Study on the molecular mechanism of rapamycin-induced autophagy in acute T lymphoblastic leukemia cells
2023-09-23XULanHUANGLiwenXIAOYishuLIUChunyaDULeRENLicheng
XU Lan, HUANG Li-wen, XIAO Yi-shu, LIU Chun-ya, DU Le, REN Li-cheng
Department of Biology,Hainan Medical University,Haikou571199,China
Keywords:
ABSTRACT Objective: To study the effect of different concentrations of rapamycin on the proliferation of acute leukemia CD4+T-Jurkat cells, and to explore its mechanism from the aspect of autophagy.Methods: The effect of different concentrations of rapamycin on cell proliferation was detected by MTT assay; Apoptosis rate and cell cycle arrest were detected by flow cytometry; The changes of autophagic lysosomes were observed by acridine orange staining;Transcriptome sequencing data were used to analyze the differential expression of autophagyrelated genes; The transcription and protein expression levels of autophagy-related genes were detected by RT-qPCR and Western Blot; The chromatin accessibility of the promoter region of autophagy gene was analyzed by FAIRE-qPCR.Results: Compared with the control group, rapamycin inhibited the proliferation of Jurkat cells in a concentration-and timedependent manner.The cell cycle was arrested in G0/G1 phase, but it could not effectively induce apoptosis.Observed by acridine orange staining, the number of autophagic lysosomes increased after administration.RNA-seq data showed that rapamycin could significantly affect the transcription level of autophagy-related genes.After Jurkat cells were treated with different concentrations of rapamycin for 48 h, 10 nM and 20 nM rapamycin could up-regulate the expression of ULK1, ATG13, ATG16L2, PI3K3R1, Raptor genes and down-regulate the expression of MAPK1 gene.At 50 nM, the expression levels of each gene were downregulated.The chromatin accessibility of the autophagy gene promoter region also changed,which was basically consistent with the trend of gene expression.Conclusion: Rapamycin can inhibit the proliferation of Jurkat cells, block cell cycle and induce autophagy.Low concentrations of rapamycin promoted the expression of autophagy-related genes, while high concentrations inhibited their expression.
1.Introduction
The mammalian target of rapamycin (mTOR) signaling pathway is a major regulator of cell growth and metabolism.The functional relationship between mTOR pathway and autophagy involves complex regulatory networks.In mammalian cells, autophagy is initiated by the ULK complex.ULK1 usually binds to ATG13,FIP200 and ATG101 to form a complex that participates in the initiation of autophagy precursor formation.On the one hand,AMP-activated protein kinase (AMPK) can activate the ULK /AGT1 complex to directly activate autophagy.On the other hand,AMPK can also indirectly activate autophagy by phosphorylating Raptor, thereby inhibiting mTOR activity.At the same time, the phosphatidylinositol-3-kinase (PI3K) complex composed of vacuolar sorting protein 34, Beclin1, Vps15 and ATG14 can also be recruited by ULK to phosphorylate phosphatidylinositol to form phosphatidylinositol triphosphate, which plays an important role in autophagy[1-3].
mTOR is a key molecule regulating autophagy and is involved in the regulation of various aspects of biological processes, including cell survival, death, proliferation, differentiation and senescence.In recent years, a large amount of evidence has shown that autophagy disorders are associated with carcinogenesis.Autophagy plays a dual role in cancer treatment, which can inhibit tumor cell growth and promote tumor cell survival[4,5].
It has been reported that the PI3K-Akt-mTOR signaling pathway is abnormally activated in about 50% to 80% of patients with acute myeloid leukemia (AML) and about 87.5% of patients with T-cell acute lymphoblastic leukemia (T-ALL) in hematological malignancies[6].Rapamycin, as an effective inhibitor of mTOR, can slow down the growth and proliferation of cancer cells.Promote autophagy of ALL cells.More and more clinical data show that rapamycin and its derivatives become potential anticancer drugs.Combining autophagy inhibition strategies with current cytotoxic chemotherapy regimens can provide new treatment opportunities for leukemia patients[7-9].
2.Materials and methods
2.1 Materials
Human acute T lymphoblastic leukemia (Jurkat) cell line was purchased from China Center for Type Culture Collection ;Rapamycin (RAPA) was purchased from Macklin; MTT purchased from Biosharp Biotechnology ; Total RNA extraction kit, reverse transcription kit Fasting RT Kit and DNA purification kit were purchased from Tiangen Biochemical Technology Co., Ltd.; TB Green® Premix Dimer Eraser TM (RR091A) was purchased from Baori Medical Technology Co., Ltd.; ATG16L2 antibody, ATG13 antibody, ULK1 antibody, PI3K3R1 antibody, MAPK1 antibody,Raptor antibody, GAPDH antibodies were purchased from Boster Biotechnology Co., Ltd.; Horseradish peroxidase-labeled goat anti-rabbit IgG (H+L) and ultrasensitive ECL chemiluminescence kit were purchased from Biyuntian Biotechnology Co., Ltd.; Cell Cycle and Apoptosis Analysis Kit and Annexin V-FITC/PI apoptosis detection kit were purchased from Shanghai Yisheng Biotechnology Company.
2.2 Methods
2.2.1 Cell culture
Jurkat cells were seeded in RPMI-1640 medium containing 10%fetal bovine serum and 1% penicillin/streptomycin, and cultured in a constant temperature incubator at 37 ℃ and 5% CO2.When the cell density in the exponential phase reached 80%, the cells were treated with RAPA at a final concentration of 10 nM, 20 nM, and 50 nM for 48 h, and the cells were collected for subsequent experiments.
2.2.2 MTT assay to detect cell proliferation inhibition rate
Jurkat cells in exponential growth phase were seeded in 96-well plates, and 100 μL (1×106cells) cell culture medium was added to each well.RAPA with different final concentrations was added to each well, and each group had 3 wells.The cells were cultured in an incubator at 37 ℃ and 5% CO2for 24 h, 48 h and 72 h.After reaching the time point, 10 μL MTT solution (5 mg/mL) was added to each well and continued to culture for 4 h.After the supernatant was removed by horizontal centrifugation, 150 μL dimethyl sulfoxide (DMSO) was added to each well and shaken at low speed for 10 min.The optical density (OD) value was measured at 490 nm.The cell proliferation inhibition rate was calculated according to the following formula : cell proliferation inhibition rate = (control group OD value - experimental group OD value ) /control group OD value× 100%.
2.2.3 Detection of apoptosis rate and cell cycle by flow cytometry
The exponentially growing cells were inoculated in 6-well plates and treated with different concentrations of RAPA.After 48h of culture in the incubator, 1×106cells were collected.Wash the precooled PBS twice, centrifuge at low speed, and discard the supernatant.Part of the cells were resuspended with 100 μL 1×Binding Buffer, followed by 5 μL Annexin V-FITC dye and 10uL PI dye, gently mixed and reacted at room temperature for 15 min, and then 400 μL 1×Binding buffer was added.The apoptosis rate of cells was detected by flow cytometry.The other part was re-suspended with 70% pre-cooled anhydrous ethanol and stood at 4 ℃ overnight.The pre-cooled PBS was washed twice, and 500 μL staining buffer was added to re-suspend, and then 10 μL RNaseA and 10 μL PI dyes were added to gently mix.After 30 min of dark staining at room temperature, the cell cycle was detected by flow cytometry.
2.2.4 Detection of autophagy by acridine orange fluorescence staining
After Jurkat cells were treated with different concentrations of RAPA for 48 h, 300 μL cell suspension was added with 3 μL acridine orange staining solution, fixed at room temperature for 15 min, and washed three times with PBS.Under the fluorescence microscope,the blue excitation filter was used to observe the morphological structure.
2.2.5 RT-qPCR and Western Blot analysis
Cell suspensions treated with different drug concentrations were collected, RNA was extracted according to the instructions of the cell total RNA extraction kit, and cDNA was synthesized by reverse transcription.The reaction program was set as : 95 ℃ 30 s, 95 ℃5 s,55 ℃ 30 s, 72 ℃ 30 s, 40 cycles.GAPDH was used as the internal reference gene, and the relative expression of mRNA was calculated by 2-ΔΔCTformula.Three replicates were set up in different treatment groups.The primer sequence was designed online by Primer 3, as shown in Table 1.The expression levels of related proteins were analyzed by Western Blot.The cells in each group were added with RIPA lysate and lysed on ice for 30 min.The total protein was extracted and the protein concentration was detected by BCA kit.After protein denaturation, the sample was separated by SDS-PAGE,transferred to PVDF membrane, sealed on a shaker for 1h, overnight at 4 ℃ with primary antibody, incubated with secondary antibody at room temperature for 1 h, developed by ECL luminescence kit, and analyzed by Image J software.

Tab 1 Primer information for RT-qPCR
2.2.6 Transcriptome sequencing analysis
The cell suspension was collected after 0 nM~50 nM RAPA treatment for 48 h, and 3 replicates were set for each concentration.The total RNA of the cells was extracted, and the integrity of the sample RNA was analyzed by 1.5% agarose gel electrophoresis.The purity of the RNA was measured by spectrophotometer.After the sample was tested, it was sent to Nuohe Zhiyuan Technology Co.,Ltd.for sequencing and analysis.
2.2.7 Formaldchyde Assisted IsoJation of Regulatory Elements ( FAIRE )
FAIRE sample preparation method referred to the method reported by Simon J M [10,11].Formaldehyde solution was added to the cell culture medium to a final concentration of 1%, cross-linked and fixed at room temperature for 10 min, added with glycine at a final concentration of 0.125 M, and oscillated at room temperature for 5min to quench formaldehyde.PBS was washed three times, 5 mL NP-40 cell lysate was added, lysed on ice for 1.5 h, and the crosslinked nuclei were collected by centrifugation, each tube containing about 107cells.The samples were ultrasonically shortened and cut to an average size of 200~300 bp.Take 10% of the above samples as Input analysis.The remaining samples were centrifuged at 4 ℃.After removing cell debris, DNA was extracted and purified by phenol chloroform to obtain FAIRE samples.The DNA enrichment efficiency in FAIRE samples was analyzed by qPCR.The reaction procedure was : 95 ℃ 30 s, 95 ℃ 5 s, 60 ℃ 25 s, 72 ℃ 20 s, 40 cycles.Quantitative PCR primer sequence is shown in Table 2.
Tab 2 Primer sequences for FAIRE
2.3 Statistical analysis
GraphPad Prism 8.0 software was used for statistical analysis and plotting.The experimental results were expressed as mean ± SD.The data between the two groups were compared by T Test.P < 0.05 was statistically significant.
3.Results
3.1 RAPA inhibits the growth of Jurkat cells
Jurkat cells were treated with 0 nM,5 nM,10 nM,20 nM,40 nM,80 nM,100 nM RAPA for 24 h, 48 h and 72 h, respectively.The optical density value at 490 nm was detected by MTT method, and the cell proliferation inhibition rate was calculated.With the increase of drug treatment time and concentration, the inhibitory effect of RAPA on the proliferation of Jurkat cells also increased.RAPA inhibited the proliferation of Jurkat cells in a concentration-and time-dependent manner (Fig.1).The proliferation inhibition curve was drawn by GraphPad Prism 8.0 software and the half maximal inhibitory concentration (IC50) of RAPA on Jurkat cells at 24 h, 48 h and 72 h was 507.20 nM, 89.51 nM and 12.01 nM, respectively.Compared with the normal control group, the proliferation inhibition rate of Jurkat cells treated with RAPA for 48h increased from (22.96 ±13.48)% to (53.80 ± 9.90)%, with significant statistical difference (P< 0.05).Therefore, in this experiment, Jurkat cells were treated with RAPA at concentrations of 10 nM, 20 nM and 50 nM for 48 h as the basic conditions for subsequent experiments.

Fig 1 Effect of RAPA on the proliferation of Jurkat cells
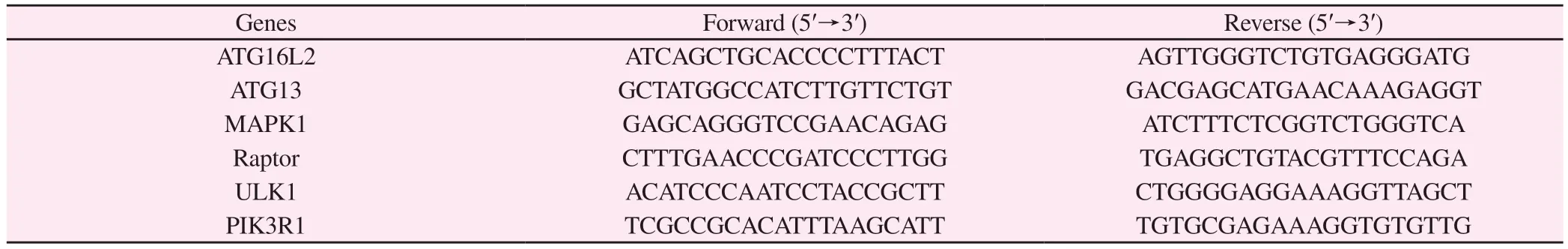
3.2 RAPA could not effectively induce apoptosis of Jurkat cells
After double labeling with Annexin VFITC and PI, the proportion of apoptotic cells in Jurkat cells treated with 0 nM, 10 nM, and 50 nM RAPA for 48 h was detected by flow cytometry.The early apoptosis rate (Q3) of Jurkat cells treated with 10 nM and 50 nM RAPA for 48 h were (0.06 ± 0.01) % and (0.14 ± 0.04) %; The late apoptosis rate (Q2) was (6.13 ± 0.24) % and (6.78 ± 0.03) %; The total apoptosis rates (Q2 + Q3) were (6.18 ± 0.25) % and (6.92 ±0.02) %; Compared with the untreated group (0 nM) (5.65 ± 0.18)%, the total apoptosis rate of each group was not significantly increased (P > 0.05).It indicated that RAPA could not effectively induce apoptosis of Jurkat cells at a concentration of 50 nM.
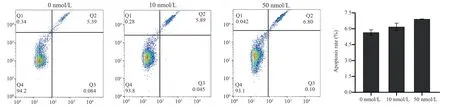
Fig 2 Apoptosis of Jurkat cells treated with RAPA for 48 h.
3.3 RAPA blocks Jurkat cell cycle
To further determine the effect of RAPA on Jurkat cell cycle.In this experiment, PI single staining method was used to detect the cycle changes of Jurkat cells treated with 10 nM and 50 nM RAPA for 48 h.The results showed that compared with the blank control group, the proportion of cells in G1 phase increased from 32.74%to 44.81%, and the proportion of cells in S phase decreased from 48.96% to 35.22%.The proportion of Jurkat cells in G1 phase increased significantly, and the proportion of S phase decreased significantly (P < 0.05).With the increase of drug concentration, the proportion of G1 phase cells gradually increased.It indicated that RAPA could block Jurkat cell cycle in G1 phase in a concentrationdependent manner.

Fig 3 Effect of RAPA on Jurkat cell cycle
3.4 RAPA induces autophagy in Jurkat cells
Acridine orange dye can penetrate into autophagic lysosomes and present red-yellow puncta after fluorescence excitation, which can indirectly reflect the autophagy level of cells.In order to explore the effect of RAPA on the autophagy level of Jurkat cells, the cell morphology was observed by acridine orange staining (Fig.4).After blue light excitation, the control group (0 nM) cells were observed under a fluorescence microscope to be spherical, green fluorescence,and uniform cytoplasm ; after treatment with 10 nM concentration of drugs, chromatin DNA began to condense, and red-yellow fluorescence appeared in the cytoplasm (shown by white arrows),indicating that autophagic lysosomes were produced.Compared with the blank group, the red and yellow fluorescence of the 50 nM treatment group was the most significant.Thus, the number of autophagic lysosomes increased in a dose-dependent manner.

Fig 4 Autophagy was detected by acridine orange fluorescence staining(×400)
3.5 Transcriptome data statistics of autophagy-related genes
After Jurkat cells were treated with 0~50 nM RAPA for 48 h, Total RNA was extracted for transcriptome sequencing.Transcriptome sequencing data were screened for differentially expressed autophagy-related genes according to the criteria of “| log2 Fold Change | > 0, P-value < 0.05 ” (Fig.5).A total of 27 expressed genes were screened, including ATG gene family (8), MAPK gene family (10), PI3K kinase family (6) and ULK kinase family (3).Among them, 17 genes were up-regulated and 10 genes were downregulated.In the ATG gene family, five genes ATG3/7/10/13/16L2 were up-regulated after RAPA induction, and three genes ATG12/14/16L1 were down-regulated.In the MAPK gene family,MAPK1/6/8/9/13were down-regulated and MAPK3/7/11/12/14 were up-regulated.In the PI3K kinase family, PIK3R1/3/4/6 was up-regulated and PIK3R2/5 was down-regulated.The ULK kinase family (ULK1/2/3) was all up-regulated after RAPA induction.In summary, through transcriptome data analysis, RAPA as a natural inhibitor of mTOR, can regulate the expression of autophagy genes,change the transcription level of genes, and then affect the role of autophagy pathway in cells.
3.6 RAPA affects the transcription level of autophagy-related genes
The transcription levels of autophagy genes ULK1, ATG13,ATG16L2, MPKA1, PIK3R1 and Raptor in Jurkat cells were detected by qPCR after treatment with RAPA at final concentrations of 10 nM, 20 nM and 50 nM for 48 h.The results showed that after treatment with different concentrations of RAPA, except that the transcription level of MPKA1 gene showed a significant downward trend, the expression of ULK1, ATG13, ATG16L2,PIK3R1 and Raptor was up-regulated by low concentration (10 nM,20 nM) of RAPA, which was up-regulated by about 2-3 times in a concentration-dependent manner.Among them, the expression of ULK1 and TG16L2 genes was particularly significant, up-regulated by about 4-6 times ; at high concentration (50 nM), the transcription level began to decrease.This shows that low concentration of RAPA can promote the expression of some autophagy-related genes after inhibiting the mTOR signaling pathway, thereby inducing autophagy ; high concentration of RAPA can inhibit the expression of autophagy-related genes, thereby blocking autophagy.
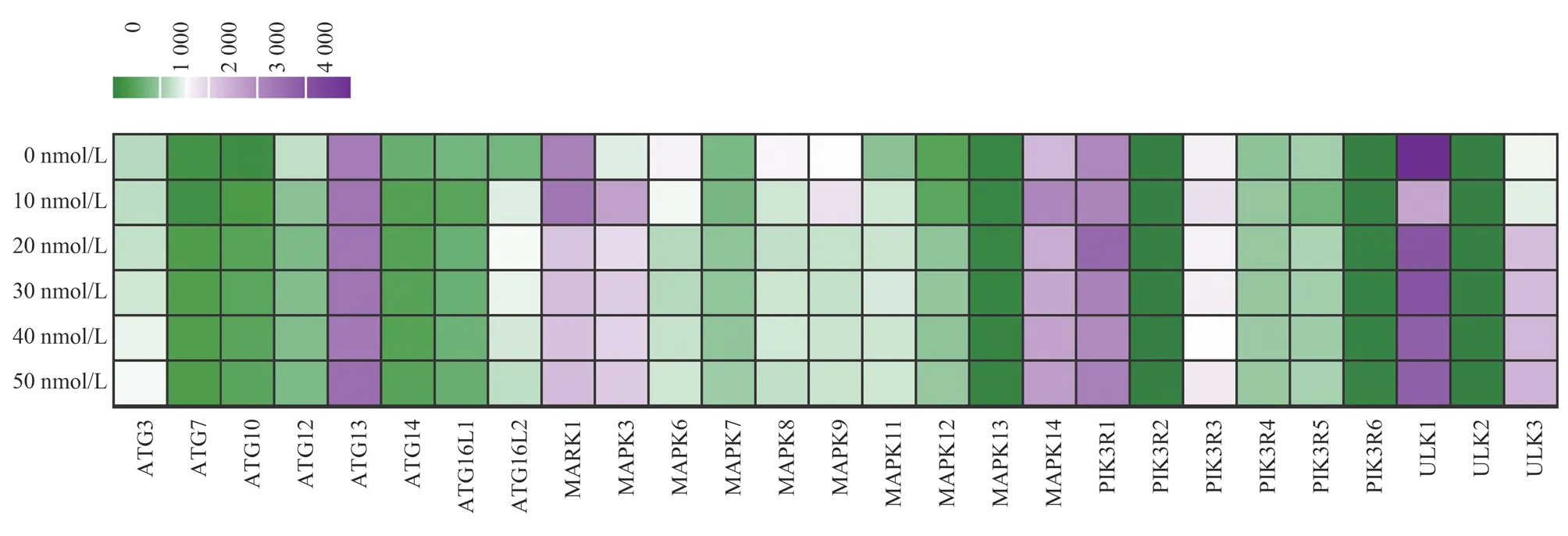
Fig 5 Heatmap of the differential expression of autophagy related genes
Fig 6 Transcript level changes of autophagy genes after RAPA induction
3.7 RAPA affects the protein levels of autophagy-related genes
Jurkat cells were treated with 10 nM, 20 nM and 50 nM RAPA for 48 h, and cell samples were collected.Western Blot was used to further analyze the change trend of autophagy gene protein expression level (Figure 7).Compared with the control group, the protein expression levels of ULK1, ATG13, ATG16L2, PIK3R1 and Raptor genes gradually increased under the induction of 10 nM and 20 nM concentrations, and the expression level began to decrease at 50 nM.The protein expression level of MPKA1 gene decreased with the increase of drug concentration.From the overall trend analysis,the protein expression level was basically consistent with the trend of qPCR results.The results of Western Blot further showed that low concentration of RAPA could induce the expression of ULK1,ATG13, ATG16L2, PIK3R1 and Raptor proteins.High concentration reduced protein expression.
3.8 RAPA induces changes in chromatin accessibility of autophagy-related genes

Fig 7 Effect of RAPA on autophagy protein expression
Gene expression is a highly regulated process that can be regulated at multiple levels.Transcription is the first step in this complex process.Chromatin plays an important role in the process of transcription, and the dynamic change of the interaction between DNA and nucleosome affects the regulation of cell transcription program[12,13].In order to further study the molecular mechanism of autophagy-related gene expression regulation, this paper analyzes the open state of chromatin in the promoter region of autophagy gene by FAIRE-qPCR technology (Figure 8).Compared with the control group, the chromatin accessibility of ULK1, ATG13,ATG16L2, PIK3R1, Raptor genes increased and the chromatin accessibility of MAPK genes decreased after treatment with 10 nM and 20 nM RAPA for 48 h.At 50 nM, due to the high concentration of drug treatment, autophagy occurred in the cells, and the chromatin structure was loose, resulting in false high experimental results.Overall, the change trend of chromatin accessibility in the promoter region of autophagy genes is basically consistent with the change trend of gene expression.The results showed that RAPA could affect the open state of chromatin of autophagy genes, change the epigenetic environment in cells, and then regulate gene expression.

Fig 8 Analysis of chromatin accessibility in the promoter region of autophagy gene
4.Discussions
mTOR regulates cell proliferation, autophagy and apoptosis by participating in various signaling pathways in vivo.Studies have shown that mTOR signaling is overactive in up to 80% of human cancers and plays an important role in maintaining cancer cell growth and tumor metabolism.Cancer is one of the major diseases associated with dysregulation of autophagy.Compared with normal cells, tumor cells are more dependent on autophagy for survival.Through the autophagy cycle, cancer cells overcome metabolic pressure during rapid proliferation.In addition, autophagy plays a ‘double-edged sword ‘ role in tumors.It inhibits tumor formation in the early stage of cancer, and promotes tumor development in the late stage.Due to its dynamic and dual role in the pathogenesis of cancer, autophagy provides a good opportunity to develop new and effective cancer therapies[14-17].
Inhibition of autophagy has become an emerging treatment option,and inhibition of autophagy by pharmacological means can promote cancer cell death.The well-known mTOR inhibitor RAPA has effective autophagy activation.A number of studies have shown that a variety of new mTOR inhibitors have shown high anti-tumor activity in clinical studies and have significant effects in combination with other anti-tumor drugs.However, the role of mTOR signaling pathway has not been clearly studied, and the molecular regulation mechanism of many autophagy-related genes in various cancer cells has not been fully deciphered.The mechanism of tumorigenesis is complex, involving multiple signaling pathways.Inhibition of some signaling pathways may lead to feedback activation of other signaling pathways.Although combination therapy is more effective, its ability to induce tumor cell death is limited.Therefore, it is very important to understand the interaction between tumor microenvironment and autophagy.Through the study of the mechanism of mTOR signaling pathway, the development of selective mTOR inhibitors is conducive to preventing tumor development and combating drug resistance[18-20].
In this study, the natural compound-RAPA was used as the target molecule to study the potential molecular mechanism of inducing autophagy genes.MTT and flow cytometry showed that RAPA had a significant inhibitory effect on Jurkat cells, and the degree of inhibition was positively correlated with drug concentration.It was found that 10 nM and 20 nM RAPA could increase the expression of autophagy-related genes ULK1, Atg13, Atg16L, PIK3R and Raptor, while 50 nM RAPA could down-regulate the expression of autophagy-related genes.We speculate that this is because in the process of autophagy, the hydrolysis of autophagosomes and the efflux of amino acids from lysosomes lead to an increase in intracellular amino acids, which are inhibited by their final products,so that the transcription level of related genes is down-regulated.
In summary, RAPA can inhibit the proliferation of Jurkat cells and induce autophagy.From the perspective of molecular mechanism,RAPA changes the open state of chromatin in the promoter region of autophagy genes, which in turn affects the epigenetic environment of genes.This change in spatial structure regulates gene expression functionally.
Author ‘s contribution
Xu Lan : Participate in some experimental operations, analyze data,write articles ; Huang Liwen and Xiao Yishu : RT-qPCR and Western Blot experiments ; Liu Chunya : culture cells and MTT method to determine cell proliferation inhibition rate experiment; Du Le :technical guidance for experimental work ; Ren Licheng : Provide technical support and guidance, and is responsible for the revision of the article.
All authors have no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Advances in the efficacy and safety of nano-silver dressings for diabetic foot ulcers (DFU) healing
- Research progress of extracorporeal shock wave therapy for plantar fasciitis
- Pathogenesis and drug resistance mechanism of Burkholderia pseudomallei
- Screening and verification of the lncRNA-miRNA-mRNA regulatory network in muscle atrophy after spinal cord injury
- Construction and biological characterization of Burkholderia pseudomallei sRNA gene deletion strain
- Clinical efficacy of dapagliflozin in the treatment of type 2 diabetes mellitus with heart failure with mildly reduced ejection fraction
