Isolation,Identification and Characteristics of Cellulose-degrading Bacteria
2024-01-08ShashaLIURuijiLUNuoCHEN
Shasha LIU,Ruiji LU,Nuo CHEN
Guangdong Provincial Key Laboratory of Environmental Health and Land Resource,School of Environmental and Chemical Engineering,Zhaoqing University,Zhaoqing 526061,China
Abstract [Objectives]To isolate and identify characteristics of the cellulose-degrading bacteria.[Methods]In view of the poor effect of crop straw composting,the strains which can degrade cellulose were isolated and purified from the soil piled with rice straw,and the strains A2 and A5 with high efficiency of cellulose degradation were finally screened through the transparent circle,cellulase activity and filter paper disintegration experiments.[Results]It was confirmed that the strains A2 and A5 could degrade cellulose well.The results of 16s rDNA showed that A2 was Pseudoxanthomonas mexicana and A5 was Bacillus cibi.[Conclusions]The results of this study are expected to provide high-quality strain resources for the degradation of cellulose in straw,and have important application value for improving the efficiency of straw composting.
Key words Cellulose,Degrading bacteria,Enzyme activity,Degradation effect
1 Introduction
The annual yield of crop straw in China is huge.At present,a large number of straws are piled up in farmland soil or burned,causing serious environmental pollution.Straw contains nitrogen,phosphorus,potassium and other nutrients,and the composting of straw can not only realize its harmlessness,but also obtain certain economic benefits[1-2].
However,the straw contains a large amount of lignocellulose,which is difficult to be completely decomposed,which hinders the composting process,resulting in low conversion efficiency of humus[3-4].Cellulose can be completely hydrolyzed into glucose by cellulase,and cellulose-degrading bacteria have a strong ability to produce cellulase.Therefore,it is urgent to separate and screen efficient cellulose-degrading bacteria to improve the composting effect of straw[5-7].
In this study,we enriched,separated and screened the strains with the ability of decomposing cellulose from the soil piled with rice straw.Through the experiments of transparent circle,we determined cellulase activity and filter paper degradation,the strains with high cellulose degradation efficiency,and identified the strains by 16s rDNA sequence.This study has important theoretical and practical significance for promoting the popularization and application of straw composting.
2 Materials and methods
2.1SoilcollectionThe topsoil (0-10 cm) of rice straw accumulation was collected by quincunx sampling method in Shuibian Village,Gaoyao District,Zhaoqing City,Guangdong Province.After the soil was naturally air-dried,the larger stones were removed and sieved.
2.2ReagentsCH3COOH-C2H9NaO5Solution: dissolved 11.57 g C2H9NaO5and 0.85 mL CH3COOH with distilled water to 1 L,and adjusted the pH to 5.5 with 0.1 mol/L CH3COOH solution[8].
CMC-Na solution: weighed 1.0 g of CMC-Na and put it into a magnetic stirrer,added CH3COOH-C2H9NaO5solution,heated and stirred until it is completely dissolved,then stopped the heating,continued to stir for 30 min,and fixed the volume to 100 mL[9].
Glucose standard solution: completely dissolved 1.0 g of glucose with distilled water,and fixed the volume to 1 L[10].
Hutchison Fluid Medium: 1.0 g KH2PO4,0.1g CaCl2,0.1 g NaCl,0.01 g FeCl3,0.3 g MgSO4·7H2O,and 2.5 g NaNO3,fixed the volume to 1 L with distilled water,adjusted the pH to 7.2-7.3,and sterilized with high pressure steam[11].
CMC-Na solid medium: 15 g CMC-Na,2 g peptone,0.5 g NaCl,0.5 g yeast extract,0.5 g MgSO4,1 g K2HPO4,15 g agar,added distilled water to fix the volume to 1 L,adjusted pH to 7.0,and sterilized with high pressure steam[12].
2.3Isolationandpurificationofcellulose-degradingbacteria
Added 10 g of soil and 90 mL of sterile water into a conical flask,and placed the flask in a shaker (30 ℃,140 r/min) and shook for 10 min.Took 10 mL of the solution and added it into a conical flask containing a Hutchison Fluid Medium (90 mL) and a filter paper strip,shook and cultured until the filter paper strip was obviously broken.Transferred 10 mL of culture solution to a conical flask (90 mL Hutchison Fluid Medium+filter paper strip),and repeated the above process for 5 times.Took the bacterial solution of the last enrichment culture and diluted it according to the concentration gradient of 10-1-10-6,transferred 0.2 mL to CMC-Na solid medium for coating,and cultured in a constant temperature incubator at 30 ℃.After the colonies grew out,picked single colonies with different morphologies and streaked to the CMC-Na solid culture medium until the single colony with stable morphology was obtained by purification.
2.4Screeningofcellulose-degradingbacteriaTransparent circle test (primary screening): inoculated a single strain to CMC-Na solid medium,after the colonies grew out,added Congo red to stain for 15 min,poured out the staining solution,decolorized with 1 mol/L NaCl for 15 min,and poured out the NaCl.If there is a hydrolysis transparent circle around the colony,it indicates that the strain can secrete cellulase.
Determination of cellulase activity (secondary screening): the single strain was inoculated into CMC-Na liquid medium,cultured with shaking (30 ℃,140 r/min) for 5 d,and the supernatant was centrifuged to obtain crude enzyme solution.1 mL of CMC-Na solution and 0.5 mL of crude enzyme solution were added into a colorimetric tube with a stopper,and the mixture was water-bathed at 50 ℃ for 30 min.Added 2 mL of 3,5-dinitrosalicylic acid into a boiling water bath for 5 min,cooled quickly,added distilled water to a constant volume of 25 mL,took the blank control without crude enzyme solution,determined the absorbance at 540 nm wavelength,and looked up the glucose content S from the standard curve.
The calculation formula of the enzyme activity is as follows:
E=1 000×S×N/(T×V)
whereEis cellulase activity,expressed in U/mL;Nis dilution times of crude enzyme solution;Sis glucose content,expressed in mg;Vis volume of crude enzyme solution,expressed in mL;Tis reaction time,expressed in min.
2.5FilterpaperdegradationtestThe bacterial solution was inoculated into CMC-Na liquid medium for 12 h for activation,and 5 mL of the activated bacterial solution was inoculated into a conical flask (95 mL of Hedgeson medium+filter paper strip) at 30 ℃ and 140 r/min for 20 d,and the disintegration of the filter paper strip was observed.
2.6MolecularbiologicalidentificationofthestrainThe screened strains capable of efficiently degrading cellulose were sent to Sangon Biotech (Shanghai) Co.,Ltd.for 16s rDNA sequencing,and the sequences were compared with BLAST homologous sequences in NCBI database to determine the species of the strains.
3 Results and discussion
3.1Preliminaryscreeningofcellulose-degradingbacteria
3.1.1Results of the transparent circle experiment.The Congo red stain turned the cellulose into a red substance,which was converted into a hydrolytic transparent ring in the presence of cellulase,and the larger the size of the ring,the stronger the ability of the strain to secrete fibers[12-13].After enrichment,isolation and purification,9 strains were obtained.After stained with Congo red,it was found that the size of the transparent circle around the colony of 6 strains was larger,indicating that these strains had a strong ability to degrade cellulose.
3.1.2Cellulase activity.Cellulase activity was measured to further determine the ability of the six strains to degrade cellulose,and the results are shown in Fig.1.The cellulase activity of strain A1 was only 22.45 U/mL; the next were strains A3 and A6,whose cellulase activities were 23.93 and 24.36 U/mL,respectively; strains A2,A4,and A5 had the highest cellulase activities,which were 25.20,25.26 and 25.20 U/mL,respectively.
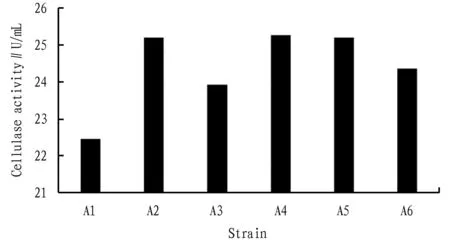
Fig.1 Cellulase activity of the strains
3.2FilterpaperdegradationexperimentAccording to the degree of disintegration of the filter paper,the ability of the strain to degrade cellulose can be evaluated."-" indicates that there is no obvious change in the filter paper,"+" indicates that the filter paper begins to be segmented; "++" indicates that the edge of the filter paper is broken and disintegration begins to occur; "+++" indicates that 1/3 of the filter paper is broken; "++++" indicates that 1/2 of the filter paper is broken; "++++" indicates that most of the filter paper is broken.The degradation of filter paper by the strain is shown in Table 1 and Fig.2.The degradation effect of strain A1 and A3 on filter paper was poor,and a large area of filter paper existed in the medium.Strains A4 and A6 degraded the filter paper into disc shape,indicating that they had a certain degradation effect on the filter paper; strains A2 and A5 had the best degradation effect on the filter paper,and the filter paper was almost completely disintegrated into paper paste.

Table 1 Disintegration of filter paper

Fig.2 Degradation effect of strains on filter paper
3.3Identificationofcellulose-degradingbacteriaAccording to the results of transparent zone,cellulase activity and filter paper disintegration test,the strains A2 and A5 were screened to have stronger ability to degrade cellulose.After 16s rDNA sequencing,two sequences of 1 469 BP (A2) and 1 445 BP (A5) were obtained.Homology analysis showed that the similarity between strain A2 andPseudoxanthomonasmexicanawas 100%; the similarity between strain A5 andBacilluscibiwas as high as 99.93%.
4 Conclusions
Nine strains capable of degrading cellulose were isolated and purified from the soil piled with rice straw.According to the results of transparent zone,cellulase activity and filter paper disintegration test,it was confirmed that strains A2 and A5 could degrade cellulose very well,and the results of 16s rDNA identification showed that A2 belonged toPseudoxanthomonasmexicanaand A5 belonged toBacilluscibi.The results can provide high quality strain resources for the degradation of cellulose in straw,and have important application value for improving the efficiency of straw composting.
杂志排行
Asian Agricultural Research的其它文章
- Research and Exploration on Ideological and Political Theories Teaching in the Course of Soil Science Experiment: Taking Pingdingshan University as an Example
- Opportunities and Challenges of Digital Development in Circulation under the Background of Rural Revitalization
- Inheritance,Innovation and High-quality Development of Herbaceous Edible Oils in Hubei Province under the Strategy of Strengthening the Country with Intellectual Property
- New Advances in Plant Biology Research on Reevesia Lindley
- Exploration of Scientific Research Mentorship on the Cultivation of Undergraduate Students Innovation Ability
- Roles of Corporate Reputation and Service Recovery in Food Safety Crises: A Case Study of Food Hygiene Incidents in Hot Pot Restaurants
