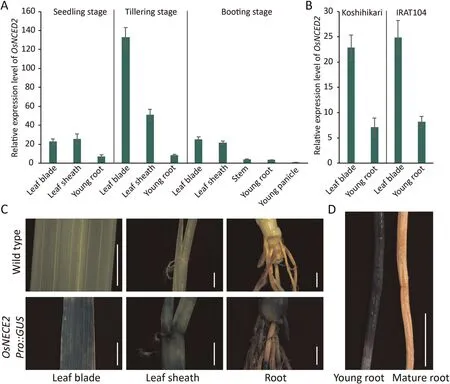
Fig.1.Expression pattern of OsNCED2 in five tissues of rice.(A) Relative expression level (2-ΔΔCT) of OsNCED2 in rice tissues at the seedling, tillering and booting stages.Young panicles were used as reference controls.(B)Koshihikari and IRAT104 exhibit similar OsNCED2 expression levels and models.Values are means±SD(n=3 biological replicates).(C)OsNCED2 expression in tissues of OsNCED2Pro::GUS transgenic rice plants determined by GUS staining.(D)Fresh young roots showed high OsNCED2 expression(left), whereas old roots showed low OsNCED2 expression (right).Scale bars, 0.5 cm.
3.5.Pathways and downstream genes involved in drought resistance mediated by OsNCED2
As shown in Table S4,respectively 200 and 546 genes were upand downregulated in the leaves of the OsNCED2T-OE3 plants compared with those of the WT plants, and 414 and 297 genes were up-and downregulated in the OsNCED2T-OE3 roots compared with WT roots under normal growth conditions (Table S5).OsNCED2Tmediated fewer upregulated but more downregulated genes in the leaves than in the roots, indicating that different pathways were regulated in a tissue-dependent manner in response to stress conditions.Many genes encoding retrotransposons and transposon proteins were highly upregulated in both leaves and roots of the OsNCED2T-OE3 lines (Tables S4, S5).
GO analysis revealed that DEGs in both leaves and roots were represented in biological process,cellular component,and molecular function pathways (Tables S6, S7).DEGs were highly differentially enriched in leaves and roots, suggesting that the ABA molecules regulated by OsNCED2 function differently among tissues.Many DEGs in leaves were enriched in ‘‘transcription factor activity”,‘‘calcium ion binding”,‘‘protein ubiquitination”,‘‘oxidoreductase activity” and ‘‘oxidoreductase activity”, whereas some DEGs in roots were enriched in‘‘response to stress”,‘‘nucleoside-tri phosphatase activity” and ‘‘actin cytoskeleton” (Tables S6, S7).Further KEGG analysis also showed that the DEGs identified in OsNCED2T-OE3 were involved in ‘‘brassinosteroid biosynthesis”,‘‘mitogen-activated protein kinase (MAPK) signaling pathway”,and ‘‘starch and sucrose metabolism”, ‘‘diterpenoid and flavonoid biosynthesis” in leaves and roots, respectively (Table S8),indicating that OsNCED2Tregulates plant growth and development and response to environmental stress via multiple pathways.These development-associated DEGs further explained the decrease in plant height and increase in tiller number of the OsNCED2Toverexpression lines (Fig.S3D–F).In particular, a phosphoprotein phosphatase 2C (PP2C, LOC_Os09g15670)-encoding gene in the MAPK signaling pathway was downregulated in the leaves of the OsNCED2T-OE3 lines(Fig.S4A),alleviating the inhibition of SnRK2s and activating ABF transcription factors to close stomata[11].Two root development-associated genes, OsAMTR1 (LOC_Os05g39770)[58] and OsMED14_1 (LOC_Os08g24400) [59] were up-regulated in roots of OsNCED2T-OE3 plants (Fig.S4B, C), and may be downstream genes of OsNCED2Tinvolved in increasing the number of lateral roots.
3.6.OsNCED2T-NILs showed more developed root systems and increased drought tolerance, resulting in higher grain yield under dryland conditions
OsNCED2T-NIL1~3 presented a deeper and denser root system than Koshihikari, which showed a greater number of crown roots and lateral roots (Fig.S5A; Table 1).To investigate the effect of OsNCED2Tand its potential application in rice breeding, we compared the grain yields of Koshihikari and OsNCED2T-NIL plants under normal and aerobic conditions with moderate drought(natural upland conditions) and severe drought (artificially controlled by rainproof installations) conditions.Under normal conditions,the two lines showed similar yields, and no differences in other agronomic traits except panicle number per plant were detected(Fig.S5B–E; Table S9).Although moderate drought significantly reduced tiller number (even panicles per plant), seed-setting rate,and 1000-grain weight in all tested plants, OsNCED2T-NIL plants showed higher grain yield per plant and yield per ha than Koshihikari plants because of these three yield factors (Fig.S5B–G).Under severe drought, more prominent physiological damage, in the form of leaf wilting and delayed flowering, was detected in Koshihikari plants than in OsNCED2T-NIL plants (Fig.S5H;Table S9).Yield per plant and yield per ha of the OsNCED2T-NILs were higher than those of the Koshihikari plants (Fig.S5F–H).Thus, under drought conditions, both the increased rooting and dehydration tolerance promoted by OsNCED2Tfacilitated increase in growth vigor and grain filling, resulting in increased yields.

Fig.2.OsNCED2 regulates root development and drought tolerance by promoting ABA synthesis.ABA content(A)and root phenotype(B)of OsNCED2T-KO plants.ABA content(C)and root phenotype(D)of OsNCED2T-OE plants.Values in(A,C)are means±SD(n=9 plants).Scale bars,2 cm.Evaluation of drought tolerance of potted WT and transgenic plants.Seedling growth phenotypes of OsNCED2T-KO mutant lines(E)and OsNCED2T-OE lines(G)under normal conditions,after 3 days of drought stress and after 7 days of rewatering.The seedling survival rates(F,H)at 7 days after rewatering.Values in(F,H)are means±SD(n=27 plants).Scale bars,5 cm.WT,wild type.In(A),(C),(F)and(H),significant differences were determined by two-tailed Student’s t-tests (*, P < 0.05; **, P < 0.01).
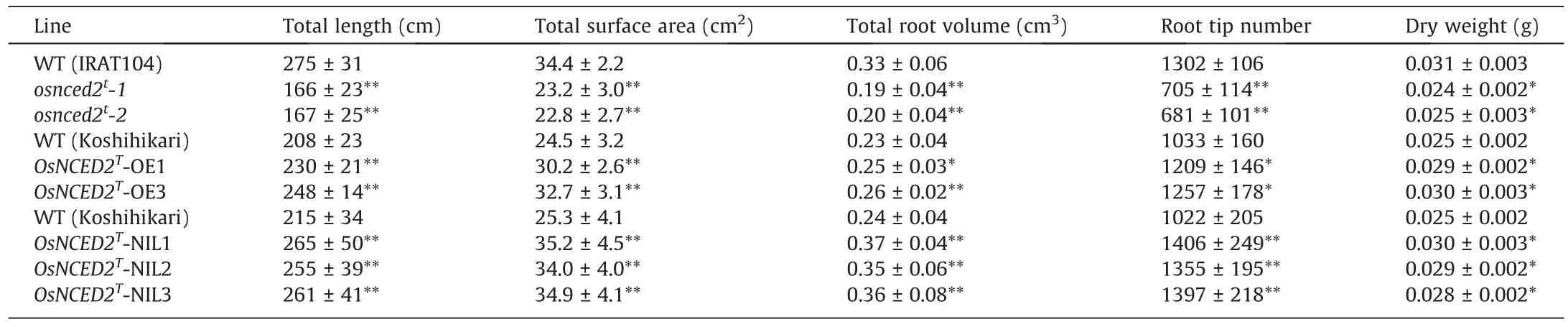
Table 1 Root phenotypes of OsNCED2T transgenic and NIL plants at the seedling stage.
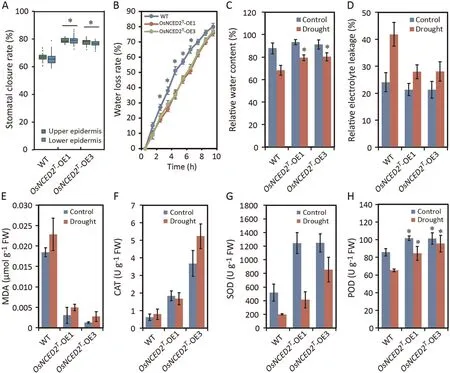
Fig.3.Physiological traits of OsNCED2T-OE lines under normal (control)and drought conditions.(A)Stomatal closure rate of leaves of OsNCED2T-OE and WT lines(both the upper and lower epidermis were counted in 10 randomly selected high-power fields).(B)WLRs of transgenic and WT lines.Thirty fully expanded leaves from each replicate at the seedling stage were used in triplicate experiments.The RWC(C)and REL(D)of OsNCED2T-OE lines under normal and drought conditions(1 d of drought treatment).(E)MDA content of OsNCED2T-OE and WT plants under normal conditions and after 1 d of drought treatment.Activity of ROS-scavenging enzymes,including CAT(F),SOD(G)and POD(H),in OsNCED2T-OE and WT plants under normal conditions and after 1 d of drought treatment.Values are means±SD(n=3 biological replicates).Differences from WT plants were determined by two-tailed Student’s t-tests (*, P < 0.05).
4.Discussion
Upland rice cultivars are regarded as promising breeding materials due to their inherent drought-tolerant characteristics [60].Upland rice plants modify their root development to adapt to the dryland conditions [61].Roots sense water deficits in soil, and the deep-penetrating and denser lateral roots of upland rice facilitate drought adaptation [3,18].Several studies have investigated the root phenotypes of upland rice[3,18,60,62],but few genes that are specifically responsible for the specialized roots in upland rice have been reported.In this study, we investigated the root developmental function mediated by OsNCED2Tin upland rice.OsNCED2T-mediated ABA accumulation increases OsAMTR1 and OsMED14_1 expression,promoting lateral root development.Similar to upland rice, deep-penetrating and thicker primary roots are known to confer drought resistance traits in several species [63–67].

Fig.4.The SNP (C to T) resulted in the functional differentiation of OsNCED2.(A) OsNCED2T-OE plants have higher ABA contents than OsNCED2C-OE plants in the same background.Values in the histograms are means±SD(n=3 biological replicates).(B)ABA content in roots of both C-type and T-type OsNCED2 edited plants.Root phenotypes of C-type and T-type OsNCED2 plants at seedling (C) and heading (D) stages.Scale bars, 0.5 cm.(E) Comparison of root phenotypes of C-type and T-type plants.The root phenotypic data,including total length,total superficial area(surface area),total root volume and root tip number,indicate that T-type plants have denser roots than C-type plants.(F) Grain yield differences between C-type and T-type lines under paddy and aerobic conditions.For the C-type and T-type rice subgroups (n = 48 plants), each subgroup contains 6 plants per line,and 8 lines were used for phenotyping.In each box plot,the centerline indicates the median,the edges of the box represent the first and third quartiles,and the whiskers extend to span a 1.5 interquartile range from the edges.Differences between C-type and T-type plants were tested by two-tailed Student’s ttests (*, P < 0.05).
Plants develop deep and dense roots to absorb water from the soil, along with rapid stomatal closure to reduce evaporation and thereby increase drought avoidance under dryland conditions[68–71].In addition to an enhanced root system due to OsNCED2Tmediated improvements in root development,this study indicated that rapid stomatal closure was regulated by OsNCED2Tin upland rice, which partially endows OsNCED2T-containing rice with improved drought avoidance ability.The regulation of stomatal closure has been studied in depth at the molecular and physiological levels.Among the pathways involved in stomatal closure,AREB/ABF transcription factors, which are regulated by active phosphatase enzymes (SnRK2s) dependent on ABA and MAPK signaling pathway, is a core pathway involved in stomatal closure[11,24,25,72].This study revealed a downregulated PP2C gene,which may result in rapid stomatal closure in OsNCED2T-OE3 plants through activation of MAPK signaling pathway.
As part of the adaptive response to stress, ABA can inhibit vegetative growth, especially the growth of aboveground tissues,increasing root/shoot ratio for further drought avoidance through inhibition of the growth-promoting properties of BRs [73,74].The expression of BR signaling-responsive genes, such as LOC_Os03g12660, LOC_Os02g11020, LOC_Os01g63260 and LOC_Os06g39880, was markedly downregulated in the OsNCED2TOE3 plants, suggesting that OsNCED2 might also participate in the inhibition of plant growth and development by antagonizing the BR pathway.Recent studies [75,76] have indicated that ABA and BR signals synergistically regulate plant growth and adaptation, but whether OsNCED2 mitigates the drought–growth tradeoff to boost yields through the synergistic interplay of ABA and BR signals needs further study.
Although ABA promotes dryland adaptation by adjusting above drought avoidance characteristics,it directly affects drought tolerance through rapid stress responses and protection under drought conditions [17].Under drought conditions, defects in osmotic homeostasis and the ion balance of plant cells trigger a signal transduction pathway for the recovery of cellular homeostasis and the repair of damaged proteins and membrane systems [77].In upland rice,OsNCED2Tstrengthened osmotic adjustment ability,and antioxidant and membrane-protection systems, which was supported by direct evidence from the measurements and RNAseq results in this study.Many enzymatic and nonenzymatic antioxidants, such as SOD, CAT, ascorbate peroxidase, and glutathione reductase, can maintain redox homeostasis by reducing excessive deleterious ROS[78,79],contributing to tolerance to various stress through ABA signaling and MAPK pathway[26,27].Consistent with these findings, the results from the present study provide further evidence supporting the role of OsNCED2 in ABA accumulation and root development as well as its downstream responses to drought stress.
ABA synthesis in plant cells is vital in control of ABA levels and subsequent downstream pathways.NCED is the rate-limiting enzyme in catalysis of ABA production [19,20].In rice, seven orthologous NCED genes originated from two ancestral genes via gene duplication may play different roles depending on the expression pattern and enzyme activity.The similar expression of OsNCED2 in the two ecotypes under both normal and drought conditions suggested that the amino acid change at this site might have resulted in functional variation.Because a direct assay of NCED activity was not successful, the notion that NCED variants of the fixed SNP contribute to enzyme activity in ABA synthesis could not be directly verified.Thus, we alternatively confirmed the fixed SNP site from C to T increasing by comparing ABA contents between plants overexpressing OsNCED2Cand OsNCED2Tin the same background as well as via the base editing assay.Variation in the protein sequence of OsNCED2 might promote dryland adaptation in upland rice.Additional studies are needed to determine whether the function of OsNCED2Tis tissue-specific and whether this gene plays a cell type-dependent role.
Unlike mobile animals, sessile plants cannot evade but rather must tolerate unfavorable environmental conditions, such as drought, salinity and cold.Our results support the key role of OsNCED2 in ABA accumulation and the subsequent ABA signaling pathway and suggest that this gene contributes to ABA-triggered drought avoidance and tolerance.ABA responses mediated by OsNCED2Tmight offer an advantageous strategy used by upland rice to survive and breed under dryland conditions.OsNCED2Tcontaining rice showed greater yield under both rainfed upland and severe drought conditions and could be used in breeding water-saving and drought-resistant rice.
5.Conclusions
OsNCED2 functions in ABA accumulation, increasing root growth and development and drought tolerance.The domesticated fixed SNP in upland rice changes OsNCED2 function and ABA accumulation,increasing dryland adaptation of upland rice by increasing survival rate and yield under dryland conditions.The gene may be used in breeding water-saving and drought-resistant rice.
CRediT authorship contribution statement
Liyu Huang:Conceptualization,Visualization,Writing–original draft, Writing – review & editing, Funding acquisition.Yachong Bao:Methodology, Writing – review & editing.Shiwen Qin:Methodology,Writing–review&editing.Min Ning:Methodology,Writing – review & editing.Shilai Zhang:Investigation.Guangfu Huang:Investigation.Jing Zhang:Investigation.Fengyi Hu:Conceptualization,Funding acquisition,Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (U1602266, 32060474, and 31601274) and grants from the Yunnan Provincial Science and Technology Department(202005AF150009 and 202101AS070001).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.12.001.
杂志排行
The Crop Journal的其它文章
- Corrigendum to ‘‘GmTOC1b negatively regulates resistance to Soybean mosaic virus”.[Crop J.11 (2023) 1762–1773]
- Decoding the inconsistency of six cropland maps in China
- Wetting alternating with partial drying during grain filling increases lysine biosynthesis in inferior rice grain
- OsDA1 positively regulates grain width in rice
- A polygalacturonase gene OsPG1 modulates water homeostasis in rice
- Trehalose: A sugar molecule involved in temperature stress management in plants
