Searching for plant NLR immune receptors conferring resistance to potyviruses
2024-03-07XinHongShufenLiXiaofeiChengHaijianZhiJinlongYinKaiXu
Xin Hong, Shufen Li, Xiaofei Cheng, Haijian Zhi, Jinlong Yin*, Kai Xu*
a Jiangsu Key Laboratory for Microbes and Functional Genomics, Jiangsu Engineering and Technology Research Center for Microbiology, College of Life Sciences, Nanjing Normal University, Nanjing 210023, Jiangsu, China
b College of Plant Protection, Northeast Agricultural University, Harbin 150030, Heilongjiang, China
c National Center for Soybean Improvement, National Key Laboratory for Crop Genetics and Germplasm Enhancement, Key Laboratory of Biology and Genetic Improvement of Soybean-Ministry of Agriculture, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China
Keywords:NLR Potyvirus resistance Genetic diversity Mapping Engineer
ABSTRACT To fight against invasion by pathogens,plants have evolved an elaborate innate immune system,of which the nucleotide-binding domain leucine-rich repeat-containing receptor (NLR) acts as the sensor and immune executor.Potyviruses, comprising one of the largest genera of plant viruses, cause severe crop yield losses worldwide.Inherited crop resistance to potyviruses can be used in breeding and plant transgenesis to control disease development.This review summarizes achievements in mapping and cloning NLR genes conferring dominant resistance against potyvirus in the families Fabaceae, Solanaceae,Brassicaceae, and Cucurbitaceae.It compares mechanisms of potyviral protein recognition and downstream signaling employed by NLRs and discusses strategies for exploiting NLRs to better control diseases caused by potyviruses.
1.Introduction
Plants have evolved immune systems to combat infection by pathogens such as fungi, bacteria, viruses, and nematodes.One such system is effector-triggered immunity (ETI) mediated by nucleotide-binding domain leucine-rich repeat-containing receptors (NLR).
Most plant NLRs contain three distinct and conserved domains:the Toll/interleukin-1 receptor (TIR) or coiled-coil (CC) domain in the N terminus, the nucleotide-binding (NB-ARC) domain, and the leucine-rich repeat(LRR)domain in the C terminus.NLRs with the TIR domain are called TNL, and those with the CC domain are CNL.NLRs perceive pathogen attacks by recognizing pathogenencoded effectors.This recognition is effector-specific and thus makes ETI a‘‘gene-for-gene”resistance.Successful ETI is characterized by a hypersensitive response (HR) in infected plant cells and induces a robust and enduring response[1–3].Because ETI confers more effective resistance than other defense mechanisms, NLR genes are targets of crop resistance breeding.
Viruses are non-cellular organisms that parasitize and multiply in host cells.The first plant virus discovered was the tobacco mosaic virus (TMV), which caused large losses in the tobacco industry in colonial Colombia in the nineteenth century [4].TMV is one of the most studied plant viruses,and the NLR gene N against TMV was the first cloned plant NLR conferring disease resistance[5].Besides TMV,more and more viruses have been found to cause crop diseases and have become a primary constraint to global agricultural development.Consequently, a growing need exists to identify NLR resistance genes in crops.
Potyviruses comprise one of the largest genera of plant viruses[6].They infect the broadest range of hosts, including Solanaceae,Fabaceae, and Brassicaceae [7], and cause damage to crops worldwide.Potyviruses are positive-sense single-stranded RNA viruses.Their genome structure is relatively simple and conserved among all potyvirus members.Due to the resemblance among potyviruses, investigations into plant resistance against them utilized common strategies and encountered similar challenges, warranting a comprehensive summary.Recently, a significant number of achievements in identifying NLR-mediated potyvirus resistance has been made, including 1) characterization of new NLR genes[8–14], 2) mapping of more resistance loci [15–18], and 3) novel ways to find and engineer functional NLRs [19–23].This review describes earlier and more recent progress in identifying host NLR-mediated resistance against potyviruses, compares strategies of NLRs for viral protein recognition and downstream signaling,and discusses prospects of engineering novel potyvirus resistances.
2.Potyvirus-recognizing NLRs in legumes (Fabaceae)
Legumes,species of the Fabaceae family,comprise about 20,000 species in 700 genera [24] and are thought to have emerged 60 million years ago[25].Around one-third of human dietary nitrogen is provided by legumes as food [26].Viral diseases are one of the main constraints to global legume production,particularly in tropical and subtropical regions [27].Potyviruses are found in almost all leguminous crops.The genetic sources of potyvirus resistance in legumes have been extensively studied.Here, we discuss those associated with NLR genes.
2.1.Soybean
Soybean[Glycine max(L.)Merr.]is a major source of dietary oil.Soybean meal, a co-product of soybean oil, feeds livestock and poultry.Cultivated soybean was domesticated from wild soybeans in East Asia 6000–9000 years ago [28] and has become a top-traded commodity crop [29].Potyviruses pose a significant threat to soybean production, with soybean mosaic virus (SMV,family Potyviridae;genus Potyvirus)being the most prevalent potyvirus in soybean, found in nearly all soybean production areas worldwide [30].Based on phenotypic reactions on soybean differential lines,SMV isolates can be classified into distinct strains.Ten differentials used in China can classify SMV into 22 strains[31,32].In the United States and the Republic of Korea, more than seven strains (primarily G1–G7, but also G5H, G7H, and others) have been identified using eight differential lines [33,34].
Several dominant resistance loci have been identified in soybean accessions resistant to various SMV strains.These include Rsv1, Rsv3, Rsv4, and Rsv5, which are based on studies of U.S.and Korean SMV strains, and Rsc3Q, Rsc4, Rsc-pm, and Rsc-p, based on studies of Chinese SMV strains.
Rsv1, the first identified SMV-resistance locus, is located on chromosome 13.It was identified in the resistant soybean cultivar PI 96983 (R/resistance) via crossing with susceptible cultivar Lee 68 (S/susceptible) [35].More than ten alleles of Rsv1 have been found in soybean accessions [36–41], making it the most frequently found among all SMV-resistance loci.Genetic mapping and resistance analysis of this locus in various soybean cultivars have revealed distinct,tightly linked resistance genes(Fig.1).First,Rsv1 recognized both P3 and Hc-Pro as effectors in a study involving the exchange of key amino acids between the avirulent SMV-N,an isolate of SMV-G2 strain,and the virulent strain SMV-G7[42].In two PI 96983-derived recombinant inbred lines(RILs),L800(carrying the Rsv1 candidate gene 3gG2)and L943(lacking 3gG2)[43],at least two genes mediating resistance could be differentiated [44].One was represented by the induction of resistance in L800 via recognition of P3 from SMV-N, and the other in L943 recognized and induced resistance to SMV-N Hc-Pro [44].Second, in F2and F2:3(F2-derived F3) lines from a cross between soybean cultivars PI 96983 (R) and Nannong 1138–2 (S), two loci that were either identical or close to Rsv1 were discovered and designated as Rscpm and Rsc-ps [45].The Rsc-pm map interval contains 3gG2 and is involved in the resistance to SMV strains SC3, SC6, and SC17.Locus Rsc-ps, at a short distance of 36.9 kb from Rsc-pm, confers resistance against the SMV SC7 strain.In addition to Rsc-ps,whose map interval does not contain the candidate gene 3gG2, the Rsv1 allelic gene Rsv1-h from cultivar Suweon 97 and Rsv1-r from cultivar Raiden have both been mapped[15]to a genomic region that is different from that containing 3gG2 and Rsc-pm(Fig.1).Rsv1-h and Rsv1-r confer resistance to SMV strains SC6-N and SC7-N.
Gene 3gG2 encodes a CNL clustered with nearby CNL-encoding genes in PI 96983[44].Virus-induced gene silencing(VIGS)of 3gG2 and its neighboring genes 5gG3 and 6gG9 compromised the resistance of L78-379, a soybean cultivar Williams-based nearisogenic line (NIL) carrying Rsv1 from PI 96983 [46].The VIGS approach also showed that EDS1 (Enhanced Disease Susceptibility 1), PAD4 (Phytoalexin-Deficient 4), JAR1 (jasmonate-amino acid synthetase), EDR1 (enhanced disease resistance 1), HSP90 (heat shock protein 90), WRKY6 (WRKY transcription factor 6), and WRKY30 (WRKY transcription factor 30) are downstream factors mediating 3gG2 signaling [46].In the Rsc-ps map interval of PI 96983, several CNL genes lie in series with nearby clustered TNL genes, as shown by the Williams 82 reference genome [15,45].Some TNL genes in the Rsc-ps map interval are also candidate genes for Rsc20 from cultivar Qihuang 1, mediating resistance to SMV strain SC20 [16].These Rsc20 candidates are Glyma.13g194700 and Glyma.13g195100,encoding two full-length TNL genes sharing high amino acid similarity [16].Additionally, two CNL genes,Glyma.13g184800 and Glyma.13g184900, are candidates of Rsv1-h and Rsv1-r.Rsv1 and the nearby loci on chromosome 13 comprise the most complex and diverse region conferring SMV resistance within the soybean genome.Although Rsv1, or neighboring loci,has been employed for breeding SMV resistance [47,48], no corresponding NLR gene(s) has been confirmed and cloned.
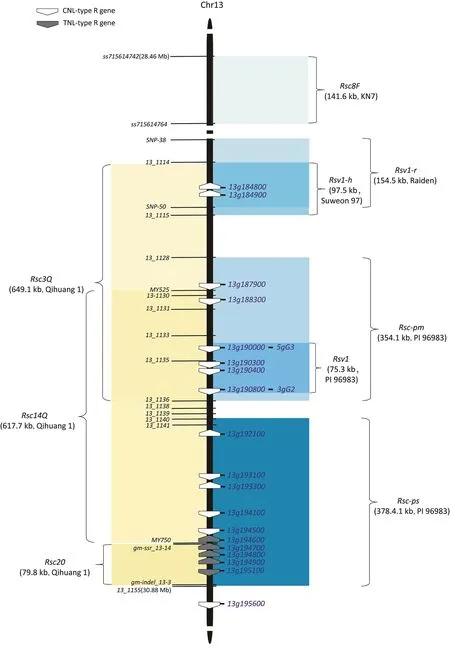
Fig.1.Rsv1 and adjacent resistance loci on chromosome 13 mapped in various soybean cultivars.The DNA markers used to map these loci are shown.The resistance gene name is followed by the physical length of the mapped interval.The cultivar name carrying the resistance gene is shown in parentheses.Empty and solid pentagonal shapes in map intervals represent genes encoding CNLs and TNLs, respectively.The corresponding gene names and physical positions are annotated based on the Wm82.a4 soybean assembly.
Rsv3 and Rsc4 have been mapped to overlapping regions on chromosome 14 [8,49,50].Rsv3 was first found in the F2-derived line OX686 carrying a resistance gene from a cross between cultivars Columbia and Harosoy [51,52].It confers resistance to SMV G5-G7[53].Rsv3 was mapped to a 154-kb region in F2populations from L29 (R) × Lee68 (S) and Tousan 140 (R) × Lee68 (S) [50], in addition to a BC3F2population of L29 (R) × Sowon (S) [54](Fig.2).This region contains five CNL genes allelic to the reference genes, Glyma.14g204500, Glyma.14g204600, Glyma.14g204700, Glyma.14g205000, and Glyma.14g205300, from Williams 82, respectively [54].The Rsc4 locus in cultivar Dabaima confers resistance to many Chinese SMV strains [8,49].The Rsc4 interval is 63 kb in length within the Rsv3 interval [49].Three CNL genes, Rsc4-1,
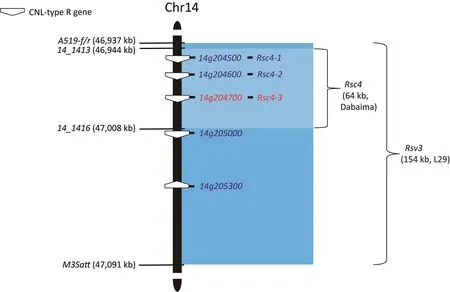
Fig.2.The Rsv3 and Rsc4 loci on soybean chromosome 14.The markers used to map the two loci and their corresponding physical position are shown.The gene name is followed by the physical length of the mapped interval.The cultivar name carrying the resistance gene is shown in parentheses.Empty pentagonal shapes represent genes encoding CNLs.The corresponding gene names and physical positions are annotated based on the Wm82.a4 soybean assembly.Rsc4-1,Rsc4-2,and Rsc4-3 are genes in the Rsc4 mapping region allelic to 14g204500, 14g204600, and 14g204700, respectively.
Rsc4-2, and Rsc4-3, allelic to Glyma.14g204500, Glyma.14g204600,and Glyma.14g204700, are located in the map interval containing Rsc4 (Fig.2).Based on CRISPR/Cas9-mediated gene knockout in Dabaima and induction of HR during transient expression upon SMV infection in Nicotiana benthamiana leaves, Rsc4-3 was shown to be the resistance gene corresponding to the Rsc4 locus.Rsc4-3 encodes a cell wall-localized CNL that recognizes SMV-encoded cylindrical inclusion (CI) protein in the apoplast and induces cell death at the front of SMV cell-to-cell movement [8].A study [55]of Rsv3 downstream signaling revealed that viral movement is inhibited by upregulated callose deposition at plasmodesmata.Rsc4-3 contains only one amino acid polymorphism relative to its allelic gene NBS_C (Glyma.14g204700) from Zaoshu 18 or L29[56].It is phylogenetically conserved with all Rsv3-type alleles[8].To date, Rsc4-3 is the only cloned NLR for SMV resistance that has also been validated using a knockout mutant [8].
Rsv4 is a locus on chromosome 2 mediating resistance to all US SMV strains[41].It was initially identified in the F3:4line LR2 from the cross of cultivar PI 486355 (R)× Essex (S) [41].The Rsv4 locus in LR2 was flanked by two markers,Satt542 at a distance of 4.7 cM and Satt558 at 7.8 cM[57].The Rsv4 locus was further shortened to 0.7 cM, representing an approximately 100-kb region, using the breeding line V94-5152 [58], an F6:7progeny derived from LR2[59].A region of about 120 kb, partially overlapping the 100-kb region [58], was located between markers Rat2 and S6ac in V94-5152 [60] (Fig.3).Similar to LR2/V94-5152, soybean landrace Kefeng 1 carries an Rsv4 allele that is resistant to most Chinese SMV strains [31].Loci in Kefeng 1 mediating resistance to SMV strains SC1 [61], SC3 [62], SC5 [63], SC7 [64,65], SC8 [66], SC9[67],SC10[68],SC13[65],and SC18[69]were mapped to genomic regions overlapping or adjacent to the Rsv4 locus in LR2/V94-5152(Fig.3).In the map intervals of these Rsv4 and their alleles,no NLR genes were found.Using five backcross populations of Peking(R)× Enrei (S) consisting of 9320 individual plants, the Rsv4 allele in Peking was mapped to a 9.8-kb region on chromosome 2 that contains only two genes in the Williams 82 reference genome[70].These two genes produce two RNase H family proteins in the susceptible cultivars Enrei and Williams 82 (NM_001249088 and NM_001253944), but have recombined into one gene via a 3.6 kb genome-deletion event.The resulting recombinant gene,named Rsv4, also encodes an RNase H that is capable of entering the viral replication complex and digesting viral dsRNA via interaction with SMV-coded P3 and NIb proteins[70].The Rsv4 allele was later reported in Kefeng 1,and silencing the allelic gene Rsc3K compromised resistance to SC3, SC5, SC7, SC8, SC10, and SC18 strains[62].The two RNase H coding genes NM_001249088 and NM_001253944 in susceptible cultivar Williams 82 also have anti-SMV activities, given that silencing either gene led to increased SMV accumulation [62].A transient overexpression assay in N.benthamiana showed that Rsv4 or Rsc3K more efficiently inhibited SMV multiplication than NM_001249088 or NM_001253944 [62,70].
In addition to Rsv1,Rsv3,Rsv4 and their alleles,some resistance loci or NLR genes have been shown to inhibit SMV infection.Rsv5 is a newly named SMV resistance locus from soybean cultivar York[71].Rsv5 was initially thought to be an allele of Rsv1 located on chromosome 13 and was previously named Rsv1-y [36].However,some segregation for resistance was observed in the F2:3populations of PI 96983(Rsv1)×York(Rsv5),supporting an inference that Rsv5 is an independent locus but closely linked to Rsv1 at a genetic distance of 2.2 cM [71].The physical map region of Rsv5 on chromosome 13 is not known.In a recent study, a membrane attack complex component/perforin (MACPF) protein GmMACPF1 encoded by a quantitative resistance locus qRsc8F in cultivar KN7[72] was mapped in a 141.6-kb interval adjacent to Rsv1-r and Rsv1 on chromosome 13 (Fig.1).GmMACPF1 belongs to a poreforming cytolytic protein superfamily sharing similarity to CNLderived resistosomes[73–75].GmMACPF1 inhibited SMV accumulation when overexpressed in hairy roots and likely conferred the resistance of cultivar KN7 [72].Besides genetic mapping of the resistance loci,two soybean genes sharing high sequence similarity to the tobacco N gene, an NLR gene mediating resistance to TMV,inhibited SMV infection in transgenic lines [19,20].GmKR3 is a TNL-encoding gene located on chromosome 6 from Kefeng 1.In its own genome context,it does not induce resistance,as suggested by genetic mapping of resistance in Kefeng 1 [62,65,66,69].However, GmKR3 transgenic expression in soybean under the 35S constitutive promoter reduced SMV accumulation in systemically infected leaves[19].A similar approach also identified a truncated TNL lacking the LRR domain, namely GmSRC7, with no sequence differences between resistant and susceptible cultivars.When expressed under the 35S promoter,GmSRC7 demonstrated the ability to inhibit SMV multiplication [20].
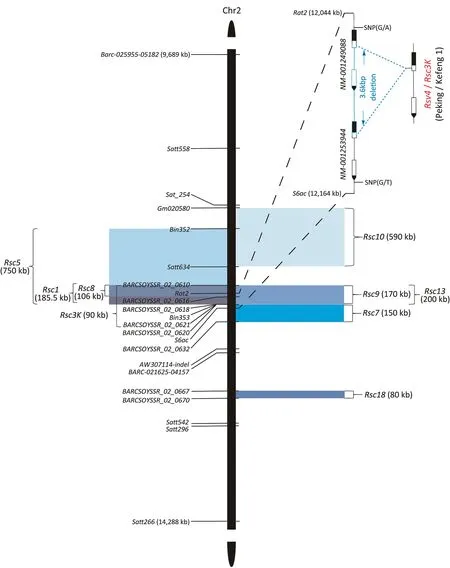
Fig.3.Rsv4 and adjacent resistance loci on soybean chromosome 2.The markers used to map these loci are shown.The resistance gene name is shown, followed by the physical length of the mapped interval in parentheses.The corresponding gene names and physical positions are annotated based on the Wm82.a4 soybean assembly.The Rsv4/Rsc3K gene structures in the Williams 82 and Peking/Kefeng 1 genomes are shown in the upper right corner.Deletions are boxed in blue,and untranslated regions are shown in black.
2.2.Common bean
Common bean (Phaseolus vulgaris L.) was domesticated in Mesoamerica and the Andes mountain range 8000 years ago,from where it spread about 500 years ago to southwestern Europe and then to the rest of the world [76–78].Common bean has genetically diverged into two major subspecies, or gene pools, corresponding to the two centers of domestication [78].Two potyviruses, bean common mosaic virus (BCMV) and bean common mosaic necrosis virus (BCMNV),which were once considered distinct strains of BCMV [79], infect common beans [80].The systemic leaf mosaic and mottling symptoms caused by BCMV and BCMNV in susceptible cultivars are similar [80].
The search for BCMV resistance in common beans began in 1918.In 1925, the variety Robust was found [81] to be resistant to BCMV.In 1934, resistance was found [82] in Corbett Refugee.Unlike Robust, which carries a recessive resistance gene, Corbett Refugee carries a single dominant gene, the I gene [83,84].The extreme resistance response conferred by the I gene against BCMV infection is low temperature- and high dosage-dependent, as increasing the temperature to 34 °C or reducing the dosage in heterozygous plants(I/i)leads to necrosis or HR in upper uninoculated leaves [85,86].The dosage-dependent quality of the I gene can be observed in protoplasts, where each I allele incrementally reduced BCMV RNA accumulation[87].Reducing the dosage while simultaneously increasing the ambient temperature reduced resistance in inoculated leaves and increased HR in upper leaves[85].In contrast,BCMNV,initially recognized as a necrotic strain of BCMV,caused a temperature-independent systemic HR,or‘‘black root”,in beans homozygous for I[88–90].The incomplete dominance of I in heterozygous plants and HR-inducing phenotype is reminiscent of the Rsv1-mediated resistance to SMV [91].
The I gene confers broad-spectrum resistance to other potyviruses besides BCMV and BCMNV.The I gene-carrying common bean cultivar Black Turtle Soup 1 induces BCMV-like temperature-dependent resistance to azuki bean mosaic virus[92] but reacts to SMV or Thailand passiflora potyvirus infection by inducing a BCMNV-like systemic necrosis phenotype [92–94].A temperature-independent complete resistance was also observed on Black Turtle Soup 1 inoculated with passionfruit woodiness virus-K and zucchini yellow mosaic virus [92].A study of a BCMV variant inducing BCMNV-like systemic necrosis attributed the different responses of I to BCMV and BCMNV to the variation in the viral-encoded P1 protein [95], which is the most variable potyviral protein and functions in viral replication and counteraction of host defense [96,97].
Several DNA markers have been developed to identify the I gene[98–100].Their use revealed that I is common in both the Mesoamerica and Andean gene pools [98,100] and may have evolved before domestication.The I gene was mapped to a region at the distal part of the long arm of chromosome 2 in an F2population of Jamapa(I/I)×Calima(i/i)[86].The mapped region of I locus contains a TNL gene cluster that likely underwent gene duplication during evolution.Southern blotting showed that the copy number of the TNL genes in the map interval from I/I genotype plants was doubled compared to that in i/i plants[86].Using the markers Bng45 and Phgp to locate the I locus in the recently released genome assembly(v1.1) of Phaseolus vulgaris 5–593 [101], which likely inherited I from Jamapa based on the pedigree, 27 TNL genes with high sequence similarity were identified as I candidates(Fig.4).
The I gene can be considered one of the most promising resources for potyvirus resistance, given its broad-spectrum resistance[92].Although not yet identified,it has already been used in many breeding practices, typically in pyramiding with recessive genes to provide the broadest possible range of resistance [102].
3.Potyvirus-recognizing NLRs in Solanaceae
The Solanaceae family comprises 3000–4000 species classified in approximately 90 genera,including Solanum tuberosum,S.lycopersicum,and Capsicum annuum,widely cultivated worldwide[103].
3.1.Potato
Potato (Solanum tuberosum L.) is a non-cereal food source in many countries.Potatoes were domesticated in the central Andes of southern Peru and northwestern Bolivia between 8000 and 6000 BCE [104].Modern potato cultivars originate from wild potato species and landraces grown in the Americas,ranging from the southwestern USA to southern Chile, and were introduced to Europe at the end of the 16th century and later to the rest of the world [104,105].
The most devastating potyvirus that infects potatoes is potato virus Y(PVY).It was first identified in the United Kingdom to cause‘‘potato degeneration” [106,107].PVY can be transmitted by tuber propagation through generations[107]and by aphids in the field in a nonpersistent manner [108].Depending on the variety and whether in a mixed infection with other viruses, PVY can cause up to 80% yield loss [109,110].Potato virus A [109] and potato virus V [111] (previously recognized as isolates of PVY strain C)are less prevalent.Resistance reactions to potyviruses are of two main types: extreme and hypersensitive [112].Genes involved in resistance reactions to PVY (Nytbr, Ncspl, Ny-1, NySmira, Rychc, Ny-2,Ryadg, Rysto, and Ry-fsto) and to PVA (Naadg) have been mapped to four loci on chromosomes IV, IX, XI, and XII [113].The name Ry indicates that the gene confers r esistance to PVY,whereas Ny indicates that it causes necrosis or hypersensitive responses to PVY infection [114].
The S.tuberosum Group Andigena carries the Ryadggene, which confers extreme resistance to prevalent PVY strains [115].Gene Ryadgwas first mapped to a distal segment of the long arm of chromosome XI flanked by markers GP125 and TG651 using an F1progeny of a cross between the haploid potato line 2x(V-2)7 carrying Ryadgand the diploid clone 84.194.30 [115].In the same year,another resistance gene Rysto(XI) was reported to lie in a map interval adjacent to Ryadgbetween markers M33b and GP259 in the parent I-1039 [116].This parent was originally thought [116]to be derived from the wild potato S.stoloniferum, accounting for the name Rystoof the resistance gene (XI).Genotyping with markers tightly linked to Ryadgor Rysto(XI)revealed the presence of both loci in PVY-resistant potato breeding lines and cultivars from Uruguay, leading to speculation that they are the same loci [117].Based on microsatellite and plastid DNA markers,I-1039 was proposed to be a hybrid of potato groups Andigena × Tuberosum rather than a derivative of wild potato S.stoloniferum [118,119].Later, S.tuberosum Group Andigena lines that carry Ryadgwere shown [118] to associate with markers M5, M6, M17, and M45,which were reported to be linked to Rysto(XI) [116].This finding showed that the previously reported Ryadglocus was identical or close to the Rysto(XI) locus, which is likely derived from S.tuberosum group Andigena[118].The Ny-2 gene conferring HR to PVY in cultivar Romula and the Naadggene for PVA resistance was also found [120,121] to be adjacent to Ryadgor Rysto(XI).Based on the S.tuberosum group Phureja DM1-3 reference genome (v4.03), the map interval of Ryadgor Rysto(XI) contains a cluster of TNLencoding genes [113].These resistance loci have not yet been characterized.
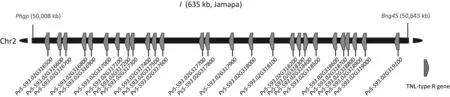
Fig.4.Physical map of the genomic region containing I on chromosome 2 of Phaseolus vulgaris.PhgP and Bng45 are markers linked to I.The 27 solid pentagonal shapes represent genes encoding TNLs.The corresponding gene names and physical positions of the two markers are based on the Phaseolus vulgaris 5–593 genome assembly(v1.1)[101].
The real wild potato S.stoloniferum carrying PVY resistance has been used in breeding various cultivars, in particular in Europe[122,123].The resistance gene Rystowas independently mapped to chromosome XII in multiple potato clones bred to inherit the PVY resistance gene from S.stoloniferum [124–127].Recently[11], the use of resistance gene capture (RenSeq) and singlemolecule real-time sequencing technology (SMRT) sequencing[128] led to the identification of 11 candidate NLR genes from the PVY-resistant diploid clone Alicja, which carries Rystofrom S.stoloniferum.When transiently expressed in N.benthamiana, one of the 11 candidate NLR genes, c630, induced HR upon PVY infection and was proposed to be the Rystogene [11].Rystoencodes a TNL gene whose function depends on the downstream EDS1 signaling module and a helper NLR NRG1 (N requirement gene 1)[11,12].Rystorecognized PVY or PVA coat protein (CP) in response to viral infection.The Rystorecognition sites were mapped to the 149-amino acid central domain of PVY CP, which forms a globular core domain containing seven α-helices and a β-hairpin.This core region is conserved among potyviruses and mediates Rystoconferred resistance to multiple potyviruses [12].
Rychcis a resistance gene from the wild species S.chacoense.Rychcfrom cultivar Konafubuki or a wild diploid S.chacoense accession 40–3 was mapped to the distal part of the longer arm of chromosome IX [13,129].Based on a bacterial artificial chromosome(BAC) library constructed from the S.chacoense accession 40–3,the sequence of the Rychcmap interval was obtained.It contains six candidate genes (C1–C6), different from those mapped in the susceptible cultivar and those from the S.tuberosum group Phureja DM1-3 reference genome [13].The C2 gene encodes an oxidoreductase.C3 and C4 encode two TNLs, whereas C1, C5, and C6 encode proteins with unknown functions.Transformation of C3 and C4 into susceptible potato cultivars showed that C4 conferred PVY resistance and confirmed it to be Rychc[13].Ny-1 and Ny-Smira were also mapped to the same region as the Rychclocus [130,131].Whether Ny-1 and Ny-Smira encode the same protein as Rychcis unknown.
In addition to the well-located loci on chromosomes IX,XI,and XII,two dominant loci,Nytbrin S.tuberosum Group Tuberosum and Ncsplin the wild species S.sparsipilum, were mapped to the short arm of chromosome IV and are likely allelic[132,133].Hc-Pro from PVY-O can be recognized by Nytbror Ncspl[132,134], of which the induced HR limits viral movement.Because the mapped genomic regions containing both loci are relatively large[132],no candidate genes of Nytbror Ncsplhave been identified.
3.2.Pepper
Capsicum spp., including chili and bell peppers, are native to southern North America,Mesoamerica,and parts of South America[135].They were domesticated by indigenous peoples over 6000 years ago.Capsicum spp.is used mainly as a food source but also for medicinal purposes and religious or ceremonial practices [136].
The Potyvirus resistance locus (Pvr) genes in C.annuum and C.chinense are well characterized and confer resistance to several potyviruses, including PVY, tobacco etch virus (TEV), and pepper mottle virus (PepMoV) [137].Among the Pvr genes, Pvr4[138,139]or its allele Pvr7[140,141]and a tentatively named gene Pvr10 are dominant, whereas pvr1 [142], pvr2 (allelic to pvr1)[143], pvr3 [142], pvr5 [138], and pvr6 [144] are recessive.
Pvr4 was identified in the C.annuum Criollo de Morelos 334(CM334), a landrace found in the Mexican state of Morelos [137–139,145].Pvr4 confers broad-spectrum resistance to all naturally occurring PVY and PepMoV strains or pathotypes while susceptible to TEV,chilli veinal mottle virus(ChiVMV),and pepper veinal mottle virus (PVMV) [138,146].Eight amplified fragment length polymorphism (AFLP) markers tightly linked to Pvr4 were developed based on bulked-segregant selection of 145 AFLP markers in the F2progeny of CM334 (R) × Yolo Wonder (S) [147].One of the closest-linked AFLP markers was converted into a cleaved amplified polymorphic sequence (CAPS) marker located 4.6 cM from CD72 [140] at the distal end of the longer arm of pepper chromosome 10 [148].The genetic location of Pvr4 was estimated as 4.6 cM from CD72 on chromosome 10 [140,149].A subsequent genetic analysis using the F2populations of CM334(R) × ECW123R (S) revealed a location of Pvr4 closer to marker TG420 [148], which is near marker CD72 on chromosome 10[150].TG420 was further used for comparative mapping in the genetic maps of pepper and tomato(S.lycopersicum L.)and helped delimit the Pvr4 locus io a 0.4 cM region [151].Screening a BAC library of CM334 followed by sequencing identified 16 genes in the Pvr4 mapping region, eight of which encode CNLs.Only one of the CNL-coding genes, CaNBARC322, induced HR when transiently co-expressed with PepMoV NIb in N.benthamiana and was assigned as Pvr4 [151].
The C.chinense accession PI 159236 and its BC3F3line 9093(developed by backcrossing C.chinense PI159236 to a C.annuum parent) were reported [140] to carry the dominant resistance gene Pvr7.Based on a few susceptible plants from the allelism test,Pvr7 was thought to be a different gene but tightly linked to Pvr4.However, in a later study, 9093 was positive for a Pvr4-specific insertion-deletion (InDel) marker not found in PI 159236[141].Three single-nucleotide polymorphism (SNP) markers from the Pvr4 mapping region cosegregated perfectly with the Pvr7 locus in 9093, suggesting that the resistance from 9093 was inherited from the C.annuum parent, not from C.chinense PI 159236, and that Pvr7 was identical to Pvr4 from CM334.
The resistance of Pvr4 is durable in the field and is often used with other recessive pvr genes in commercial hybrid cultivars to obtain broad-spectrum resistance [147].In the laboratory, resistance to PVY conferred by Pvr4 was broken in grafted pepper plants, with the rootstocks from susceptible plants and the scions from CM334 [146].The virulent PVY mutant contains a K472E mutation in the C terminus of the viral encoded NIb protein.This mutation imposes a high fitness cost on the mutated virus such that it is not competitive with other PVY isolates and never accumulates naturally [152].Transient overexpression of individual PepMoV proteins in CM334 leaves revealed that NIb is the avirulence factor recognized by Pvr4 [153].
Another dominant gene Pvr10 is carried by the commercial hybrid cultivar Magli-R (Sakata Seed Sudamerica Ltda., Brazil)and an inbred line, PIM-025 [17].An allelism test of PIM-025 (R)and C.annuum line Myr-29–10 (R) carrying Pvr4 suggested that the resistance from PIM-025 differs from that of Pvr4.The gene was tentatively named Pvr10 [17], and its genome location is unknown.
Besides mapping resistance genes in resistant cultivars, a homology search method has been used to identify a CNLencoding gene Pvr9 in C.annuum cultivar Floral Gem.Pvr9 is orthologous to the late blight resistance gene Rpi-blb2 from the wild potato species S.bulbocastanum [154].Overexpression of Pvr9 in the presence of PepMoV infection or during viral NIb coexpression induced HR in N.benthamiana leaves [155], a reaction similar to that of overexpression of Pvr4 [151].Pvr9 is transcriptionally upregulated upon PepMoV infection.However, probably owing to a lack of other components of the signaling module needed for resistance, Floral Gem is susceptible to PepMoV [155].Pvr9 is located on chromosome 6 based on an alignment search of Pvr9 sequence against pepper reference genomes.The genomic region containing Pvr9 was also found [9] to contain a dominant resistance gene Cvr1 in the commercial C.annuum varieties CV3 and CV8.Whether Cvr1 is identical to Pvr9 awaits study.
4.Potyvirus-recognizing NLRs in Brassicaceae
Brassicaceae was formerly named Cruciferae, a Latin name meaning ‘‘cross-bearing”, for its characteristic four petals.It is speculated to have originated in the Irano-Turanian region and has spread to most of the world [156].It contains vegetables such as Brassica rapa, B.napus, B.juncea, B.oleracea, and the model species Arabidopsis thaliana.Many of these species are natural hosts of turnip mosaic virus (TuMV) [157].
4.1.Brassica spp.
Brassica is the representative genus of the Brassicaceae family.Brassica species contain one or two of the three genome types A,B, and C.They include B.rapa (with genome formula AA), B.nigra(BB),B.oleracea(CC),B.juncea(AABB),B.napus(AACC),and B.carinata (BBCC).TuMV is the only known potyvirus that naturally infects Brassica species.It was first described in 1921 in the USA as causing mosaic disease on turnips[158].An epidemiology study[159] on the phylogeny of the TuMV isolates suggested that the ancestor of TuMV was a virus of wild orchids.It adapted to Brassicaceae plants and initially occurred in southern Europe, Asia Minor,and the Middle East,and then spread to East Asia, Oceania,the Americas, and other parts of the world.Infected crops show symptoms including leaf mosaic,stunting,and vein necrosis.Yield losses of crops infected by TuMV were as high as 70%[160].Several dominant TuMV resistance genes have been identified in Brassica spp., most in the A genome [161].
TuMV RESISTANCE IN BRASSICA 01 (TuRB01) is the first mapped TuMV resistance locus[162].It confers extreme resistance to infection by TuMV pathotype 1 (isolate UK1) in B.napus spring oilseed rape doubled haploid(DH)line N–o–1.Following a cross of N–o–1(R)with winter oilseed rape DH line N–o–9(S),a segregating population of DH lines derived from F1pollen cells was used to map TuRB01 to a 7.2 cM region between markers pN101a and pW137cNM on the long arm of chromosome A6 [162] (Fig.5).The presence of TuRB01 on the chromosome A6 of the Brassica genome type A in N–o–1 indicated its origin in B.rapa.Indeed,TuRB01b,conferring extreme resistance to TuMV-UK1 in the B.rapa Chinese cabbage cultivar Tropical Delight, was later mapped on chromosome A6 to the exact interval to which TuRB01 was mapped[163].A naturally occurring TuMV-UK1 variant carrying an N459D mutation in the viral-encoded CI protein (or N1686D mutation in the TuMV-encoded polyprotein) broke the resistance of both TuRB01 and TuRB01b genotypes [164,165], supporting the hypothesis that these two genes are identical [163].In the B.napus line 22S resistant to TuMV pathotype 4 (isolate CDN1), a dominant resistance gene TuRB03 was identified and mapped to the same interval as TuRB01 and TuRB01b between positions 105.5 and 116.2 cM on chromosome A6 [18,166] (Fig.5).TuRB03 is susceptible to all TuMV pathotype 1 isolates and is different from TuRB01 and TuRB01b.P3 seems to be the avirulent factor for TuRB03-triggered immunity, given that an amino acid mutation I153F in P3(or I973F in polyprotein)converted virulent TuMV-UK1 to avirulent in B.napus line 22S.A P3 F153I mutation in TuMV-CDN1 led to viral infection [167].Although TuRB03 was proposed to be located in the same gene cluster as TuRB01 and TuRB01b, they may be different genes in view of the differences in recognition of viral virulence factors.TuRBCH01,conferring resistance to TuMV strain C5 (isolate Hu1) in B.rapa subsp.chinensis (pak choi), was also mapped to this region containing TuRB01, TuRB01b, and TuRB03 [168].Candidate genes for these loci have not been identified.
TuMV-R is a dominant resistance locus identified in the B.rapa DH line VC40 in a cross with susceptible DH line SR5[169].It confers resistance to the TuMV C4 strain and is located in an 0.34-Mb interval on the short arm of chromosome A6 between SNP marker N0343 and inDel marker CUK_0040i (Fig.5).In the reference genome, the map interval contains 56 genes, four encoding CNLs and two encoding pathogenesis-related protein 1.Besides TuMV-R,another C4 strain-resistance locus, TuRB07, was mapped and located on the short arm of chromosome A6 in the B.rapa DH line VC1 [170] (Fig.5).The TuRB07 locus lies in a 0.13-Mb physical interval flanked by simple sequence repeats (SSR) markers H132A24-s1 and KS10960.This region contains a cluster of NLR genes, of which only Bra018863 encodes a full-length CNL and was accordingly proposed as a candidate for TuRB07 [170].The mapped regions of TuMV-R and TuRB07 both contain CNL gene clusters [171] (Fig.5).Recently [18], a new dominant resistance gene to TuMV strain C4 was mapped to chromosome A6 in B.rapa line B80124 in a cross with susceptible line B80450 (Fig.5).The map interval of 0.33 Mb is flanked by SNP markers A06S11 and A06S14 and differs from the previously reported lociTuRB01,TuRB01b, TuRB03, TuMV-R, and TuRB07 on chromosome A6.This new dominant resistance locus has 67 candidate genes, including three genes encoding CNLs.Based on transcriptome and sequence differences between resistant and susceptible alleles, one of the three CNL encoding genes, BraA06g035130.3C, was considered the likeliest candidate.The three B.rapa loci for resistance to TuMV strain C4 (TuMV-R, TuRB07, and BraA06g035130.3C) were all mapped to chromosome A6 and likely encode CNLs, but they appear to be different genes, as there is no overlap of their map intervals.
TuRBCS01 is a dominant resistance locus from the Chinese cabbage line BP8407 that confers resistance to the TuMV C4 strain[172].It is located in a 1.98 Mb-long region in chromosome A4 flanked by markers SAAS_mBR4055_194 and BrID10723.TuRBCS01 is the only known dominant resistance locus on chromosome A4 for TuMV.The gene corresponding to TuRBCS01 is unknown.
Some Brassica resistances have been distinguished based on varying reactions to viral genotypes.Two such loci, TuRB04 and TuRB05,were reported[173]to be present in the Brassica A genome of B.napus line 165 as two loci segregating in a BC1S1population of 165 (R) × N–o–9 (S).TuRB04 was found to confer extreme resistance by recognizing P3 encoded by TuMV-UK1 or TuMV-CHN12(pathotype 3).A point mutation on P3 (F312L) broke resistance in the TuRB04 genotype [173].TuRB05 was shown to recognize CI, and an M589T mutation in CI was found to compromise resistance.Unique amino acid requirements of viral proteins for the induction of TuRB04- or TuRB05-mediated resistance suggest that TuRB04 or TuRB05 differ from previously characterized resistance loci in the Brassica A genome.The locations of these loci and the corresponding genes are not known.
4.2.Arabidopsis thaliana
Arabidopsis thaliana has a short lifecycle and a small genome of about 135 Mb.As a model organism, it has been used for genetic,cellular, and molecular biological studies of flowering plants.Although no potyvirus has been found to infect this species naturally,it has been challenged by artificial methods.A few dominant loci, including NLR genes, have been found to confer resistance against potyviruses.
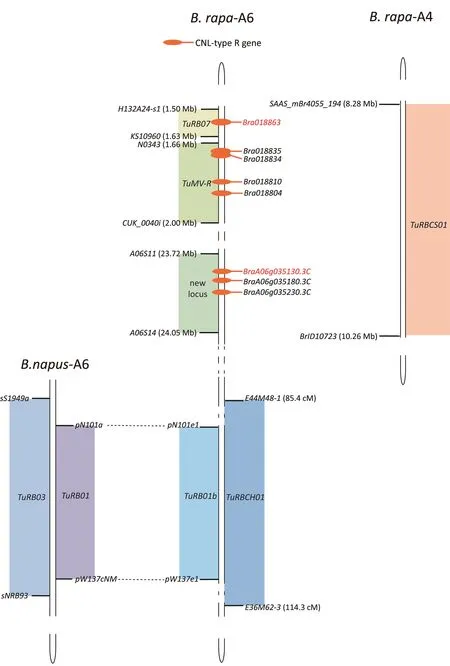
Fig.5.TuMV resistance loci on chromosomes 4 and 6 of the type A genome of Brassica.The markers flanking these loci are shown.Known physical positions are shown in parentheses for some markers.Mapped regions for each resistance gene are highlighted in color.Empty pentagonal shapes represent genes encoding CNLs.Each candidate NLR gene is highlighted in red.Dotted lines connecting the mapped regions of TuRB01 and TuRB01b suggest that these two genes are the same gene [163].
TuNI is a dominant locus on chromosome 1 of the Arabidopsis ecotype Landsberg erecta (Ler), conferring the HR-type resistance to the TuMV isolates Azu and TuR1 [174].The mapped region obtained via crossing with the susceptible ecotype Col-0 is about 105 kb long and flanked by markers mXF41 and mRF28.This interval contains 19 genes, of which five encode CNLs.Through gene swapping between the genomes of TuMV isolates TuR1(avirulent)and TuC(virulent),P3 was identified as the symptom determinant[175].Expression of P3 in transgenic Ler plants or transient expression in Ler protoplasts was sufficient to induce cell death.By primer walking in a BAC clone BIBAC2 of Ler, the TuNI locus was localized to a small region in chromosome 1 containing three CNL candidate genes [176].These CNLs likely synergistically contribute to the resistance responses.Arabidopsis thaliana ecotype Ler is also resistant to local infection by the plum pox potyvirus(PPV) isolate SoC [177].Because the local resistance, restricting viral infection to inoculated leaves, depends on the expression of SGT1 and RAR1, an NLR gene is speculated to mediate the resistance.
A genome-wide association study with 450 natural A.thaliana accessions was performed to identify the genes responsible for infection by the TuMV-G and TuMV-S strains [178].Variation among AT2G14080 alleles, encoding a TNL, was associated with plant symptom severity upon infection by both TuMV-G and TuMV-S.
5.Potyvirus-recognizing NLRs in Cucurbitaceae
Cucurbitaceae,the gourd family,is a large family with 130 genera and 800 species worldwide[179].The most common cucurbits are squash or pumpkin (Cucurbita spp.), watermelon (Citrullus spp.), cucumber (Cucumis sativus L.), and honey melon (Cucumis melo L.).The original habitats of various cucurbits are in the Americas,Africa,and Asia[180].Cucurbita spp.was domesticated in the Americas, whereas Citrullus spp.originated in Africa and Cucumis spp.in Asia and Melanesia.Viruses can cause devastating diseases in cucurbits and include potyviruses such as papaya ringspot virus(PRSV), watermelon mosaic virus (WMV), and zucchini yellow mosaic virus (ZYMV).
Cucumis melo accession PI 180280, collected in the Indian state of Gujarat, carries a dominant gene, Wmv-1 or Prv1, conferring extreme resistance to PRSV (formerly called watermelon mosaic virus 1) [181–183].Melon line PI 180283 from India also carries a dominant gene for resistance to PRSV [184].Cantaloupe lines B66-5 [185] and WMR 29 [186] were selected to carry the Wmv-1/Prv1from PI 180280, and Charentais melon breeding line 72,025 was generated to carry the resistance gene from PI 180283 [187].These two genes have been shown [184] to be allelic,as no segregation for susceptibility was observed in the F2progenies of B66-5 × 72025 or WMR 29 × 72025.
PI 414723 is a C.melo breeding line derived from PI 371795,collected in Mussoorie (Uttar Pradesh, India).It is resistant to PRSV[188], WMV [189,190], and ZYMV [188].The resistances to these potyviruses in PI 414723 have been shown [191] to be controlled by different genes.Among them, Wmv or Prv2confers dominant resistance to PRSV[183,188]and is allelic to Prv1in WMR 29[183].
Gene Prv2was located[183]within a 10,057-bp region between markers RG10-1 and RG-A in the linkage group IX based on a BC1S1mapping population developed by backcrossing the F1hybrid of PI 414723 (R) × Védrantais (S) to PI 414723.The interval contained only one TNL-type gene RGH10 designated Prv.Prv alleles in both resistant cultivars PI 414723 and WMR 29, namely Prv2or Prv1,and those from susceptible cultivars MR-1, Dulce, and Védrantais share the same domain structure, and all contain an extra copy of NB-ARC domain at the C terminus [183].The NB-ARC domain represents a molecular switch that triggers downstream signaling.Thus, the extra copy may contribute to another level of effectortriggered activation of the defense signal.In a recent study [192],CRISPR/Cas9-mediated knockout of the WMR 29-origin Prv1allele in resistant melon line Charéntais Prv-R compromised PRSV resistance, confirming the previous genetic mapping results that identified Prv [183].
The resistance to ZYMV strain E15 in PI 414723 has been shown[188,193] to be controlled by a single dominant gene Zym.This gene reacts to ZYMV infection without symptoms or by causing top necrosis or plant death.Based on its linkage to gene Female flower form (or gene a), Zym was located on chromosome 2 [194](linkage group IV in the formerly used reference map [195]).In another study, three complementary dominant genes in PI 414723 (Zym-1, Zym-2, and Zym-3) conferred resistance to an aphid-nontransmissible, highly virulent ZYMV NAT strain [193].Among these genes,Zym-1 was mapped to the same linkage group as Zym, whereas Zym-2 was located on chromosome 10 (formerly linkage group VII [195]).Recently [14], locus Zym or Zym-1 in PI 414723 was confined to a 17.6-kb region on chromosome 2 based on newly developed markers derived from two BAC clones of the susceptible cultivars MR-1 and WMR 29.Based on the differences from the susceptible genotype, the likely candidates for the Zym locus include genes encoding an NAC-like transcription factor and two or three CNLs (NBL-1, NBL-2, and NBL-3) depending on the cultivar[14].Whether these Zym candidate genes confer resistance to ZYMV is unknown.
PI 414723 also displays incomplete resistance to WMV (formerly watermelon mosaic virus 2), represented by symptom development from WMV-infected plants at an early stage of infection and no virus detected in newly emerged leaves [190].The resistance to WMV has been shown [190] to be controlled by a dominant gene Wmr.In addition to C.melo, dominant resistance genes against potyviral infection have been reported for cucumbers [196–198], citron melon [199], and Cucurbita moschata[200–202] but have not been characterized.
6.Plants have evolved divergent mechanisms for recognizing potyviruses
The speciation of potyviruses started 7250 years ago in southwest Eurasia or North Africa,likely from an ancestor virus infecting monocots[203].Since then,potyviruses have evolved the ability to infect many plant hosts, especially dicots, and expanded their diversity as one of the largest viral genera [6,203].A potyvirus encodes 11 functionally conserved and structurally similar viral proteins in its ~10 kb-long single RNA genome.At the initial stage of plant resistance response mediated by NLRs,these viral proteins can potentially serve as avirulent factors recognized by NLR immune receptors [3].It is recognized that some pathogen effectors are selectively and convergently recognized by plant immune receptors.Pseudomonas syringae cysteine protease AvrPphB can be recognized by Arabidopsis CNL RPS5 and barley (Hordeum vulgare L.) CNL PBR1, which are not orthologous [204].Two rice NLR Pik-1 alleles independently evolved to recognize the same effector AVR-PikD of the blast fungus [205].Thus, a few potyviral proteins may be preferentially recognized by multiple plant immune receptors, owing to the function and sequence similarity of potyviral proteins among potyviruses.
Based on genetic studies of potyvirus-plant pathosystems, five of 11 viral proteins can be recognized by various NLRs (Fig.6).Of these five, CP is the structural protein that is present at the time of viral infection and accumulates to a high level in the form of assembled virions at a late stage of viral infection[206].The other four are all nonstructural proteins that are produced during infection.CI likely assists viral RNA replication via its RNA helicase activity and forms intracellular cylindrical inclusions to facilitate the intracellular movement of the virus [207,208].CI is also found in the apoplast, where it can be recognized by the soybean CNL Rsc4-3 [8].HC-Pro is an RNA silencing suppressor localized to the cytosol and occasionally associates with microtubules and the cortical endoplasmic reticulum [209].P3 and NIb are components of the viral replication complexes derived from the endomembrane system [210,211].These five potyviral proteins are distinct in their functions, subcellular localizations, or structures.Although three or four NLRs can recognize P3 or CI,no apparent strong preference for specific potyviral proteins in the NLR recognition is currently observed (Fig.6), suggesting that recognition of potyviral proteins by NLRs evolved divergently.
7.Requirements of downstream signaling components
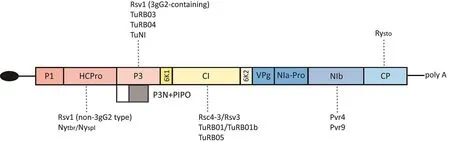
Fig.6.Recognition by NLRs of different potyviral proteins.Colored boxes in the long open reading frame represent mature proteins produced by proteolysis during potyvirus infection.The viral protein genome-linked (VPg) is shown at the 5′-end of the viral genome as a black ellipse.The dotted lines represent the recognition of NLRs to viral proteins.
Despite their diverse recognition mechanisms, evidence from genetics studies on the signaling requirements of these NLRs suggests that relatively conserved downstream factors are needed.First, the solanaceous plants N.benthamiana and N.tabacum have been shown [8,10–12,155] to be capable of displaying HR when a heterogeneously expressed NLR recognizes the avirulent potyvirus or the viral effector.These NLRs include potato TNL Rysto[11,12],pepper CNL Pvr4 [10] and Pvr9 [155], and a non-solanaceous CNL Rsc4-3 from soybean[8],suggesting that downstream components are conserved across species.Second, some key downstream factors are involved in resistance to multiple potyviruses(Table 1).RAR1 (Required for Mla12 resistance), SGT1 (Suppressor of G-two allele of Skp1), and HSP90 are molecular chaperones that help maintain the inactive form of NLR in the absence of pathogen infection [212] and assist in NLR oligomerization [213].The successful immune responses conferred by Rsv1, Pvr4, and the PPV-resistance gene in Arabidopsis ecotype Ler required some or all of the RAR1-SGT1-HSP90 components [10,46,177,214].
EDS1 functions in the plant immune system.It forms heterodimers with PAD4 or SAG101 (Senescence Associated Gene 101)upon the activation of upstream TNL and induces Ca2+channel activity in downstream helper NLR ADR1 (activated disease resistance 1) or NRG1, respectively [74,215,216].Potato TNL Rystorequires EDS1, PAD4, SAG101, and NRG1 for its function [11,12].It is not known whether ADR1 is also required for Rysto-mediated immunity.Rsv1 from soybean line L78-379 likely encodes a CNL[44,46].A VIGS experiment showed that EDS1 and PAD4 are required for Rsv1-mediated resistance.However,based on previous studies [217–219] on HRT-mediated resistance to turnip crinkle virus, RPS2-mediated resistance to P.syringae, and RPP8-mediated resistance to Hyaloperonospora arabidopsidis biotype Emco5, it has been suggested that EDS1 works redundantly with the salicylic acid pathway to regulate CNL-mediated immunity in plants.An eds1 single knockout usually does not lead to compromised resistance by CNLs in Arabidopsis [217,220,221].Elucidating how EDS1 and its binding partner PAD4 are involved in SMV resistance in soybean line L79-379 will help characterize the complex and still-mysterious Rsv1 or the neighboring genes on chromosome 13 (Fig.1).
8.Exploring and engineering new NLR resources
Decades of genetic studies on different pathosystems of potyvirus infections have revealed limited resistance gene resources in various plant species (Table 2).However, resistance-breaking isolates of many potyviruses have been continuously reported and endanger the use of resistance genes in breeding practices[8,42,70,152,173].There is a demand for innovative approaches.
There is evidence that well-characterized NLRs could be used across plant species.Nicotiana tabacum plants transformed with potato Rystodisplayed resistance to PPV and TuMV[12].Transgenic Arabidopsis expressing Rystowas resistant to TuMV infection [12].Pvr4 transformed into potatoes conferred immunity to PVY [227].Given that many potyviruses can infect more than one crop,deployment of NLR genes across plant species could increase their utility.
As in most examples in this review,NLR genes are usually found in selected plant accessions with qualitative resistance.In a search for genes homologous to the tobacco N gene in susceptible soybean cultivars or in genome regions not containing a resistance locus,GmKR3 and GmSRC7 induced SMV resistance when overexpressed under a 35S constitutive promoter [19,20].Similarly, pepper Pvr9 was identified based on screening for PepMoV resistance conferred by those homologs of the late blight resistance gene Rpi-blb2[155].These identified NLRs are normally nonfunctional, as their native promoters probably cannot drive enough gene expression to induce resistance.Numerous NLRs are likely concealed from mapped resistance loci, awaiting exploration and utilization.
Engineering a well-characterized NLR is a proven strategy for achieving new effector recognition [228].It is plausible that wellcharacterized NLRs conferring potyvirus resistance,especially their LRR domains, can be improved for extended recognition of more potyviruses, given that potyviral proteins are functionally and structurally conserved among species[7].Alternatively, the proteolytic activity of potyvirus NIa protease (NIa-Pro) has been successfully exploited in a decoy-based system [21,23,229], in which degradation of a modified decoy protein PBS1 harboring a NIa-Pro cleavage site was triggered upon potyviral infection.NIa-Pro cleavage sites comprise seven amino acids displaying consensus within each potyviral species[230].This system allows the internal bacterial protease AvrPphB cleavage site in PBS1 to be replaced with one NIa-Pro-cleavage consensus sequence.The PBS1 cleavage caused by NIa-Pro expression leads to CNL RPS5 activation and species-specific resistance to potyviruses.
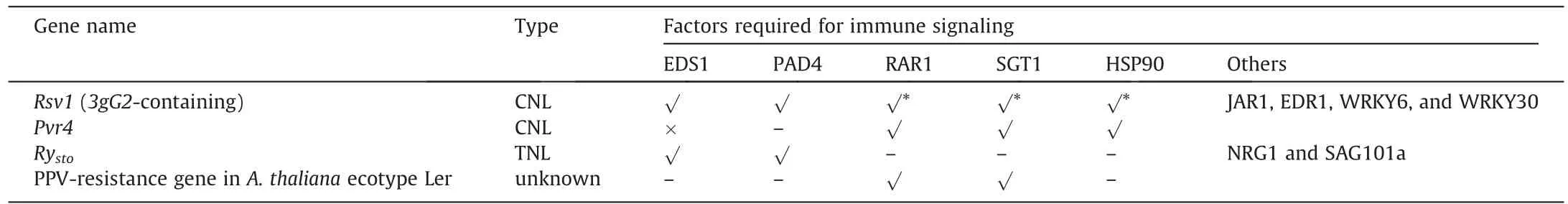
Table 1 Downstream signaling components required by NLRs.
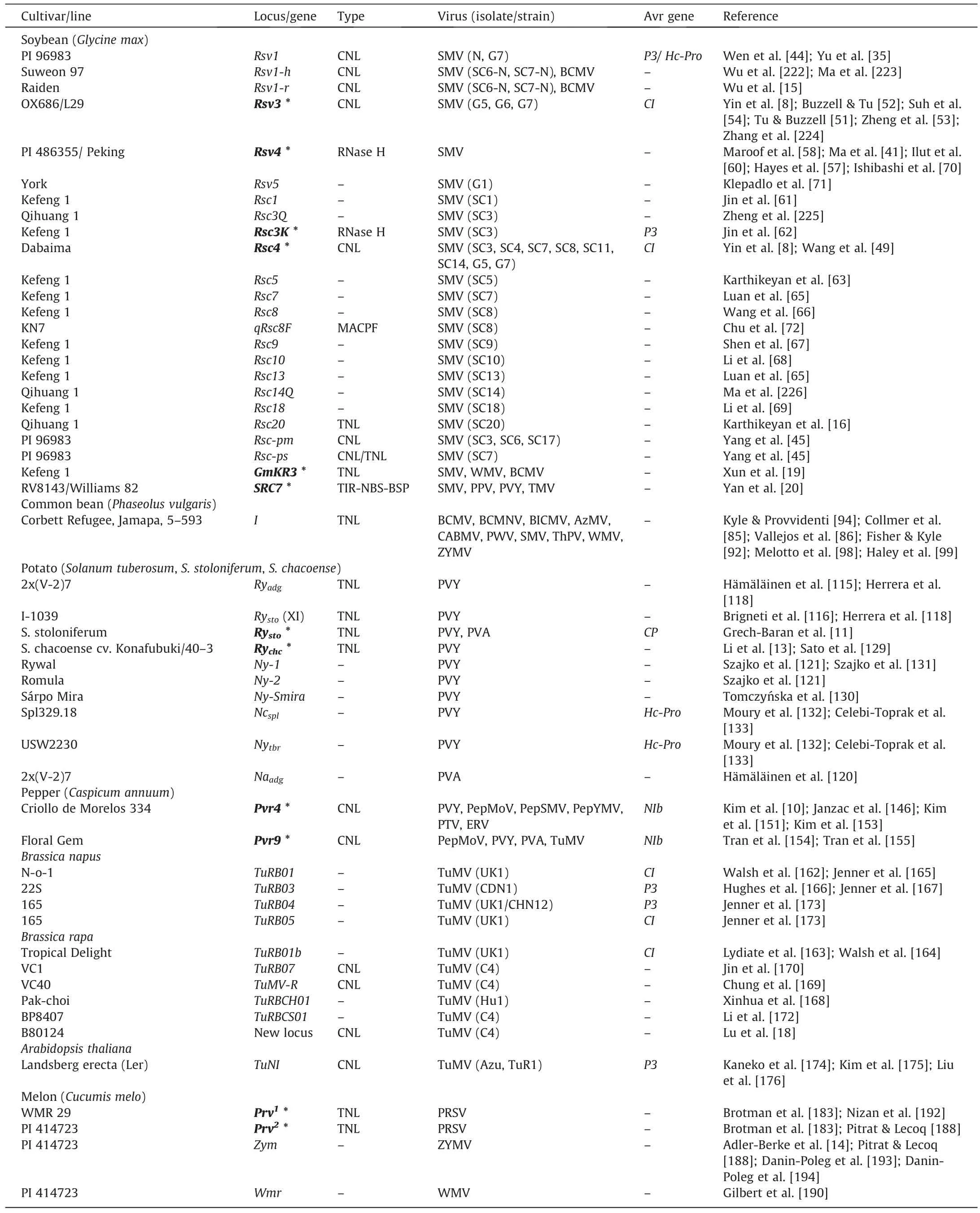
Table 2 Potential dominant resistance genes involved in potyvirus resistance.
9.Concluding remarks
Recent discoveries in NLR-mediated plant immunity have deepened our understanding of the plant immune system and provided new ideas for crop disease resistance improvement.Recently described NLR genes required for potyvirus resistance include soybean Rsc4-3,pepper Pvr4,and potato Rysto.Many potyvirus-specific NLRs in crop plants have not been fine-mapped and cloned.Noncrop plants likely harbor larger, unstudied NLR gene reservoirs.Future steps in mining and cloning new NLR genes in crop plants and related wild species,designing accompanying molecular markers,and finding novel approaches to achieving potyvirus resistance are needed to ensure a dominant position in winning the perennial plant–potyvirus arms race.
CRediT authorship contribution statement
Xin Hong:Visualization, Writing – original draft.Shufen Li:Writing–original draft.Xiaofei Cheng:Writing–review&editing.Haijian Zhi:Conceptualization,Writing–review&editing.Jinlong Yin:Writing – original draft, Writing – review & editing, Project administration.Kai Xu:Conceptualization, Funding acquisition,Project administration, Writing – original draft, Writing – review& editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge Dr.James C.Nelson from Kansas State University for proofreading and English editing and Dr.Guanzhu Han from Nanjing Normal University for helpful suggestions.This work was supported by the National Natural Science Foundation of China (31770164) and Jiangsu Province’s Innovation Program(JSSCTD202142).
杂志排行
The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
