Weakly supervised machine learning
2023-12-01ZeyuRenShuihuaWangYudongZhang
Zeyu Ren | Shuihua Wang | Yudong Zhang
School of Computing and Mathematical Sciences, University of Leicester,Leicester,UK
Abstract Supervised learning aims to build a function or model that seeks as many mappings as possible between the training data and outputs,where each training data will predict as a label to match its corresponding ground-truth value.Although supervised learning has achieved great success in many tasks,sufficient data supervision for labels is not accessible in many domains because accurate data labelling is costly and laborious, particularly in medical image analysis.The cost of the dataset with ground-truth labels is much higher than in other domains.Therefore,it is noteworthy to focus on weakly supervised learning for medical image analysis, as it is more applicable for practical applications.In this review, the authors give an overview of the latest process of weakly supervised learning in medical image analysis, including incomplete, inexact, and inaccurate supervision, and introduce the related works on different applications for medical image analysis.Related concepts are illustrated to help readers get an overview ranging from supervised to unsupervised learning within the scope of machine learning.Furthermore, the challenges and future works of weakly supervised learning in medical image analysis are discussed.
K E Y W O R D S deep learning, unsupervised learning
1 | INTRODUCTION
In artificial intelligence,an agent tries to improve performance by discovering the surroundings, and we call it machine learning when the agent is a computer [1].In the last decades,machine learning had significant achievements in many areas,and it is ubiquitous in our daily lives.Typically, machine learning can be categorised into four types: supervised learning, weakly supervised learning (WSL), unsupervised learning, and reinforcement learning, as Figure 1 shows.Supervised learning gets more achievements compared with others.Formally, supervised learning learns a function that maps from training examples to outputs.Every training data contains a feature vector to describe the object itself, and a label shows the ground-truth output of the object.There are two types of supervised learning tasks:supervised classification and regression.In detail, the label of the classification task indicates the category of the example, and the regression task label shows the output's value.However, for most domains,especially for medical image analysis, it is challenging to guarantee all the training images with ground-truth labels.Data labelling is expensive and biased towards manual labelling.
In contrast, unsupervised learning extracts the inherent information directly from the data without ground-truth labels and represents their understanding of the data to the outputs.There are five major types of tasks: self-supervised learning, clustering, density estimation, dimensionality reduction, and association rules [2, 3].Unsupervised learning does not need much human intervention like supervised learning.In contrast, the performance of unsupervised learning usually is lower than supervised learning.Furthermore, it is more computationally expensive than supervised learning when extracting deeper information from the dataset.Meanwhile, it also needs human intervention to validate the results [4].Thus, it is ideal to use machine learning with weak supervision for medical analysis by comparing the advantages and disadvantages of supervised and unsupervised learning.
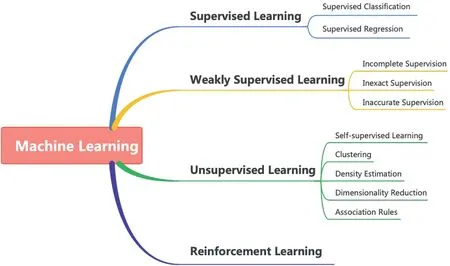
F I G U R E 1 Taxonomy of machine learning.
There are three weak supervisions of weakly supervised learning by comparing the label's confidence: incomplete,inexact, and inaccurate supervision.An intuitive illustration is shown in Figure 2.For incomplete supervision, most training examples do not have labels and only the remaining examples with ground-truth labels.This is a common situation in medical analysis.For instance,a doctor can label a small subset of lung cancer cell scans.Then the machine learning model can be more accurate in predicting the severity of the patients and making further diagnoses.The second one is inexact supervision.All the training examples only have a coarsely-grained label.The coarsely-grained label can only describe the imagelevel label instead of the object-level label [5], that is, an MRI image of the patient diagnosed with a‘lung cancer’label.However,the‘lung cancer’label cannot precisely describe how many sections and types of lung cancer are in this MRI image.The last one is inaccurate supervision.It always happens in the labelling process, and annotators might provide a biased(incorrect)label to the images and not all the labels are ground truth for training examples, for example, when the annotators are not well-trained to label the class of blood cells, or the doctor is tired of labelling the images.
The last type of machine learning is reinforcement learning.Reinforcement learning works great for games and simulations.In detail, there is an agent in the environment, and the agent should efficiently interpret the perceptions of the environment and then take related actions or decisions based on the perceptions.However, reinforcement learning has limited real-world application and adoption.It needs lots of trials and tests before being applied to real-world applications, and it is not easy to keep security because the environment in the real world is much more complex than the simple virtual world.
In the last decade, exhaustive reviews and surveys on medical image analysis have been presented [6–10].However,the dedicated review on weakly supervised learning for medical image analysis is not yet done as far as we know.As shown in Figure 3, weakly supervised learning has become a hot topic since 2015.Besides, Rony et al.[11] focus on the weakly supervised learning methods for histology image classification and localisation.Hassan et al.[12] analysed the weakly supervised learning-based transfer learning and data augmentation methods for the COVID-19 CT diagnosis.However,they have not presented the comprehensive aspects of weakly supervised learning.The work presented by Qu et al.[13] summarise the weakly supervised learning-based methods into three types:instance-based, bag-based, and hybrid methods, but their taxonomy has not covered all the categories for weakly supervised learning-based methods.Although Zhou et al.[5] present a whole picture of weakly supervised learning, they have not introduced more details in each category, and the entire scope does not focus on the medical images.Overall, only some studies use weakly supervised learning on specific tasks or diseases[11,12],our review covers most of the comprehensive aspects of weakly supervised learning, and there are more details regarding the medical image analysis.
Although supervised learning has significant achievements in many areas,getting strong supervision for all the training data is challenging.Moreover,the labelling process is easily biased by human intervention,and there are always inaccurate labels that we cannot avoid.Especially in medical image analysis,on the one hand,the costof the labelling process usually is muchhigher than in other domains,and only well-trained specialists can finish the annotation task.On the other hand, biased information or inaccurate label for the medical image will cause unexpected accidents in the prediction of disease.Summarising both previous works and existing challenges,we comprehensively review WSL-based methods for medical image analysis within the scope of machine learning.There are more than 300 papers reviewed on a wide variety of applications based on weakly supervised learning in medical image analysis.The contributions of this review show as follows:
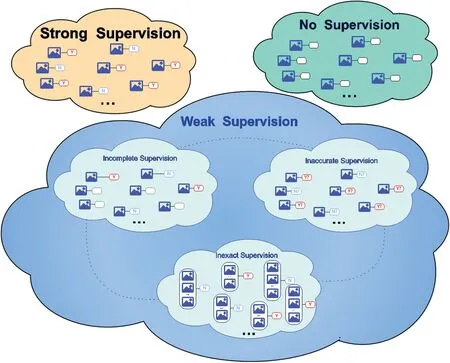
F I G U R E 2 Taxonomy of weakly supervised learning.The image icon represents the feature vector or training instance; the red ‘Y’ and blue ‘N’ denote labels;‘?’shows that the label might not be the ground truth;the empty shape represents no label;the dotted line depicts the transitions between each type of the weak supervision.
• We introduce a comprehensive overview of weakly supervised learning for medical image analysis within the scope of machine learning.
• Summarise the state-of-art approaches to different weak supervision tasks for medical image analysis.
• Present the newest research process of the various applications based on weakly supervised learning for medical image analysis.
• Conclude the challenges and potential solutions on weakly supervised learning for medical image analysis.
• Give a detailed discussion based on the existing works and plan the future research interests of our work.
In this review, all the concepts are presented by a simple binary classification problem.The symbols and their meaning are shown in Table 1.
This review has six sections in total.In Section 2, we introduce the essential concepts in weakly supervised learning and give intuitive examples to illustrate the ideas behind these concepts.In Section 3, we shortly introduce the related concepts of unsupervised learning.In Section 4,we summarise the latest WSL-based applications in medical image analysis.In Section 5, we discuss the challenges of weakly supervised learning in medical image analysis and describe our future work.Finally, we end up this review with a summary.
2 | WEAKLY SUPERVISED LEARNING FOR MEDICAL IMAGE ANALYSIS
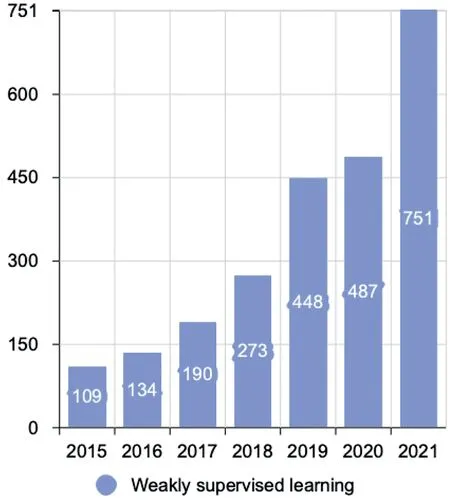
F I G U R E 3 The number of publications on weakly supervised learning(the data from Web of Science).
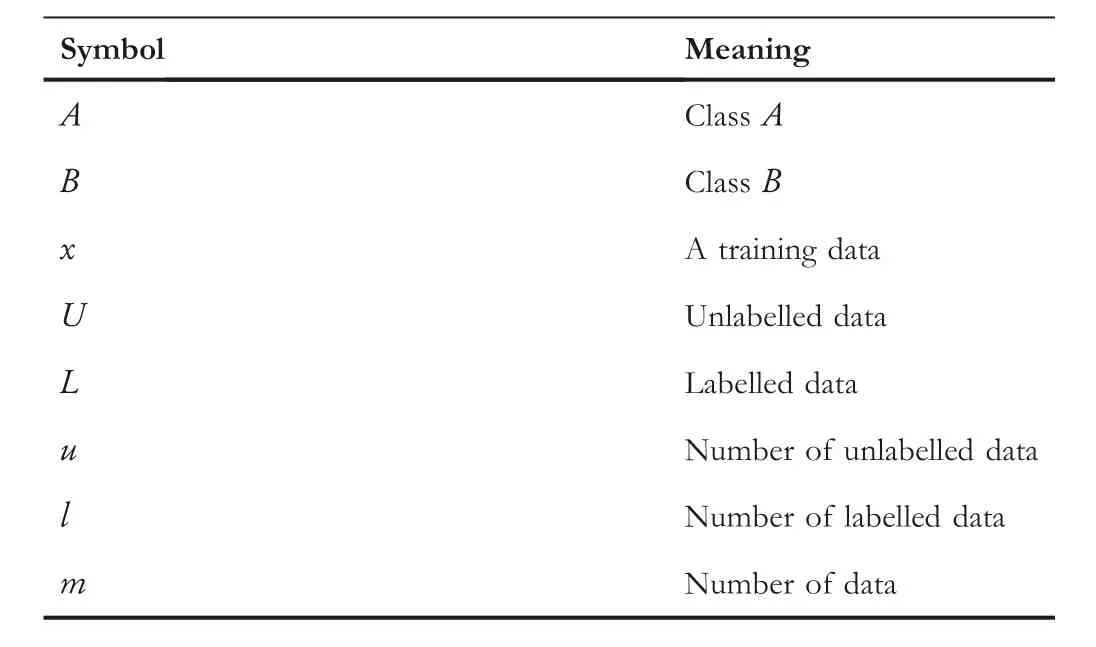
T A B L E 1 Symbols and meaning.
Weakly supervised learning is one category of machine learning which tries to build a predictive model with weak supervision.Weakly supervised learning does not like supervised learning and does not need strong supervision for all the data.Especially in practical tasks, the labelling process costs a lot and is time-consuming.Usually, the performance of weakly supervised learning is better than unsupervised learning, and the computational power of weakly supervised learning is less than unsupervised learning.Therefore, weakly supervised learning has recently become a hot paradigm for machine learning,and it is desirable to work with weakly supervised learning in many domains.Weakly supervised learning mainly contains three types depending on the type of label: incomplete,inexact, and inaccurate supervision.A more detailed introduction to weakly supervised learning is proposed by Zhou[5].In this review,we no longer discuss details of weakly supervised learning instead of briefly introducing all the concepts and techniques related to medical image analysis.
2.1 | Incomplete supervision
Incomplete supervision only has a small part of ground-truth labels, which is insufficient to construct a predictive model.In contrast, there are abundant unlabelled training examples provided.There are two types of incomplete supervision concerning human intervention: active learning [14, 15] with human intervention and semi-supervised learning [16, 17]without human intervention.
The main idea behind active learning is that the active learner will query the selected unlabelled training example to get the ground-truth labels by an‘oracle’.The ‘oracle’denotes the annotator who can assign the ground-truth label to the training example.Instead, semi-supervised learning tries to learn the information from the labelled and unlabelled training examples without human intervention.The semi-supervised learning has two types.The first type is inductive learning,where unlabelled data are not the test data.In simplest terms,the test data are unseen during the training process.The second type is transductive learning, in which the unlabelled data is the test data, and the predictive model aims to increase performance with unlabelled (test) data.The differences between these types are shown in Figure 4.
2.1.1 | Active learning
Active learning [15] aims to find a subsetL′of the entire datasetL.The performances obtained fromL′andLare the same.For instance,p(x|L′) ≈p(x|L), here theUrepresents the performance training on the dataset,xis a training example.In order to find the subsetL′,active learning needs to pick the most useful training examples in theLand queries the ground-truth label from an ‘oracle’ or group of ‘oracles’.Therefore, the purpose of active learning is to minimise the cost of the labelling process.For simplicity, train a machine learning model which can minimise the number of queries.Settles [15] provided a comprehensive review of active learning,and Budd et al.[14]presented a survey containing upto-date active learning approaches for medical image analysis.
There are three main scenarios of active learning trying to query the label from the ‘oracle’:membership query synthesis,stream-based selective sampling,and pool-based sampling[15].
For membership query synthesis, the active learning will query any unlabelled training example as well as the generated samples.The indexes of the selected data points are the most valuable data for the current model.Then it annotates by the‘oracle’ [18].The main drawback of membership query synthesis is when the model does not have prior knowledge of unseen data distribution.It will request some queries which cannot be recognised and correctly labelled by the annotators[19].However, with the development of Generative Adversarial Networks(GANs),the quality of synthetic images is more promising than previous methods, and it will solve the challenges for the membership query synthesis.This scenario is suitable for data distribution with precise semantic meanings,or the models can automatically answer the queries rather than human annotators [20].
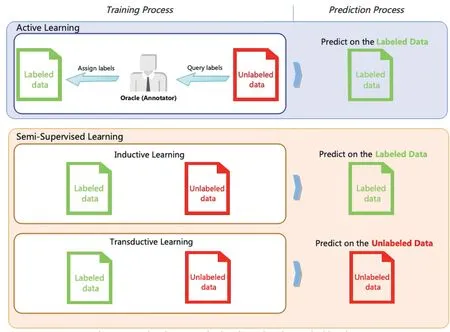
F I G U R E 4 Comparison between active learning and semi-supervised learning.
For stream-based selective sampling, a sequence of data points without labels is provided to the model, and then the model labels or drops the provided data points based on their information [21].However, one disadvantage of stream-based selective sampling is that the selected data distribution sometimes loses the actual underlying distribution.Another disadvantage is that we cannot discard any incoming data points once we calibrate the threshold.Then we might lose valuable data points [14].
For pool-based sampling, it relies on the fact that the data can be collected at once in real-world applications,and then it assumes that the model can select the queries from a large pool of unlabelled data distributionU[22].Usually, the pool-based sampling method will use a metric to rank all the training examples in theU.Then,‘oracles’ will label the top-ranked data points.In this method,we denote an unlabelled dataset with?instances asUn={X,Y} whereXrepresents the training instance, andYrepresents the ground-truth label.The potential data distribution is defined asU(x,y),andx∈X,y∈Y.The labelled training dataset denotes by theLm={X,Y}wheremmeans m instances in this dataset.Then the poolbased sampling will sample a queryQfrom the unlabelled dataset.Then it needs to optimise the formula,as Equation(1)shows.In Equation (1), we denote the loss function asℓ(·)∈R+,and we assume that them≪nand try to minimise the value ofmwith higher performance.
It is widely used with different machine learning applications based on the pool-based scheme.However, the iteration process of the pool-based sampling method is expensive.Therefore, stream-based selective sampling and membership query synthesis are more suitable to show their advantages for applications or hardware with lower computational powers[15].An intuitive illustration of the differences between these scenarios is shown in Figure 5.
Once the queryscenario is decided,active learning also needs a selection criterion.Basically, there are two types of selection criteria:informativenessand representativeness[23].Theformer evaluates the effectiveness of unlabelled data in decreasing the uncertainty of a statistical model.The latter evaluates whether the unlabelled instance can represent the inherent data distribution of input vectors.There are two main approaches to informativeness:uncertainty and query-by-committee.The uncertainty assumes that the instance will provide more information if a prediction is uncertain.For this reason,all the queries will start to label the instance with the least confidence[24].For the uncertainty approach, entropy [25] can be used as an uncertainty measure, as Equation (2) shows.In this equation,yidenotes the potential labelling, and the entropy depicts the information which is necessary to‘encode’as a data distribution.
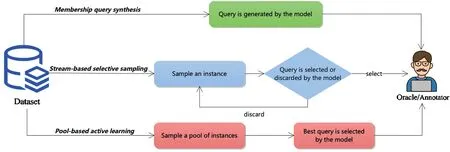
F I G U R E 5 Three scenarios of active learning.
Another choice of entropy with a more complex setting is least confident, where it will query the instance with the most confident labelling as shown in Equation (3).
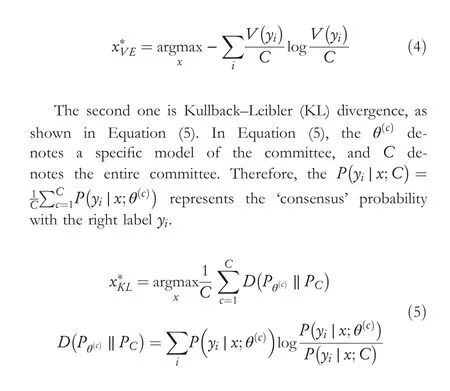
The query-by-committee involves multiple committees to vote on unlabelled instances.The most disagreed instance is the most valuable instance, and it contains more information than others.Representativeness methods usually focus on using different approaches to sample different representations or areas of the data distribution and design algorithms to increase unlabelled data's diversity or cluster structure [14, 26, 27].There mainly has two methods to measure the level of disagreement.The first one is vote entropy [28], as demonstrated in Equation (4), whereyidenotes all potential labelling, and theV(yi)is the amount of ‘votes’ which derives from the predictions made by the committees.
2.1.2 | Semi-supervised learning
The semi-supervised learning [29, 30] differs from active learning.Semi-supervised learning tries to extract knowledge with unlabelled training examples without human intervention.For semi-supervised learning, one obligatory condition is that an underlying data distribution should exist for any examples of the input space.Therefore, the underlying data distribution of unlabelled data can be extracted.Otherwise, it cannot improve the predictive model's performance with unlabelled data[30].Based on this obligatory condition,the basic intuition behind semi-supervised learning is shown in Figure 6.We assume that there are three groups of data points: positive and negative data points with ground-truth labels and unlabelled data points without labels.If some unlabelled points are located in the middle of the positive and negative data points,it is difficult for us to identify the ground truth of these data points.However, if we can draw the position of all the unlabelled data points, the locations of labelled data points can predict the locations of unlabelled data points.So semisupervised learning can make predictions based on the underlying data distribution presented by the unlabelled data.
The goal of semi-supervised learning can be described as Equation (6), whereLsrepresents the supervised loss of each instance,Lurepresents the unsupervised loss of each instance,and R represents the regularisation of each example.Theθdenotes all the parameters of the model.It needs to be mentioned that there are no obvious differences between unsupervised loss and regularisation terms, and different choices of these terms can generate different semi-supervised models.
There are different assumptions of data structures to apply semi-supervised learning: smoothness assumption, cluster assumption, and manifold assumption [29].The definition of assumptions lists as follows:
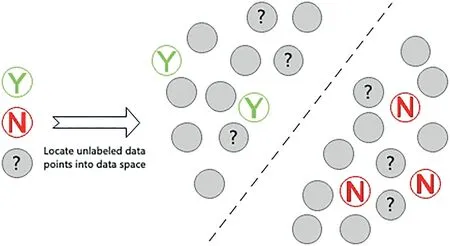
F I G U R E 6 Usefulness of unlabelled data for semi-supervised learning.Three circles on the left denote the instances with labels(green‘Y’and red‘N’).The grey circles with ‘?’ represent the test instance.The grey circles on the left without ‘?’ represent the unlabelled instances.
• Smoothness assumption: if two pointsx1andx2reside in a high-density region, and their locations are close.Then the locations of the corresponding outputsy1andy2should also be close to each other[29].Otherwise,their outputs should not be the same if they are not close to the low-density region.The smoothness assumption works both on the classification and regression tasks.
• Cluster assumption:if points are in the same cluster,they are likely to be of the same class.The cluster assumption is a particular case of smoothness assumption when all the data points of the same cluster represent the same class.Additionally, the cluster assumption is equivalent to the lowdensity separation: The decision boundary should lie in a low-density region[29].Overall,if all the data has the same inherent cluster structure, it belongs to the cluster assumption.
• Manifold assumption: the (high-dimensional) data lie(roughly) on a low-dimensional manifold.If all the data points lie on a manifold or have multiple clusters, all the nearby data points should belong to the same predictions or classes.
Semi-supervised learning is composed of five crucial types regarding the involvement of model designs and loss functions:consistency regularisation methods,graph-based methods[31–35], generative methods [36, 37], pseudo-labelling methods,and hybrid methods [17].The taxonomy of these categories illustrates in Figure 7.As Figure 7 shows that the generative methods have two types: semi-supervised generative adversarial networks (GANs) such as SGAN [38], AC-GAN [39],Triple GAN [40], and SCH-GAN [41]; Semi-Supervised Variational Autoencoders (VAEs) such as SSVAEs [42],ADGM [43], Infinite VAE [44], PepCVAE [45], and SDVAE[46].For consistency regularisation methods, there are some representative works such as Mean Teacher [47], VAT [48],Dual Student[49],Semi-Supervised Ladder Network[50],and Temporal Ensembling [51].The graph-based methods also
have two types: autoencoder-based methods such as SDNE[52] and GAE [53], and GNN-based methods such as GCN[54], and GraphSAGE [55].The third type is pseudo-labelling methods.It includes disagreement-based methods (for example, Tri-Net [56], DAS-CIDS [57], and Deep Co-Training[58]) and self-training methods (for example, Noisy Student[59], MPL [60], and SimCLR [61]).The last type is hybrid methods,such as MixMatch[62],ICT[63],FixMatch[64],and DivideMix [65].Moreover, some works focus on medical report generation [66–68].A comprehensive review of semisupervised learning is proposed by Yang et al.[17].The definitions of these categories are as follows:
• Generative methods assume that labelled and unlabelled data can be produced by the same model, and all the data can represent the inherent data structure.Therefore, the missing parameters obtained from all the missing labels of unlabelled data points will be applied to the model.Then the missing parameters can be estimated by specific algorithms,such as the expectation-maximisation algorithm, abbreviated as the EM algorithm [69].Semi-supervised GANs and semi-supervised VAEs are the most common types of generative methods.
• Consistency regularisation methods are based on the manifold or smoothness assumptions.The basic idea is that any data perturbation will not influence the predictions[70].Basically,if there are different data augmentation techniques implemented to the same data points, as predicted, the augmented data points should be the same as the original data points.
• Graph‐based methods aim to build a similarity graph.The graph's nodes denote the training instances, and edges represent the relations between the node pairs.Then the model inferences the labels for the unlabelled instances based on the labelled samples.

F I G U R E 7 Taxonomy of semi-supervised learning based on the different model architectures and loss functions.
• Pseudo‐labelling methods try to assign pseudo-labels for all the unlabelled instances.It highly relies on the confidence of pseudo-labels.There are two major types,the first one is trying to exploit the disagreement from multiple learners during the training process (disagreement-based models),and another one is the self-training model, which produces the pseudo-labels by the model.However, the pseudolabelling methods cannot make sure all the pseudo-labels are ground-truth labels.It inherently has a bias in the confidence of the pseudo-labels.
• Hybrid methods will combine several methods above to make the final prediction.
2.2 | Inexact supervision
Inexact supervision concerns that each data point only has part of supervision information that cannot describe the exact supervision information.For inexact supervision, each instance only has a coarse-grained label, that is, for COVID- 19 prediction on 3D volumetric CT images, we aim to predict whether the patient gets COVID-19 or not by analysing his 3D volumetric CT images.This 3D volumetric CT image can convert into different slices.If a slice of a 3D volumetric CT image is diagnosed as COVID-19, we can say that this patient gets COVID-19, and this 3D volumetric CT image contains COVID-19.However, we cannot define that each slice of this 3D volumetric CT image is a COVID-19 slice.If there is at least one slice predicted as COVID-19,the 3D volumetric CT image is also predicted as COVID-19, and this image predicts health if all the slices are healthy.Therefore for inexact supervision, the label can only describe the image-level information,and it cannot contain further detailed information(i.e.some regions of interest in medical image analysis) about the image.The most representative technique of inexact supervision is multi-instance learning (MIL) [71, 72].
MIL [73] is a variant of inductive machine learning, and each training example of MIL represents a group of feature vectors.The training example is called a bag(an image or a text file).There are multiple vectors in the bag, and they are called instances.The major goal of MIL is predicting labels for unseen bags.Here, we use a binary classification to explain the standard assumption of MIL.We define that MIL aims to learn a projectionp:X →Y with the datasetD= {(X1,Y1),(X2,Y2),…,(Xm,Ym)} whereXi={xi1,…,ximi}⊆X is named as a bag, there aremiinstances in the bagXiwherexij⊆X is an instance andj∈{1,...,mi}.Yi∈Y={T,F} is the label for the bag,andTrepresents the true(positive)label,Frepresents the false(negative)label.Assume that the label ofXiisT, then there exists an instance labelled asT, and the labels of the remaining instances in thisXiare unknown.Then the goal of MIL is to predict the labels for unseen bags.It also needs to mention that the bag is positive if at least one of the instances has a positive label, and the bag is negative if all instances have a negative label.Figure 8 shows an intuitive comparison between traditional machine learning and the MIL process for image classification, and we can find that the training process of traditional machine learning only uses images as training datasets.In contrast, the MIL can use images(bags) or instances to train the model.

F I G U R E 8 Comparison between traditional machine learning and MIL for the image classification.

F I G U R E 9 Taxonomy of MIL applications.
MIL recently gained more attention in many real-world applications because it can fit various problems.Here we can summarise it with four different tasks, as Figure 9 shows:classification, regression, ranking, and clustering.The classification can be bag-level and instance-level classification.Most of the tasks in MIL are bag-level classification,and the labels of instances are not important compared with the labels of bags.Therefore, the incorrect labels of instances are not important and cannot directly affect the performance of bag-level classification, and most of the bag-level classification cannot benefit the instance-level classification.Instead, instance-level classification trains on the different sets of data and evaluates the labels for each instance [71].For MIL classification,multi-label scenarios exist, meaning that a bag or an instance can have different labels[74].The second task is the regression task which aims to predict a value for the bag or instance.There are different ways to assign the value, such as getting bag-level value from instances that are close to the defined constraints [75] and using weighted instance-level features to get the bag-level value[76].The next task is ranking,unlike the regression task, which needs a concrete value.Instead, it aims to sort the bags or instances based on predefined criteria.The last one is clustering which tries to identify clusters or a structure of the underlying data distributions by the unlabelled bags or instances.In bag-level clustering, some methods use standard algorithms or set-based distance measures to do clustering.In instance-level clustering, the clustering can be done by applying maximum margin clustering to the most representative instance of bags [77].
Based on the idea from Quellec et al.[78], we can categorise MIL into three types: global detection, local detection,and false positive reduction.Global detection aims to find the certain data pattern of the image,that is,,the data pattern refers to a class or a segmentation mask of the image.Local detection can be regarded as instance classification, which aims to find the data pattern of each instance.For the false positive reduction, the number of negative instances requires to decrease.Meanwhile, the false positive reduction assures at least one positive instance exists.Several reviews or surveys illustrate a better understanding of MIL [71, 72].
There are many effective algorithms and approaches have been implemented for MIL.The most representative one aims to use supervised learning algorithms to transfer the singleinstance to the multi-instance representations [79].Some algorithms do it reversely by representing transformations from the multi-instance to the single-instance representations [80].Amores and Jaume [81] propose a categorisation that groups all the algorithms into paradigms of instance-space,bag-space,and embedded-space.Moreover, some approaches focus on the implementation of the bag generator: a bag needs to generate instances by a bag generator, and the bag generator specifics the approaches about how to generate instances.The bag generator just recently gained attention and reports by Wei et al.[82].Furthermore,some approaches aim to determine the key instance.It is useful when there are no fine-grained labelled training instances for the tasks of locating regions of interest in images[83].Some methods describe the relationships between the instances and bags, a comprehensive review of this can be found in the review presented by Foulds et al.[84].
MIL has already been victoriously implemented in various tasks: image classification, image annotation, regression tasks,spam detection, medical analysis, object detection, object tracking,image retrieval,and so on.The unique features of the bag and instance can fit well into the medical analysis.
2.3 | Inaccurate supervision
The labels of inaccurate supervision are not always accurate and might have some errors.This situation is common in practical applications.For example,the annotator makes some errors due to the lack of domain knowledge,or a deep learning model generates inaccurate pseudo-labels.Moreover, some techniques,such as crowdsourcing[85]and collecting labels by web searching[86],can also introduce inaccurate labels for the data.For example, crowdsourcing aims to outsource labelling work to individuals or companies[85].It is a prevalent method in machine learning, and it assigns all the unlabelled training examples to a group of workers or companies, such as Amazon Mechanical Turk(AMT).However,some‘spammers’or ‘robots’ try to assign random labels to the tasks, and it is difficult for workers to assign ground-truth labels to some domain-specific datasets.Therefore, the crowdsourcing method is not easy to maintain.For collecting labels by web searching, the situation is even worse than crowdsourcing because of the redundant information that can be provided for the image, and it is also difficult to maintain without data filtering or human intervention.In most of the situation mentioned above are easy to obtain large amounts of labels at low cost.However,there are also label noises introduced at the same time.
Label noise is a key problem in many classification tasks.The most apparent weakness is that it will directly decrease the model's accuracy and increase the training time and computational power [87].Many works have been proposed to eliminate the label noise [88].By analysing the related reviews of inaccurate supervision [5, 89, 90] and methods, we summarise these works into five types: label filtering, label correction, label inference, noise-robust models, and costsaving methods.
Label filtering methods try to filter inaccurate labels in different ways.The most basic one uses data pre-processing to define the noisy data points and removes them before the training process [91, 92].Some works also use ensemble with different classifiers to define the inaccurate labels and remove them [93, 94].The widely used ensemble-based method is the majority voting ensemble.It bases on disagreements from different classifiers with weights to predict the noisy labels.Moreover, some works use graph-based methods to map the relationship of each training data and remove the noisy labels[95].These graph-based methods can efficiently extract the neighbouring information between the nodes and locate the inaccurate labels.
Label correction will identify the potentially inaccurate labels first, then try correct these inaccurate labels into correct labels [96].The data-editing [95] is one of the label correction methods.It maps each example with a label into a relative neighbourhood graph.In this graph, the nodes represent different training data with labels, and a cut edge is defined when two neighbouring nodes with different labels.Then using cut-edge weight statistic measures to define the suspicious data points which link to the multiple cut edges.Finally, these suspicious nodes are either removed or corrected.The entire process can describe in Figure 10.One shortcoming of the data-editing method is that it relies highly on low-dimensional feature spaces and cannot perform well when data are sparse.To identify the potential inaccurate labels, Lawrence et al.[97]propose an EM algorithm in the probabilistic model of a kernel Fisher Discriminant to find the inaccurately labelled instances.
Label inference tries to deduce the ground-truth label through the crowd.There are different methods of label inference, for example, the majority voting strategy from the ensemble methods [98], which consists of different models together, and using vote strategy to infer the final prediction,an intuitive representation shown in Figure 11.Firstly,it trains different models with the same dataset, then uses generated predictions with a voting strategy to get the final prediction.Different approaches are proposed to assign the weights of the model, such as applying the EM algorithm to probabilistic models[99,100],spammer elimination in probabilistic models[101], and low-quality workers elimination [102].
Noise-robust models aim to improve the robustness of the model against inaccurate labels.Various changes can improve the robustness of the model, such as designing robust loss functions, model architectures, and data augmentations.For example, Wu and Liu [103] design a truncated hinge loss for support vector machines against noisy labels in outliers.Bouveyron et al.[104] propose mixture models with a classifier to extract information with uncertain labels.Many researchers also suggest using ensemble-based methods to improve the robustness of the model [105, 106].
Some works concern the cost-saving effect, such as the minimal cost[107], non-adaptive task assignment mechanisms[108],adaptive mechanisms[109],and the Dawid-Skene model[110].
3 | UNSUPERVISED LEARNING

F I G U R E 1 0 Graph-based label correction.
Unsupervised learning can be regarded as an extreme case of weakly supervised learning.Some weakly supervised learning methods also use the unsupervised learning-based model for feature extraction.For example, combining unsupervised learning and supervised learning to construct a semi-supervised learning-based model.Therefore, it needs to mention unsupervised learning in this review and help the reader to get a comprehensive overview of machine learning methods with different data supervision.Unsupervised learning retrieves the data pattern or information from the dataset without the ground-truth labels.Sometimes, it can be regarded as a data processing approach before constructing a supervised learning model.Due to the ability to find the differences and similarities without human interventions, unsupervised learning becomes the ideal solution for data analysis, object recognition, and so on.Compared with supervised learning,unsupervised learning has advantages as follows: saving the cost of the labelling process,reducing the bias produced by the supervised process or human interventions, finding hidden data patterns, and deducing the scalability of the target function [111].There are five major categories of unsupervised learning:self-supervised learning, clustering, density estimation, association, and dimensionality reduction.
3.1 | Self‐supervised learning
Self-supervised learning has drawn massive awareness in the last decade,and many works are presented following this paradigm.The concept of‘self-supervised learning’was first proposed in the robotics domain,then on the AAAI 2020,in Yann LeCun,any perceived part could be applied as the machine's input[112].Basically, self-supervised learning gains the supervision information based on the data itself and predicts the results from the existing input or variants of the original data [113].Here, the variants of the original data could be any augmented, transformed or corrupted variant of the original data.The motivation behind self-supervised learning is that it exploits a new way to extract underlying data structure from the tremendous amount of unlabelled data itself.The supervision comes from data itself instead of traditional human supervision.It is noteworthy to mention that the self-supervised learning process is like humans and animals trying to understand the world through our observations.Self-supervised learning has four major methods:generative,contrastive,adversarial,and predictive methods.
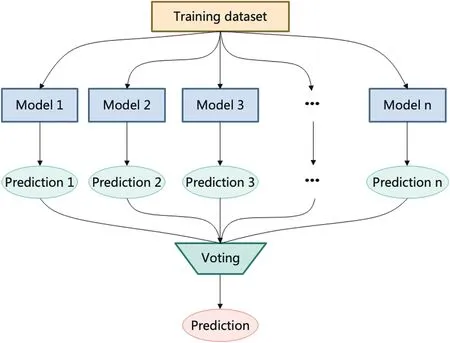
F I G U R E 1 1 Majority voting strategy.
• Generative Methods aim to train an encoder to map the inputxto the outputy, then train a decoder to retrievexfrom the outputy.
• Contrastive Methods measure the similarities or differences between the different variants of inputs.
• Adversarial Methods train two models: encoder-decoder and discriminator.The encoder-decoder can produce artificial data instances, and the discriminator can differentiate the instances generated from the encoder-decoder, whether real or fake.
• Predictive Methods learn the latent representations from the unlabelled instances by regarding the pretext task as a classification or regression task.
There exist several reviews that express the most recent research on self-supervised learning [113–115].
3.2 | Clustering
Clustering aims to classify unlabelled data into different classes based on their similarities and differences,and it naturally finds the inherent data pattern from the raw data [116].Exclusive,overlapping, hierarchical, and probabilistic clusterings are the most commonly used methods of clustering.
3.2.1 | Exclusive clustering
Exclusive clustering aims to assign the training instance with a single cluster.It is also named‘hard clustering’[3]or‘partitional clustering’[117].It decomposes the data points into a group of disjoint clusters and then optimises the criterion function based on the disjoint clusters [118].The most popular exclusive clustering algorithm is K-means clustering, where all the training examples are divided into K groups.These K clusters are based on distance from the centroid of the current cluster,and if the training instances are close to the same centroid,they are assigned to the same groups.The K-means algorithm aims to minimise the squared error of K clusters[119].
3.2.2 | Overlapping clustering
Overlapping clustering is different from exclusive clustering.Multiple clusters can have the same data points, and it is also called ‘soft’ clustering.Overlapping clustering is more applicable for practical applications that tolerate overlapping clusters to fit more complex data patterns.In the last 4 decades,overlapping clustering developed into various types of methods, such as k-means-based, hierarchical, generative, and graphical approaches.A comprehensive review is done by N’Cir et al.[118].
3.2.3 | Hierarchical clustering
The term hierarchical clustering is also named HC, and hierarchical cluster analysis is named HCA, which tries to define clusters recursively.In other words, it finds clusters based on the existing clusters.Agglomerative and divisive clustering are the two categories that belong to hierarchical clustering.Agglomerative clustering follows a ‘bottoms-up’ mode.In the beginning, all the data points are spread into several groups randomly.Then the agglomerative clustering will merge them together and filter these data points based on the similarities until the first cluster has been extracted.There are four primary methods to evaluate the similarity:ward's linkage,average linkage, complete (maximum) linkage, and single (minimum)linkage.The most common metrics to calculate the distance are Euclidean and Manhattan distances [3].Divisive clustering is the opposite of agglomerative clustering and follows a ‘topdown’ mode.It begins with a cluster that contains each data point.Then it divides the cluster into smaller clusters.However, divisive clustering is not commonly used.There are several reviews that discuss the HC approaches [120, 121].
3.2.4 | Probabilistic clustering
Clusters in the probabilistic clustering are divided by the likelihood of the data distribution.The Gaussian Mixture Model(GMM) [122] is the most common probabilistic clustering method.GMM is proposed based on the assumption that the data points can be generated by a finite number of Gaussian distributions with unknown parameters.Then it tries to determine the Gaussian or normal probability distribution of an existing data point by the mean or variance.The GMM is a parametric model of probability distribution commonly used in biometric systems.The Expectation-Maximisation algorithm is abbreviated as EM and Maximum A Posteriori, also known as MAP,are the most commonly used algorithms to estimate the parameters of GMMs [122].
3.3 | Density estimation
Density estimation aims to estimate a probability density function (PDF) from the existing data points.Density estimation mainly works on data visualisation tasks,but it also can be regarded as an element of analysis methods.There are also other analysis methods, such as kernel regression [123],anomaly detection [124], and clustering [116].There are two types of density estimation: parametric and non-parametric approaches.The parametric approach concerns the situation that all the data points are given by an existing distribution.The non-parametric approach assumes that the data distribution has a probability densitypand estimates thepby using all the data points instead of deciding whetherpbelongs to any existing parametric group in advance [125].For parametric methods, the most common method is GMM which we already mentioned in Section 3.2.4.Non-parametric methods mainly include three types of methods:histogram[126],kernel density estimation (KDE) [127], and k-Nearest neighbour estimator [128].
3.4 | Dimensionality reduction
With the development of various sensors and devices,the raw data is always high-quality and can help machine learning models get more accurate results than before.However,redundant information is always included in the raw data,which will influence the performance of machine learning models.One possible solution is dimensionality reduction which can remove the redundant information or dimensions of raw data.Then the filtered data will keep the most important data feature or information equivalent to the raw data.Therefore, the size of the training data has been reduced, and the raw data's integrity has been preserved at the same time.The dimensionality reduction techniques are widely used in the data preprocessing step.The most common methods are principal component analysis [129], factor analysis [130], singular value decomposition [131], autoencoders [132], and kernel PCA [133].
3.5 | Association rules
Association rules analysis uses rule-based methods to define the relationships between the data points and clusters.It is a common method for analysing market-basket data and data mining.The goal of association rules analysis is to find joint values of the variablesV=(V1,V2,…,Vn).For example, if we want to analyse the products sold in the supermarket, the variableVrepresents the products listed in the store.Each variableVassigns a valueVn∈{0,1}: ifVn=1 means that the item is sold in this transaction, and ifVn=0 means that the item has not been sold.By analysing existing information,we can summarise many conclusions, such as which combination of products is most frequently purchased together or which is most popular on certain days or months.There are lots of works that research the association rules analysis that can be found in refs.[2, 134, 135].
4 | APPLICATIONS IN MEDICAL IMAGE ANALYSISBASED ON THE WEAKLY SUPERVISED LEARNING
In this section, we summarise the four major tasks in medical image analysis based on weakly supervised learning with related datasets.In medical image analysis, image classification is a fundamental task that plays an important role in computeraided diagnosis.Most of the classification tasks in medical image analysis try to assign image-level labels to the given medical images.The labels of these medical images can either belong to one (or different) disease or a healthy case.We explore the related classification applications in Section 4.1.Segmentation is different from classification tasks in medical image analysis, and it aims to assign pixel-level labels to the images.The segmentation tasks in medical image analysis can draw the contour of the disease in the organs or anatomical structures.We explore the related segmentation applications in Section 4.2.Object detection in medical image analysis focuses on identifying and localising diseases.It is different from classification and segmentation, and it will assign an object bounding box to localise the position of the disease and give a label for the predicted bounding box.We explore the related object detection applications in Section 4.3.The next task is image registration which transforms multiple medical images or different medical image modalities into an optimal spatial transformation.The optimal spatial transformation generated by medical image registration can efficiently help doctors to analyse comprehensive information for patients' health conditions.We explore the related registration applications in Section 4.4.Moreover,we also introduce other related tasks for medical image analysis in Section 4.5.
4.1 | Classification
Image classification plays a crucial role in analysing medical images.The objective of image classification is to classify the training examples into different image-level classes.Since the development of deep learning has grown rapidly, image classification tasks can achieve excellent results since 2012 [136].Various methods could be applied to ameliorate the neural network's performance, such as data augmentation [137–140],transfer learning [141–143], and so on.The comprehensive reviews and surveys of medical image classification can be found in [144–146].In spite of the fact that numerous applications have been developed in this domain, not having enough ground-truth labels for the training datasets is still a common problem.Therefore,the existing works involving the weakly-supervised learning approaches to achieve medical image analysis have been summarised and revised in this section.
Although there are a large number of cutting-edge techniques and methods which are fully automatic, medical image analysis still has some challenges that human interventions can solve.Here, we list some works related to active learning.
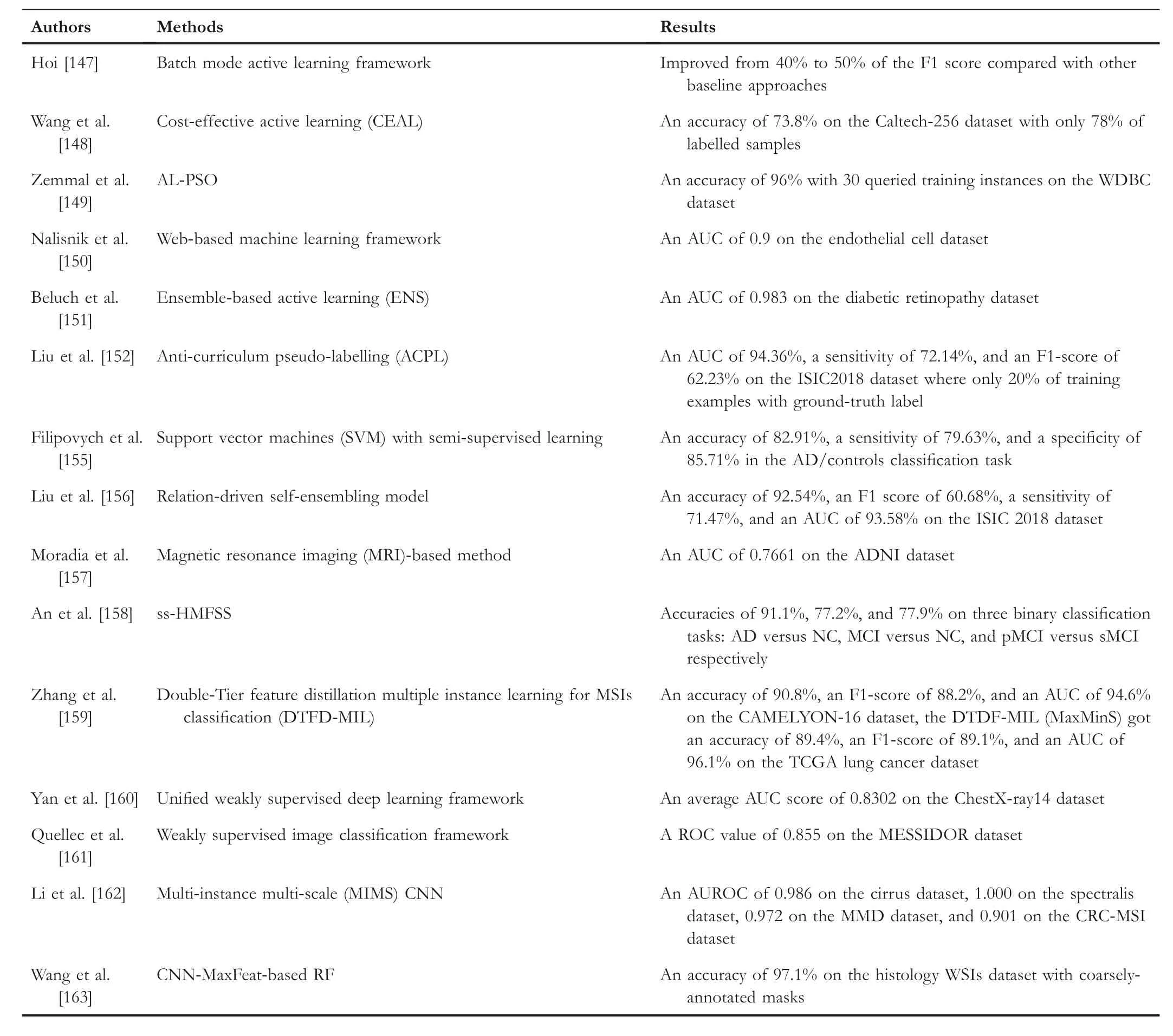
Hoi and his colleagues [147] have suggested a framework that uses the fisher information matrix to simultaneously filter the most informative training instances, which is named as Batch Mode Active Learning framework.For the purpose of determining the unlabelled instance to reduce the Fisher information, they designed a greedy algorithm to locate them.The proposed framework is different from traditional active learning methods.It can query multiple training examples at the same time.As a result, their framework improved from 40% to 50% of the F1 score compared with other baseline approaches for the medical image dataset.
Wang et al.[148] introduced an active learning framework that uses an optimal feature representation to build the model,named Cost-Effective Active Learning (CEAL).In the proposed model, the model can simultaneously learn and update from the manual labels with the most confident annotations.The approach reaches cutting-edge performance with no more than 60% data for the datasets CACD (face recognition task)and Caltech-256 (object classification task) datasets.Specifically,the proposed CEAL framework can achieve an accuracy of 73.8%on the Caltech-256 dataset with only 78%of labelled samples.
Zemmal et al.[149]proposed a framework called AL-PSO,which amalgamates the active learning and the particle swarm optimisation (PSO) algorithm in order to minimise the spent on the labelling process.In this framework, they design a bioinspired optimisation algorithm of the framework and a particle swarm optimisation algorithm with the uncertainty measure to extract the most useful training examples.Finally,they evaluate their framework on 18 benchmark datasets.In the WDBC dataset,the AL-PSO got an accuracy of 96%with only 30 queried training instances.The results show that their performance is close to the fully supervised and other semisupervised approaches.
Nalisnik et al.[150] proposed a web-based machinelearning framework that can help users construct classifiers based on active learning.The proposed framework can efficiently deduce the labelling effort and achieves an AUC of 0.9 on the endothelial cell dataset.
In 2018, Beluch et al.[151] used ensemble-based active learning (ENS) to classify medical images.Their method combines the ensemble approaches with the uncertainties'consistency mechanism together, and their method is better than other uncertainty estimation methods.The MNIST and CIFAR-10 datasets have been evaluated through Beluch's method.They tested their model on the two datasets mentioned above, and they reached an AUC of 0.983 as their best-performance version of ENS called ENS-VarR by applying the diabetic retinopathy data group.
The aim of semi-supervised learning is to resolve two medical image analysis-related problems: the model performs well on multi-class and multi-label tasks (an image might include multiple classes or has several labels).Solve the imbalance data distribution problems (the data distribution of different classes might be different, and the overfitting problem of training a model would occur due to the distribution of unbalanced data) [152].There are some common methods of the most advanced semi-supervised learning: consistency learning [62, 64], self-supervised pre-learning [153], and pseudo-labelling learning [59, 154].In the remaining part of this section, we will show related works based on semisupervised learning and then introduce other weaklysupervised learning-based works related to classifying medical images.
Liu and his colleagues [152] designed an Anti-curriculum Pseudo-labeling (ACPL) method, which introduces a new semi-supervised learning algorithm to choose the most useful unlabelled training example.The proposed method has also enhanced the capability of the model to work on the unbalanced data distribution, and it also works well on multi-label and multi-class tasks.Moreover, they use an ensemble-based method which would help to enhance the accuracy of pseudo-labels.This proposed method tests on two datasets:Chest X-Ray14 and ISIC2018.They reach an AUC of 94.36%,a sensitivity of 72.14%, and an F1 score of 62.23% on the ISIC2018 dataset, with only 20% of training examples having ground-truth labels.
Filipovych's research [155] obtained the pattern classification of mild cognitive impairment (MCI) by using support vector machines with semi-supervised learning.They conclude that semi-supervised learning could overcome the challenges in the field of progressive disorders caused by the uncertainty of the diagnostic results.To prove the effectiveness of the currently used semi-supervised learning approach, they also utilise fully supervised methods to compare with the proposed method.The proposed method tests in the AD/controls classification task and reaches superior results in accuracy,sensitivity, and specificity with values of 82.91%, 79.63%, and 85.71% respectively.
Liu et al.[156] classified the medical images via relationdriven self-ensembling.The consistency-based method combines various perturbations and a self-ensembling model for the unlabelled data.Moreover, they design a sample relation consistency(SRC)paradigm to map the semantic relationships between different samples with perturbations.Then the model can extract more semantic information from the unlabelled data.This approach gets an accuracy of 92.54%,an F1 score of 60.68%,a sensitivity of 71.47%,and an AUC of 93.58%on the ISIC 2018 dataset.The result exceeds most of the high-tech methods applied to the same datasets.
Moradia et al.[157] developed a magnetic resonance imaging (MRI) based method to predict the conversion from MCI to AD.There are two parts to this method.In the first part, they design the MRI biomarker to predict MCI-to-AD conversion based on semi-supervised learning.Secondly, they propose an aggregate biomarker that integrates the first part with age and cognitive measures to get the result.Finally,the MRI biomarker gets an AUC of 0.7661 on the ADNI dataset.
An et al.[158] introduced a method that uses a semisupervised hierarchical multimodal feature to diagnose Alzheimer's disease (ss-HMFSS).In this method, they extract valuable and complementary data features from structural magnetic resonance imaging (MRI) and single nucleotide polymorphism (SNP) to improve the effectiveness of the diagnosis.During the learning process, they integrate a semisupervised mechanism with an SVM-based model to classify the disease.AD versus NC,MCI versus NC,and pMCI versus sMCI are the three binary classification tasks that have been tested by this method.For these tasks, it gets accuracies of 91.1%, 77.2%, and 77.9% respectively.
Histopathology whole slide images (WSIs) classification tasks are widely deployed by the MIL.However, there are still some problems with the small sample cohorts.Normally, too many patches are cropped to form a single histopathology whole slide image with high resolution.To resolve related problems,Zhang et al.[159]developed a Double-Tier Feature Distillation Multiple Instance Learning for MSIs Classification(DTFD-MIL).They first introduce the pseudo-bags to increase the size of the bags,and the feature distillation uses the derivation of the instance probability.Finally, they evaluate their framework on the CAMELYON-16 and TCGA lung cancer dataset, and the framework outperforms the other upto-the-minute methods.This framework has four feature distillation strategies: MaxS, MaxMinS, MAS, and AFS.The DTDF-MIL (AFS) got an accuracy of 90.8%, an F1 score of 88.2%, and an AUC of 94.6% on the CAMELYON-16 datasets.The DTDF-MIL (MaxMinS) gets an accuracy of 89.4%,an F1 score of 89.1%, and an AUC of 96.1% on the TCGA Lung Cancer dataset.
Yan et al.[160] proposed a unified, weakly supervised deep-learning framework to classify and localise chest diseases.The proposed framework uses multi-map transfer layers to learn the data features and squeeze-and-excitation blocks to recalibrate the cross-channel features.They tested the performance of the framework on the ChestX-ray14 dataset and received 0.8302 as the average AUC score.
Quellec et al.[161] presented a weakly supervised image classification framework that can automatically detect the relevant pattern of unlabelled images based on the given reference images.They get a ROC value of 0.855 on the MESSIDOR dataset.
Li et al.[162] proposed a multi-instance multi-scale(MIMS) CNN in order to classify medical images.In the proposed MIMS, they design a multi-scale convolutional layer that can fuse the data patterns from different receptive fields.They also present a ‘top-k pooling’ to integrate feature maps from multiple spatial dimensions.This method test both the 3D and 2D datasets, and it gets an AUROC of 0.986, 1.000,0.972, and 0.091 on the dataset of Cirrus, Spectralis, MMD,and CRC-MSI respectively.
Wang et al.[163] designed a weakly supervised learning method called CNN-MaxFeat-based RF.This method retrieves the discriminative block via a fully patch-based convolutional network to produce holistic WSI descriptors.They apply the aggregation strategies as well as the feature selection with a context-aware technique.They tested this method on different datasets and got an accuracy of 97.1% on the histology WSIs dataset with coarsely-annotated masks.
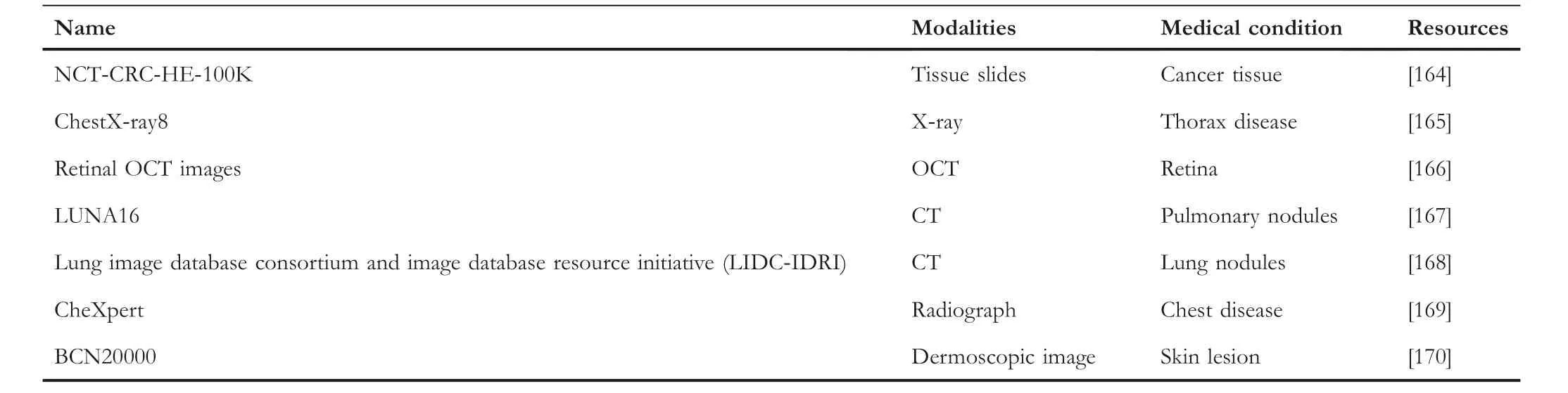
As described above, we summarise these works in Table 2 and the related datasets in Table 3.By analysing these works,we found that although the active learning-based methods need annotations from humans, the active learning-based methods still can get good performance, such as approaches proposed by Hoi [147], Zemmal et al.[149], and Beluch et al.[151].However, most of the works currently focus on semi-supervised learning-based methods such as Anticurriculum Pseudo-Labeling (ACPL) [152], Relation-driven self-ensembling model [156], ss-HMFSS [158].Moreover,some works use MIL methods,such as DTFD-MIL[159]and MIMS [162].Overall, the semi-supervised learning-based method is a promising way and widely used in classification applications.
4.2 | Segmentation
Image segmentation is a vital function in image analysis and processing that can improve the effectiveness of subsequent image processes [171].It seeks to categorise each individual component of the image or volume into separate groups,with each group's components sharing the same set of attributes[172].With regard to analysing the medical image, it has a property that is always associated with an organ or a tissue type.As deep learning develops over time, various methodshave existed and been proposed to use for segmentation,such as threshold-based segmentation [173], region-growing method [174], morphological image processing [175],watershed-based segmentation[176],active contours[177]and level set methods[178].Recently,there are also some effective segmentation architectures like FCN[179],U-Net[180],Mask R-CNN [181], RefineNet [182], and DeconvNet [183].Existing reviews and surveys also review the recent progress of segmentation in medical image analysis[7,184–186].However,the cost of pixel-level annotation of medical images is high and tedious.In order to reduce the cost,weakly supervised learning method could be the optimal choice,and it does not need too many human interventions.We will show related works in this section.
T A B L E 2 Summary of weakly supervised learning-based classification applications on medical image analysis.
T A B L E 3 Overview of datasets used for medical image classification.
Gorriz and Carlier [187] did the semantic segmentation task for the medical images by using a Cost-Effective Active Learning framework.In this framework, the pixel-wise uncertainty is calculated using the dropout approach as Monte Carlo sampling.The dataset's intrinsic information is then extracted to enhance the framework's performance.The framework tests on the ISIC 2017 Challenge dataset for the Skin Lesion Analysis and reached a Dice Coefficient of 74%with nine queries and two training epochs.
Yang et al.[188] proposed a suggestive annotation-based deep active learning framework that integrates the fully convolutional network (FCN) and active learning.The active learning in this framework aims to make precise suggestions to the annotation areas with the highest confidence.The framework can efficiently reduce the annotation cost.They evaluate the performance of two datasets which are called the 2015 MICCAI Gland Challenge and the second one is named the lymph node ultrasound image dataset.The result shows that they reach the Mean IU of 87%and F1-score of 87.4%for the lymph node ultrasound image segmentation task.
Gu et al.[189] coped with the homogeneous related areas of medical images with low confidence problems by forestoriented superpixels (Voxel).By collecting the additive feature maps of the random forest, the suggested forestoriented superpixels approach can enhance the performance of the default random forest classifier.It also can automatically identify the area which can mislead the classifiers.Additionally,a semi-supervised learning-based Bayesian strategy is applied to leverage the confusing parts of the medical image.Finally,they reach apvalue that is less than 0.0000005 on the DRIVE dataset.Their segmentation performance indeed improves for the various sizes of the training dataset.
Jia et al.[190] proposed a constrained, deep, weak supervision system for histopathology image segmentation (DWSMIL).The proposed system is based on MIL,and it follows an original formulation called deep weak supervision(DWS).The histopathology image segmentation runs with fully convolutional networks (FCNs).Moreover, to improve the system's performance, they introduce constraints of positive instances to help the model explore extra hidden data features.In order to validate the effectiveness of the system,they test the system on large-scale histopathology datasets as well as other medical datasets.They reach the F-measure of 0.835 and an ODS of 0.559 on a histopathology image dataset of colon cancer.
Nie et al.[191] designed an attention-based semisupervised deep network called ASDNet, which addresses the insufficient training data with ground truth label problems in medical image analysis.The proposed ASDNet contains two parts: the fully convolutional adversarial learning trains the segmentation model, and a confidence network aims to build the confidence map with a region-attention mechanism to train the unlabelled instances.They evaluate the performance of the ASDNet with five-fold cross-validation and get 0.900 in terms of DSC to segment the prostate.
Kervadec et al.[192] presented a Constrained-CNN Loss for the weakly supervised segmentation task.Their method introduces a new penalty that can bring inequality constraints to the loss function.The loss function avoids using the Lagrangian dual optimisation and proposal generation.They test this method on different segmentation tasks, and their experiments only pay attention to the basic linear constraints.It also can easily be deployed on other non-linear constraints.The best results are a mean dice score of 0.8604 on the vertebral body dataset and a mean dice score of 0.8298 on the prostate dataset.
A collaborative learning approach was suggested by Zhou et al.[193]in 2019.In this approach,they use semi-supervised learning with an attention mechanism to segment the image.Firstly, they design a multi-lesion mask generation model to implement the segmentation task.Secondly, a disease grading model is proposed to classify the severity.At the same time, a lesion attention-based model also refines the feature maps of the lesion by using image-level information with semisupervised learning.This approach evaluates diabetic retinopathy (DR) problems with three datasets: IDRID Dataset,EyePACS Dataset,and Messidor Dataset.Finally,their method reaches an AUC of 97.6% and an accuracy of 93.9% for the referable settings of the Messidor.For the normal settings of the Messidor, this method gets an AUC of 94.3%, and an accuracy of 92.2%.
Xu et al.[194] suggested a framework called multiple clustered instance learning (MCIL) for the classification, segmentation, and clustering of medical images based on weakly supervised learning.In their framework, it only needs the image-level labels, and the framework can implement classification tasks at the image level, segmentation tasks at the pixel level, and clustering tasks at the patch level simultaneously.They evaluated the framework on the cytology dataset and got an F-measure of 0.673.
Rajchl et al.[195] proposed DeepCut, which can obtain pixel-level object segmentations.They regard the segmentation task as an energy minimisation task with the densely-connected conditional random field.And they develop different versions of DeepCut methods.Finally, the proposed DeepCut gets a mean DSC of 74.9%on the foetal magnetic resonance dataset.
Wang et al.[196] designed a Mixed-Supervised Dal-Network called MSDN, which contains two neural networks to implement the detection and segmentation tasks respectively.In this framework, a set of connection modules are designed between two models.These modules can transfer valuable knowledge from the detection task to the segmentation task.Moreover, a ‘Squeeze and Excitation’ technique is also used to accelerate the knowledge transfer process.They also proposed a variant of MSDN called MSDN- and tested MSDN-on the lung nodule dataset and got a mean dice score of 84.90 ± 0.60.
Meier et al.[197]developed a fully automatic segmentation method named SSDF for postoperative brain tumour segmentation.In this method,they try to fuse the preoperative and postoperative scans of the patient to boost the performance of segmentation of the postoperative image.They also use the semi-supervised decision tree in the method.This approach evaluates a cohort of 10 high-grade glioma patients'dataset.Finally, they get a result shown in a tuple pattern(median, range), and their results are the sensitivity of(0,16,0.27),the specificity of(0.99,0.08),the positive predictive value of (0.24,0.93) and the absolute volume error in [ml](0.24,4.72).
Li et al.[198]presented a shape-aware semi-supervised 3D semantic segmentation method.The developed approach can obtain useful information from the unlabelled data and generate a geometric shape constraint for the output.During the training process, they propose an adversarial loss between labelled and unlabelled SDMs.The method evaluates the Left Atrium (LA) dataset from Atrial Segmentation Challenge, and it gets a Dice of 89.54% and a Jaccard of 81.24%.
Luo et al.[199] proposed a dual-task-consistency semisupervised framework to segment medical images.A dualtask neural network is used to predict the pixel-wise segmentation map and represent the geometry-aware level set of the object.They also use a dual-task consistency regularisation in the framework for both labelled and unlabelled data.This framework validates on the Left Atrium MRI dataset and gets a Dice of 89.42% and a Jaccard of 80.98%.
Bortsova et al.[200] designed a series of methods to segment the medical images with learning consistency under transformations.They dub the consistency-regularised version as SupTC.All of the methods are based on semisupervised learning that only uses the unlabelled images of the training set (SemiTC), and a SemiTC + method uses additional unlabelled examples as validation and test set.In these methods, they use a Siamese architecture with two branches to accept transformed images and a loss function that can check the segmentation consistency of two branches.These methods validate on the Japanese Society of Radiological Technology (JSRT) dataset, and the SemiTC + got an IOU of 95.5 ± 1.9% for the lungs, 88.8 ± 4.9% for the heart, and 88.1 ± 4.4% for clavicles with only 124 (or 123)labelled images.
Xia et al.[201] proposed an uncertainty-aware multi-view co-training (UMCT) framework to segment the volumetric medical images.Firstly, they transform the 3D volumes into different views and train a 3D neural network for every view.Secondly, they use co-training to force the consistency of unlabelled data to the multi-view.The proposed framework gets a DSC of 81.18% with 20% labelled data and a DSC of 78.77% with 10% labelled data on the NIH dataset.
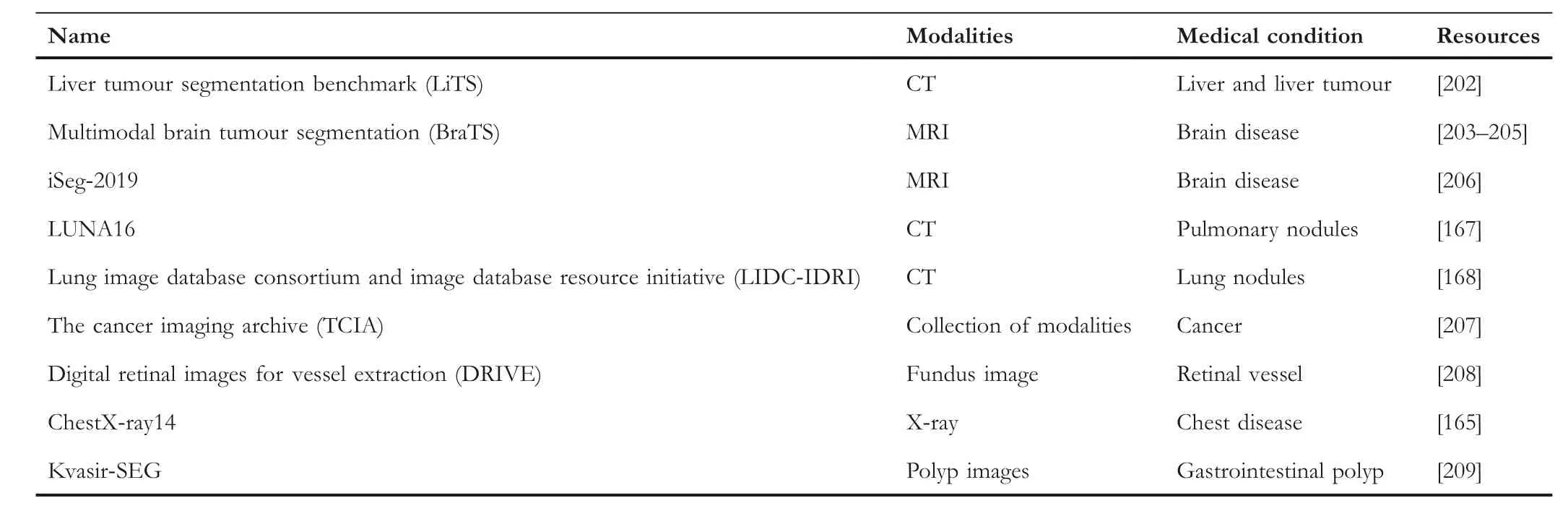
Table 4 summarises the existing works mentioned above,and we provide related datasets in Table 5.As Table 4 shows,the methods proposed by Gorriz and Carlier [187] and Yang et al.[188] apply the active learning mechanism.Most of the other works use semi-supervised learning to do the segmentation task.For example, the works proposed by Luo et al.[199] and Li et al.[198] use semi-supervised learning as the backbone and train on the Left Atrium(LA)dataset,and both works reached a Dice score of around 89%and a Jaccard score of around 81%.Moreover, the MIL-based method is also proposed by Xu et al.[194].Overall, we found that the semisupervised learning-based methods can achieve excellent performance on different segmentation datasets, and they can combine the advantages of different supervised and unsupervised models or algorithms.
4.3 | Object detection
Object detection can help humans to locate instances in videos and images, especially for advanced driver assistance systems(ADAS).It plays an important role in maintaining the safety of drivers.Moreover, it is also useful for other domains and has recently attracted much attention [210].Concretely, object detection mainly focuses on classifying training examples and identifying the location and concepts contained in each training example [211].With the development of deep learning, object detection no longer heavily relies on handcrafted features and shallow neural network architectures.Instead, it uses deep learning to learn the semantic, deeper features of the training examples.Object detection can be divided into two types:classical object detection and deep learning-based methods.There are some classical object detection methods have already developed for years,such as Viola-Jones(VJ)[212],Histogram of Gradients (HOG) [213, 214], and Sliding Window (SW)[215].Deep learning-based methods have one-stage detectors and two-stage detectors.One-stage detectors aim to predict object bounding boxes directly from an image without other intermediate tasks.Two-stage detectors aim to generate the proposals and then extract the features based on the generated proposals.There are some popular approaches of one-stage detectors like overfeat [216], YOLO [217, 218], Single Shot Detector (SSD) [219], RetinaNet [220, 221], and Cornernet[222].For two-stage detectors, there are also plenty of the methods such as R-CNN [223], SPP-Net [224], Fast R-CNN[225], FasterR-CNN [226], R-FCN [227], Mask R-CNN[181], and Feature Pyramid Networks (FPN) [228, 229].The related reviews and surveys for object detection in medical image analysis are summarised in refs.[230–232].We will discuss the weakly-supervised learning-based disease detection methods in the remaining part of this section.
Smailagic et al.[233] presented a sampling approach(MedAL) for the Diabetic Retinopathy detection task.Firstly,they query the unlabelled instances by maximising the average distance.Then use Oriented FAST and Rotated BRIEF(ORB)feature descriptors to detect the disease.Finally, they reached an accuracy of 80% with only 425 annotated examples, and there is a 32%cost reduction in the labelling process compared with uncertainty sampling and a nearly 40% cost reduction compared with random sampling.
Zhu et al.[234] developed a DeepEM framework for the pulmonary nodule detection of the lung CT dataset.It is a deep 3D ConvNet framework with an expectation-maximisation algorithm.The pulmonary nodule detection of lung CT images is commonly using fully supervised approaches, and itnormally needs lots of training images with ground-truth labels.In this work, they try to extract information from the electronic medical records (EMR), which includes partial information about the image.Their results got an FROC of 76%with DeepEM (MAP) and an FROC of 76.4% with DeepEM(sampling) on the Tianchi dataset.

T A B L E 4 Summary of weakly supervised learning-based segmentation applications on medical image analysis.
T A B L E 5 Overview of datasets used for medical image segmentation.
Costa et al.[235] designed a weakly supervised framework for interpretable diabetic retinopathy detection on retinal images.This framework uses the MIL, which can extract the hidden information from the image-level annotations.It also uses the joint optimisation technique in instance encoding.Moreover, it introduces a new loss function to enhance the explainability of the framework.The framework got an AUC of 90% on the Messidor dataset, an AUC of 93% on the DR1 dataset, and an AUC of 96% on the DR2 dataset.
Mohammed et al.[236] developed a weakly supervised COVID-19 detection method called ResNext+.This method only needs volume-level image labels to predict the slice-level information.To extract spatial features from the dataset, they combine a lung segmentation mask with the spatial and channel attention mechanism.A Long Short-Term Memory(LSTM) is also used to get the axial dependency of the slices.Finally, they designed the ablation study to evaluate the effectiveness of their approach and got a precision of 81.9%,the F1-score of 81.4% on the COVID CT dataset.
Hu et al.[237]proposed a novel weakly supervised paradigm to detect COVID-19 infection and classify the different classes of COVID-19 from the chest computerised tomography scans.This paradigm reduces the cost of the human annotation process for COVID-19 CT scans, and it also can implement infection detection and classification tasks.This method can classify the COVID-19 dataset into three classes of COVID-19:COVID-19(coronavirus disease 2019),NP(Non-Pneumonia),and CAP (Community Acquired Pneumonia).They evaluate this method on the dataset from two participating hospitals and the TCIA dataset [238].The “NP”, “COVID-19” and “CAP”class get a sensitivity of 91.3%,87.6%,and 83.0%,and a mean AUC of 0.901,0.923,and 0.864 respectively.
Wang et al.[163] used a weakly supervised framework to detect lesions automatically.This framework only needs a group of normal and abnormal retinal images without the annotations to implement the detection task.In detail, they regard fundus images as a superposition of background.The background is regarded as a low-rank structure by applying different data preprocessing techniques.This framework is tested on Kaggle and Messidor datasets.Overall, the framework gets an AUC of 0.9907,a MAP of 0.8394 on the Kaggle dataset, and an AUC of 0.9974, a MAP of 0.9091 on the Messidor dataset.
Chamanzar and Nie [239] designed a weakly supervised multi-task approach to detect and segment the cell.It combines different point-label encoding approaches and a multitask scheduler to train the model.They evaluate their method on the PMS2 stained colon rectal cancer and tonsil tissue datasets and get a precision of 94.1%,a recall of 92.5%,and a concordance correlation coefficient (CCC,α=0.05) of the detected cell count is 0.998.
Li et al.[240] tried to detect mitosis in breast histopathology images using a weakly supervised method, which combines with a novel loss function called ‘concentric loss’.Specifically,they use semantic segmentation with the deep fully convolutional network as the backbone to detect mitosis.In the proposed method, the label of the training instance is a centroid pixel.Moreover, they design their label with concentric circles.Inside the concentric circle, it is a mitotic region,and there is also a ring around the circle called‘middle ground’.They get the F-score of 0.562, 0.673, and 0.669 for the ICPR 2014 MITOSIS, AMIDA13, and TUPAC16 datasets respectively.
Sebai et al.[241] proposed a multi-task deep learning framework that integrates object detection and instance segmentation Mask RCNN(MaskMitosis)to detect the mitosis in histopathology images.The object detection part will localise the mitosis and classify the mitosis classes.The instance segmentation Mask RCNN will try to estimate the mitosis mask labels for the weakly labelled mitosis training instances.They get an F-score of 0.475 for the 2014 ICPR dataset.
Kuo et al.[242] presented a cost-sensitive active learning ensemble method to detect intracranial hemorrhage.The method is based on query-by-committee active learning.Their experiments were tested on the head computed tomography(CT) dataset and got a stack AP of 95.6 ± 0.9.
Dubost et al.[243] introduced a GP-Unet with weak supervision to detect lesions.The GP-Unet uses the image-level label to generate the 3D output with the lesion count.The backbone of the GP-Unet is a fully convolutional architecture with a global pooling layer.The experiment first trains the model to calculate the number of lesions and compute the localisation maps of input data by deleting the pooling layer.The approach achieves a sensitivity of 62% on the 3D PDweighted MRI scans.
Li et al.[244] proposed a MIL approach to detect cancer.They solve the submodular set cover problem by searching a group of region-level prototypes and then using regularised regression trees integrated with prototypes based on the multiple instance boosting (MILBoosting) framework to detect cancer.The proposed method gets an AUC of 0.93 on the breast cancer TMA images and an AUC (image-level) of 0.79± 0.01%, and an F-measure (instance-level) of 0.45 ± 0.03%on the OPT images of colorectal polyps.
Wang et al.[245] presented a two-stage hip and pelvic fracture detection approach which uses weakly supervised ROI mining to localise fracture classification.They use a fully convolutional network at the first step, then use a small capacity model with mined RUIs from the whole PXR of the training data to localise fracture classification.This method validates on 44100 PXRs and gets a ROC value of 0.975.
Costa et al.[246]introduced weakly supervised pre-trained convolutional neural networks (EyeWeS) to detect Diabetic Retinopathy(DR).The proposed method can convert any pretrained models to a weakly supervised manner with high performance.A set of methods can adapt to this method to detect DR automatically and locate the regions of the images that contain lesions.This method tests on the Messidor dataset and gets an AUC value of 95.85%.
Zhou et al.[247]proposed a weakly feature encoding with autoencoders to detect the anomaly.This strategy uses an autoencoder to encode the input data with a hidden vector,reconstruction residual vector, and reconstruction error to a new representation of the input data.Moreover, newly designed architecture can seamlessly integrate three factors.This method gets an AUC-ROC value of 0.999 ± 0.001 on the Cardio dataset.
We conclude these works in Table 6, and Table 7 lists related datasets for object detection in medical image analysis.By analysing these works,the weakly supervised learning-basedmethods can reach a better performance than the supervised learning-based methods, such as approaches proposed by Zhou et al.[247], Chamanzar and Nie [239], Wang et al.[163]and Wang et al.[245].Some of the works try to extract semantic information from different modalities: Zhu et al.[234]use EMR records to get hidden information to improve the performance with images.Mohammed et al.[236] extract information from volume-level to slice-level.Overall,the weakly supervised learning-based object detection methods in medical image analysis can either extract hidden knowledge from limited labelled datasets or different modalities to improve the performance, and some of the works can reach or be further improved than the supervised learning-based methods.

T A B L E 6 Summary of weakly supervised learning-based detection applications on medical image analysis.
4.4 | Registration
Image registration focuses on transforming different scenes into a single integrated image that can align the anatomical structures of existing scenes[253].In medical image analysis,it is common to have multiple images or different imaging modalities at the same time,so it is desirable to align the different images into an optimal spatial transformation.Image registration has been widely implemented in many tasks.For example,image guidance[254–260],image segmentation[261–263], dose accumulation [264–267], image reconstruction[268–271] and so on.There are some reviews and surveys specific to image registration in medical image analysis [253,272–274].We will list the weakly supervised learning-based registration works in the remaining part of this section.
Blendowski et al.[275]illustrated a weakly-supervised deep learning-based feature extraction method to align the different medical images to an integrated image.They propose a Supervised iterative descent algorithm (SUITS) with modifications to deploy their method.This method gets a Dice score of 51.3% and a Hausdorff distance of 33.8 mm.
Nguyen-Duc et al.[276] designed a deformable image registration method with a weakly supervised mechanism to analyse the ssEM images.This method improves slice interpolation when images have structural differences, and it onlyneeds roughly aligned data to train the deformation model.The proposed method is validated on the ssEM datasets and gets the best dice coefficient of 0.798 on the CREMI dataset[277].
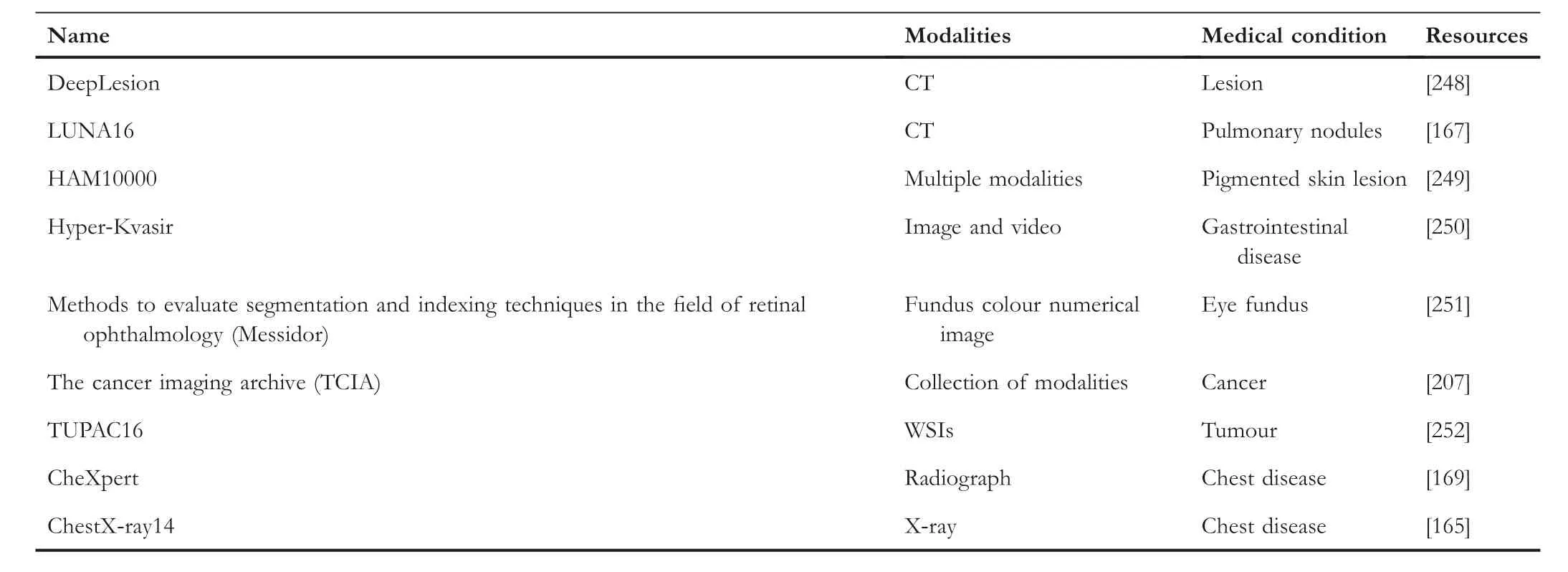
T A B L E 7 Overview of datasets used for medical image object detection.
Ding et al.[278] proposed a weakly supervised and annotation-efficient framework to find the vessel in ultrawidefield (UWF) fundus photography (FP).The framework includes two steps:the first one is image registration,and they use a pre-trained model registered with parametric chamfer alignment.The second step is the weakly supervised learning step, they train the model with noisy labels, and then the detected FP vessel maps are used in the following registration iterations.They also introduce a new dataset called PREIMEFP20 to evaluate their framework.This framework gets an AUC PR of 0.841,a Max DC of 0.771,and a CAL of 0.730 on the noisy PREIME-FP20 dataset.
Ferrante et al.[279] designed a weakly supervised method to aggregate the domain-specific conventional metrics with anatomical segmentations.This approach used the support vector machine as the backbone and evaluated the method on different CT and MRI datasets.Compared with the single metric approach,it improves by 0.033,0.082,and 0.037 of the Dice coefficient value for RT Parotids, RT Abdominal, and IBSR datasets respectively.
Xu and Niethammer [280] proposed a joint semisupervised learning approach called DeepAtlas for image registration and segmentation tasks.The segmentation task can boost the performance of the registration process, and conversely, the registration task can benefit the segmentation task.The experiments validated the knee and brain 3D magnetic resonance (MR) images, and it gets a Dice of 85.66%,which only uses 10 of 200 labelled images.
Tony and Albert [281] introduced the Coarse-to-Fine Vision Transformer (C2FViT) for 3D affine registration.The proposed method learns global affine registration by combining the global connectivity with the locality of the vision transformer and multi-resolution strategy.This method tests on the LPBA dataset and gets an average Dice score of 0.53 and HD95 of 4.06.
Hu et al.[282] presented a weakly supervised and labeldriven formulation to learn the correspondence between the 3D voxel and higher-level labels.They optimise the model by generating a dense displacement field(DDF)to correspond to the counterparts of the fixed image.In the registration process,the model uses a pair of new images to output an optimal DDF.The experiment tests 111 pairs of T2-weighted magnetic resonance images and 3D transrectal ultrasound images with over 4000 anatomical labels.Finally,this method gets a median target registration error of 4.2 mm on landmark centroids and a median Dice of 0.88 on prostate glands.
Zeng et al.[283] designed a weakly supervised learning method (SR-L and SR-I) for the label-driven magnetic resonance imaging (MRI)-transrectal ultrasound (TRUS) registration.This method includes three sub-networks:the first part is two whole-volume-based 3D FCNs to segment the prostate;the second part is a 2D CNN to segment MRI with US prostate labels; the last part is a 3D UNET-like model to generate a non-rigid registration model.All these parts are trained independently.These methods test on the MRI-TRUS dataset and produce a mean target registration error (TRE) of 2.53 ± 1.39mmand a mean Dice loss of 0.91 ± 0.02.
Wang et al.[284] proposed a content-adaptive multimodal retinal image registration for the retinal images.This method uses globally coarse alignment with several weakly supervised models for the different tasks.The first one is vessel segmentation,the second task is feature description and detection,and the last task is outlier rejection.This proposed method uses infrared reflectance and fluorescein angiography images to register colour fundus images.To evaluate the performance of the method, they compare it with different methods and validate it on the JRC CF-IR dataset and get a Dice value of 0.6306 ± 0.1256.
Meng et al.[285] introduced a weakly supervised 2D/3D vascular registration framework to register the preoperative 3D computed tomography (CT) with intraoperative 2D digital subtraction angiography (DSA) in order to provide useful information from 3D-2D projection to the missing parts of 2D vessels.Firstly, a CNN regressor with a 3D-2D mask projection module is proposed, and a supervised pre-training and weakly supervised fine-tuning are implemented.They evaluated this framework on different datasets.They got a Dice of 88.66%, a Recall of 68.49%on the complete vessel dataset,and a Dice of 37.64%, a Recall of 74.55% on the incomplete vessel dataset.
Jia et al.[286]proposed a two-stage registration framework with weakly supervised learning for medical images(SeRN).It consists of two components: unsupervised registration and weakly supervised registration models.For the weakly supervised registration model, they based on the semi-supervised learning framework and trained the model with intensity information from the dataset.In this framework, only partial images have the ground-truth label, and the remaining images are labelled with pseudo-labels.They test the effectiveness of the framework on two datasets: cardiac and abdominal datasets, and the weakly supervised model (SeRN-wsup) gets a Dice of 85.23±4.14%on the myocardium(MYO),a Dice of 87.91 ± 3.47% on the left ventricle (LV) with only four labelled images of 40 images.
These works mentioned above are listed in Table 8,and an overview of datasets used for medical image registration is shown in Table 9.Within these works,the researchers combine supervised, weakly supervised, and unsupervised components to implement the task: Blendowski et al.[275] combine the weakly supervised and supervised mechanisms.Ding et al.[278]use a supervised model to do registration, then use a weakly supervised process in the following registration iterations.Jia et al.[286]combine an unsupervised registration with a weakly supervised registration model to do the object detection task.The work illustrated by Xu and Niethammer [280] uses registration and segmentation tasks to provide useful information on both sides and improve the performance on two sides.Overall,the weakly supervised learning-based registration methods in medical image analysis can utilise the information from different modalities and also can integrate supervised, weakly supervised,and unsupervised components into the pipeline.
4.5 | Other applications
In addition to the different applications that we mentioned above,there are also various applications of weakly supervised learning in medical image analysis.For example, weakly supervised augmentation [297–299], weakly supervised medical image retrieval [300, 301], weakly supervised disease recognition[302,303],weakly supervised medical diagnosis[304–307],and so on.
5 | CHALLENGES AND FUTURE WORKS
This review summarises the recent research on weakly supervised learning in medical image analysis.We can see that lots of researchers have already developed different methods andapplications in this domain.However, shortcomings still need to be fixed and further studied.We summarise these shortcomings as follows:
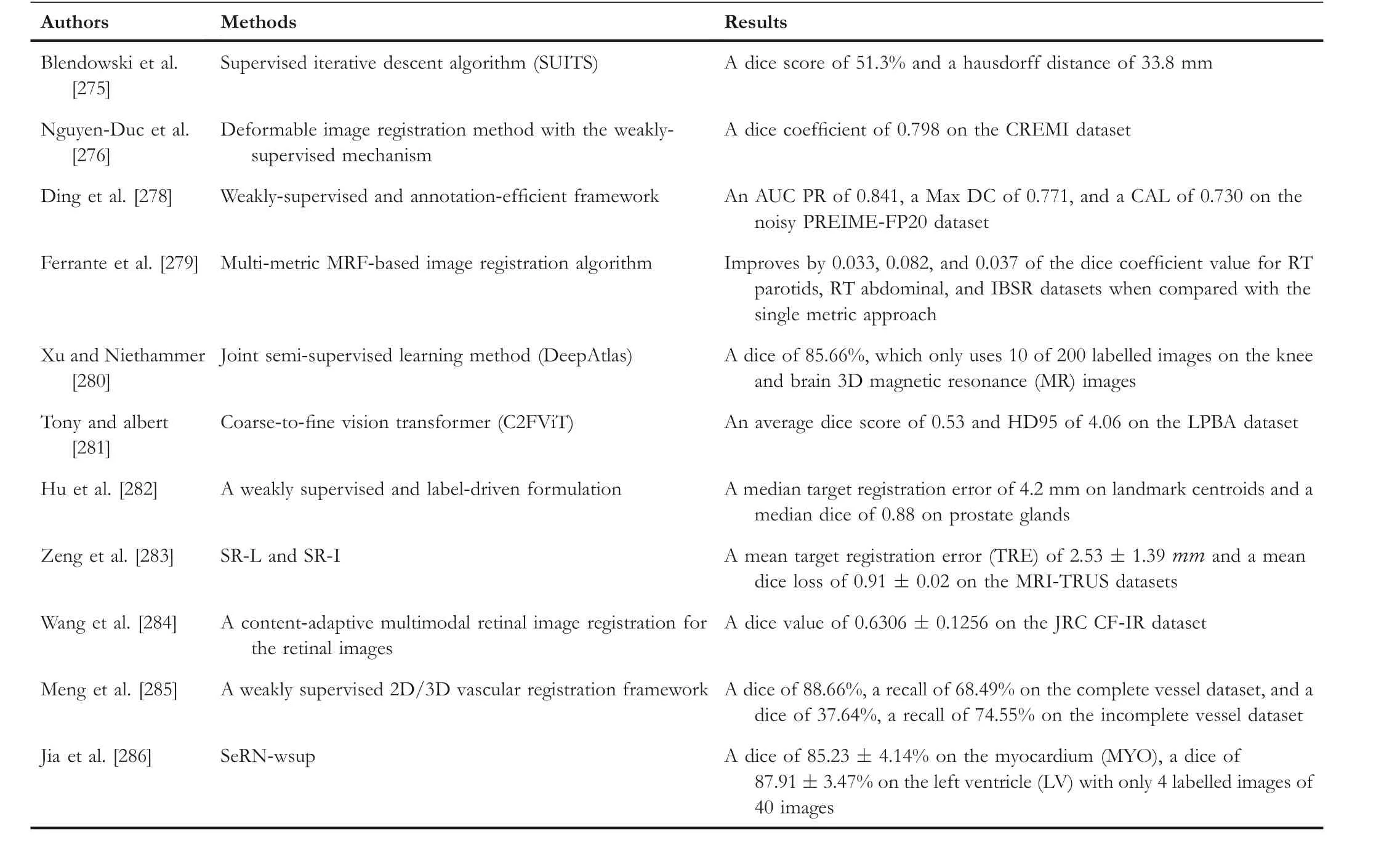
T A B L E 8 Summary of weakly supervised learning-based registration applications on medical image analysis.

T A B L E 9 Overview of datasets used for medical image registration.
• Reduce the degree of supervision: most weakly supervised methods can achieve higher performance by adding extra mechanisms or complex model architectures.The outcomes from existing approaches still have plenty of room to improve to fit real-world applications.Therefore,the researchers need to pay more attention to how to get the same or higher performance of the same dataset with lower supervision.
• Enhance the data augmentation process: in medical image analysis, it is common to use raw data with different data augmentation techniques to do medical analysis.However, the raw datasets always come with noises and redundant information, which can lead to the poor performance of models.It is important to simplify and remove the unuseful information and noises for all the training examples.Especially on weakly supervised learning methods for medical image analysis, effective data augmentation techniques can noticeably increase the performance of models.
• Multimodel fusion: compared with the images in other domains, the medical image always suffers from issues of low contrast and limited texture.One possible solution is getting spatial information from different organs or lesions or using other complementary data to provide reliable information to the model.This extra information can guide the learning process of weakly supervised learning in medical image analysis.
• Combination of domain knowledge: although various models or approaches use weakly supervised learning mechanisms for medical image analysis, applying these approaches in practical applications is still difficult.The main reason is that most of the methods are evaluated on the ideal assumptions.Based on these assumptions, models can always learn the valuable data feature from the dataset.However, the datasets or situations in the real world always cannot meet the ideal assumptions.Therefore, if the data distribution cannot give valuable data patterns to the model,the performance will be worse if more unlabelled data is used.Then it is necessary to incorporate domain knowledge into the model, especially for medical image analysis.
• Explore multi‐task applications: Cholakkal et al.[308]proposed an approach that combines counting and segmentation in a single framework.These two sub-tasks can be done at the same time.For weakly supervised learning in medical image analysis,it is beneficial to apply two or more subsequent tasks to a single model or framework.For example, it is possible to implement image segmentation and text interpretation in a single model.In general, exploring multi-task applications on weakly supervised learning for medical image analysis can produce more practical value and benefits.
• Realisation of unsupervised learning:at the beginning of machine learning, researchers implemented supervised learning for different tasks.With the development of realworld applications, researchers aim to design effective and cost-economy methods based on weakly supervised learning.We can inspire some knowledge from weakly supervised learning, and these ideas can pave the way for unsupervised learning.As far as we know, unsupervised learning cannot perform well as supervised learning and weakly supervised learning.Then,it is possible to implement unsupervised learning based on the ideas from weakly supervised learning.Therefore, the machine learning approaches can act as a human recognising the environment by them self without any ground-truth labels.
• Safe weakly‐supervised learning: the unlabelled data can help the weakly supervised learning models to learn the inherent feature map of the dataset.Nevertheless, there are some empirical studies that also prove that unlabelled data will decrease the performance of the model in certain situations.Sometimes the performance is even worse than the full supervision models.Therefore, it is desirable to secure the performance of weakly supervised learning for medical image analysis.
The next stage of our work will use various data augmentation techniques to remove redundant information and noise in the datasets.Secondly, an efficient feature extraction method is also important for selecting the most representative data features in the medical image.Thirdly, we can also try to use multimodel fusion techniques to extract more inherent information from different sources to increase the performance of weakly supervised learning models.Fourthly, we can also use generative models to generate training examples with the ground-truth label.Finally, we can explore the multi-tasks as we mentioned above in a single model.In summary, weakly supervised learning in medical image analysis has already attracted lots of work in recent years,and there are many potential methods to explore.Although some methods have been got great success in supervised learning, it is still a desirable domain to focus on, and it will produce lots of practical value in real-world applications.
6 | CONCLUSION
This review summarises the weakly supervised learning approaches in medical image analysis,and it mainly pays attention to the core concepts of weakly supervised learning and different applications in medical image analysis, like image classification, segmentation, object detection, registration, and so on.It also introduces related concepts to help the reader get an overview of machine learning methods with different data supervision.In the last decade, supervised learning methods have gained many achievements in many domains because they can provide strong supervision information for all of the training data with ground-truth labels.However, in most realworld applications, especially in medical image analysis, it is challenging and expensive to ensure that all the datasets have ground-truth labels.People prefer to use more effective and low-budget methods to do medical analysis.Therefore,weakly supervised learning is a desirable method for medical image analysis, and the related works in this domain have soared in recent years.Lots of researchers pay attention to weakly supervised learning approaches for medical image analysis.Nevertheless, weakly supervised learning has become an essential method for medical image analysis, and many challenges still exist in the future.
ACKNOWLEDGEMENTS
This paper is partially supported by MRC,UK(MC_PC_17171);RoyalSociety,UK(RP202G0230);BHF,UK(AA/18/3/34220);Hope Foundation for Cancer Research, UK (RM60G0680);GCRF, UK (P202PF11); Sino-UK Industrial Fund, UK(RP202G0289); LIAS, UK (P202ED10, P202RE969); Data Science Enhancement Fund,UK(P202RE237);Fight for Sight,UK (24NN201); Sino-UK Education Fund, UK (OP202006);BBSRC,UK(RM32G0178B8).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
ORCID
Yudong Zhanghttps://orcid.org/0000-0002-4870-1493
杂志排行
CAAI Transactions on Intelligence Technology的其它文章
- Fault diagnosis of rolling bearings with noise signal based on modified kernel principal component analysis and DC-ResNet
- Short‐time wind speed prediction based on Legendre multi‐wavelet neural network
- Iteration dependent interval based open‐closed‐loop iterative learning control for time varying systems with vector relative degree
- Thermoelectric energy harvesting for internet of things devices using machine learning: A review
- An embedded vertical‐federated feature selection algorithm based on particle swarm optimisation
- An activated variable parameter gradient-based neural network for time-variant constrained quadratic programming and its applications
