Genome-Wide Association Study for Milled Grain Appearance Traits Using Multi-Parent Advanced Generation Intercross Population in Rice
2023-09-05LiXiaoxiangLiuJindongGuoLiangWeiXiucaiWangYameiPanXiaowuDongZhengLiuWenqiangLiuLichengMinJunLiuSanxiongYeGuoyouLiYongchao
Li Xiaoxiang, Liu Jindong, Guo Liang, Wei Xiucai,Wang Yamei, Pan Xiaowu, Dong Zheng, Liu Wenqiang,Liu Licheng, Min Jun, Liu Sanxiong, Ye Guoyou, 4, Li Yongchao
Letter
Genome-Wide Association Study for Milled Grain Appearance Traits Using Multi-Parent Advanced Generation Intercross Population in Rice
Li Xiaoxiang1, #, Liu Jindong2, 3, #, Guo Liang1, #, Wei Xiucai1,Wang Yamei3, Pan Xiaowu1, Dong Zheng1, Liu Wenqiang1,Liu Licheng1, Min Jun1, Liu Sanxiong1, Ye Guoyou3, 4, Li Yongchao1
(Key Laboratory of Indica Rice Genetics and Breeding in the Middle and Lower Reaches of Yangtze River Valley, Hunan Rice Research Institute, Hunan Academy of Agricultural Sciences, Changsha 410125, China; Crop Science Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China; Agricultural Genomics Institute, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China; Rice Breeding Innovations Platform, International Rice Research Institute, Metro Manila DAPO Box 7777, the Philippines; )
The identification of loci and markers associated with milled grain appearance traits is essential for breeding high-yielding and good-quality rice variety. To detect stable loci for these characteristics, grain length (GL), grain width (GW), grain length/width (GLW), chalkiness degree (CD), chalky-grain rate (CR) and translucency degree (TD) of 378 rice lines were evaluated in three seasons. These lines were derived from a multi-parent advanced generation intercross (MAGIC) population,and genotyped using genotyping-by-sequencing, which identified 52 503 single nucleotide polymorphism (SNP) markers for genome-wide association study (GWAS). A total of 24 loci for milled grain appearance traits were detected, explaining 3.6%–27.7% of the phenotypic variations, among which, 10 were identified in the similar locations to known genes or QTLs, while 14 were believed to be novel. In addition, 15 loci exhibited effects across two or more traits, and 18 loci were stable across two or more seasons. Further, we identified four candidate genes possibly involved in signal transduction pathways of plant hormones and starch synthesis for milled rice using qRT-PCR validation. This study provides novel insights into the genetic basis underlying milled rice, and the significantly associated SNP markers and accessions with a larger number of favorable alleles could prove useful in improving grain yield and quality through breeding efforts.
Rice (L.) is a staple food for more than half of the world’s population (Bai et al, 2018). The worldwide objective of rice breeding is to improve both grain yield and quality, with grain appearance being primary quality factors (Liu et al, 2020). Grain appearance makes significant contributions to the market value of rice, including grain size, shape, chalkiness and translucency (Zhao et al, 2018; Zhou et al, 2019). Despite more than 200 QTLs for grain appearance have been detected on all 12 rice chromosomes using various mapping populations, the genetic basis of these traits in rice is not yet fully understood. Several major QTLs controlling grain appearance have been identified and functionally characterized, including,,,,,,and(Shomura et al, 2008; Ying et al, 2018; Zhao et al, 2018). GWAS using high-density markers has become increasingly popular in crop genetics (Liu et al, 2017; Wang et al, 2021; Tatiana et al, 2021). The use of MAGIC population provides an alternative approach to both natural population-based GWAS and linkage mapping. MAGIC population offers better control over population structure and kinship, making it an effective tool in identifying major genes through association mapping (Bandillo et al, 2013; Ponce et al, 2018). In this study, GWAS was conducted in a MAGIC population, tested across three seasons, to identify loci associated with the grain appearance traits. The results of this study could provide valuable information for understanding the genetic basis of rice grain appearance and facilitating marker-assisted breeding.
The grain shape factors (GL, GW and GLW) of a MAGIC population consisting 378 lines appeared to be normally distributed, whereas most of the grain appearance factors (CD, CR and TD) showed biased distribution (Fig. S1). The mean values of CD, CR and TD were 10.8% (1.7%–32.5%), 32.4% (5.3%–86.2%) and 2.5% (1.7%–4.0%), whereas the mean values of GL, GW and GLW were 6.2 mm (5.4–7.5 mm), 2.0 mm (1.6–2.5 mm) and 3.1 (2.3–4.4). Pearson’s product- moment correlation coefficient between the tested traits is presented in Table S1. The CD was positively correlated with CR (= 0.911,< 0.01) and TD (= 0.634,< 0.01). Additionally, CR was positively correlated with TD (= 0.652,< 0.05). Unsurprisingly, GLW was positively correlated with GL (= 0.717,< 0.01), and negatively correlated with GW (= -0.738,< 0.01). The broad sense heritabilities (b2) estimated for CR, CD, TD, GL, GW and GLW were 0.56, 0.52, 0.55, 0.65, 0.57 and 0.58, respectively, indicating that the total phenotypic variances for all the six traits were significantly influenced by genotype, environment and genotype × environment interaction (Table S2).
A wide diversity of phenotype was observed in the MAGIC population for all the measured traits, indicating the emergence of transgressive segregants (Fig. S1). This segregation is of particular interest to breeders because it provides an excellent source for breeding materials to improve crops (Descalsota et al, 2018; Ponce et al, 2018). The substantial variability in grain appearance-related traits among the MAGIC lines will enable breeders to select superior lines with improved grain size. Additionally, the high level of variation observed suggests that the MAGIC population can effectively be utilized to identify allelic variants responsible for grain appearance differences (Bandillo et al, 2013; Descalsota et al, 2018).
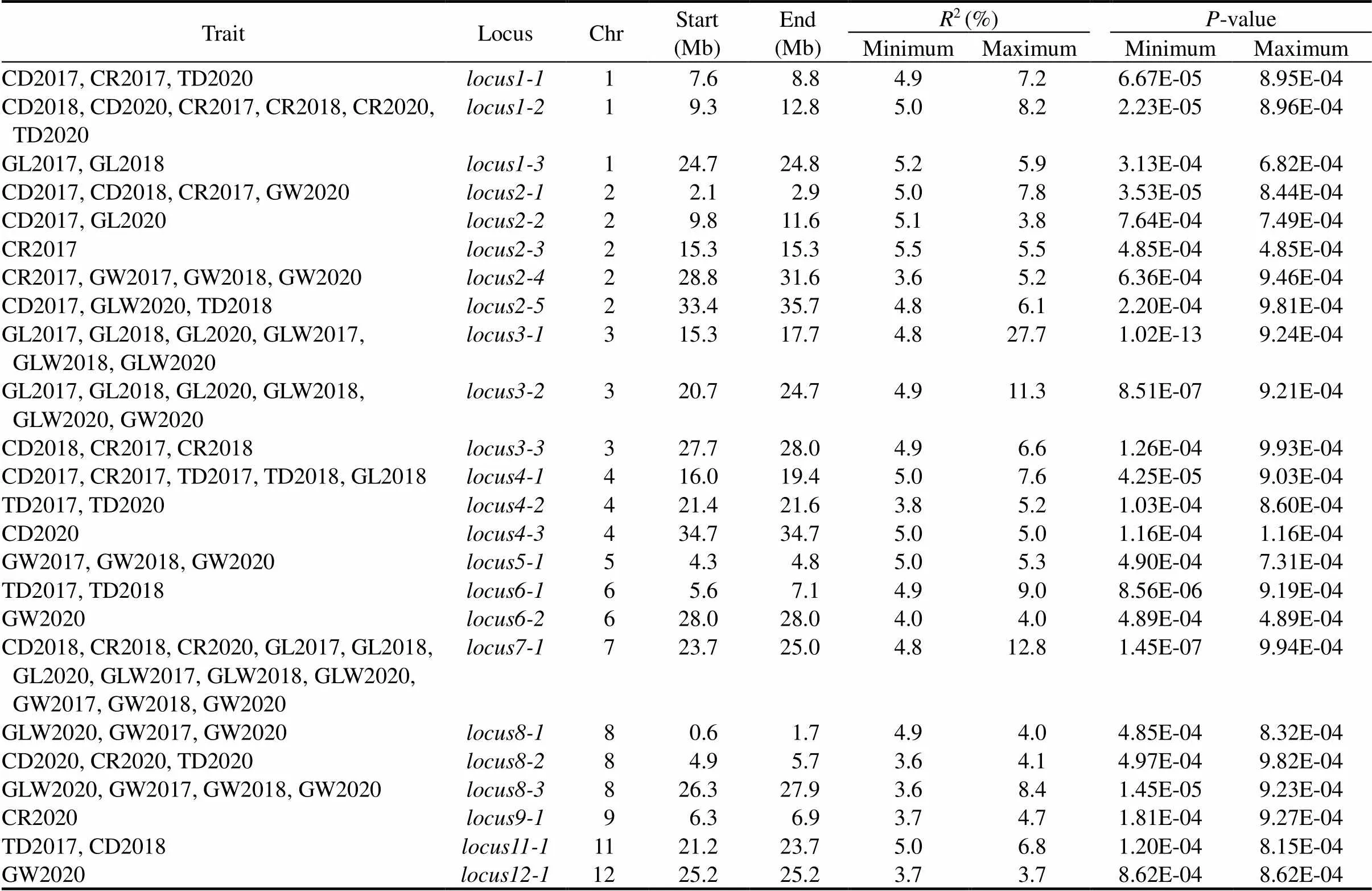
Table 1. Loci for milled grain appearance traits in Bandillo indica multiparent advanced generation intercross population by genome-wide association study.
Chr, Chromosome; CD, Chalkiness degree; CR, Chalky-grain rate; TD, Translucency degree; GL, Grain length; GW, Grain width; GLW, Grain length/width;2, Proportion of phenotypic variance explained by the QTL effect; 2017, 2018 and 2020 represents the cropping seasons of 2017, 2018 and 2020, respectively.
As reported in Ponce et al (2021), 27 042 SNPs were employed for the construction of physical map, with an average of 14 kb per SNP. Principal component analysis (PCA) indicated that no population structure was observed in the association panel, and the top three PCs only explained about 2.7%, 2.3% and 2.2% of the variances, respectively (Fig. S2). The decline of linkage disequilibrium (LD) to 50% of its initial value was at 1.70 Mb for the BandilloMAGIC (BIM) population. The mixed linear model (PCA + K) was used to conduct association analysis. A total of 24 loci were identified associated with GL, GW and CD, explaining 3.6%–27.7% of the phenotypic variation (Table 1, Figs. S3 and S4). Of these, 6 loci for GL were identified on chromosomes 1–4 and 7, 9 loci for GW were identified on chromosomes 2, 3, 5–9 and 12, 6 loci for GLW were distributed on chromosomes 2, 3, 7 and 8, 11 loci for CD were detected on chromosomes 1–4, 7, 8 and 11, 10 loci for CR were identified on chromosomes 1–4 and 7–9, 8 loci for TD were identified on chromosomes 1, 2, 4, 6, 8 and 11 (Table 1). Of these loci,,andwere significantly associated with CD, CR and TD,showed effects on CD, CR and GW,was significantly associated with CD and GL,was associated with CR and GW,had effects on CD, GLW and TD,showed effects on GL and GLW,was associated with GLW, GL and GW,andhad effects on CR and CD, andwas associated with GLW and GW,showed effects on GW and GLW, andhad effects on both TD and CD. The most two important considerations in QTL detection studies are the genetic background effect and QTL-by-environment interaction, which refers to whether the identified QTLs remain consistent across different populations and environments. Out of the 24 QTLs identified,,,,,,,,andwere stably expressed across all the three seasons, making them important in marker-assisted breeding to improve grain appearance.,,,andwere only detected in one of the testing seasons, suggesting that these five loci were more sensitive to the environment, leading to phenotypic plasticity.
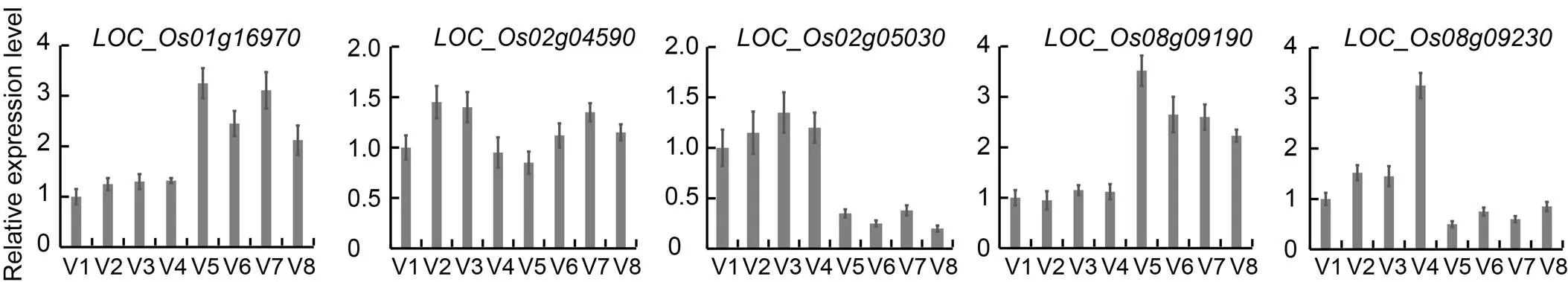
Fig. 1. Quantitative real-time PCR for candidate genes related to grain appearance traits in eight parents.
V1, Fedearroz 50; V2, Shanhuangzhan 2; V3, IR64633-87-2-2-3-3; V4, IR4630-22-2-5-1-3; V5, IR45427-2B-2-2B-1-1; V6, IR84196-12-32; V7, IR77298-14-1-2-10; V8, IR77186-122-2-2-3.
Ten of the QTLs were previously detected, including,,,,,,,,and.(9.8–11.6 Mb) was nearby(8.11–8.12 Mb), which encodes E3 ubiquitin-ligase enzymes and plays a crucial role in grain width and weight (Choi et al, 2018).(28.8–31.6 Mb) was coincided withor(32.1 Mb) (Ruan et al, 2020).(15.3–17.7 Mb) was coincided with(16.72 Mb), which encodes a putative transmembrance protein, playing an essential role in grain weight and length (Fan et al, 2006).(20.7–24.7 Mb) was overlapped with(Ponce et al, 2021).(34.7 Mb) was coincided with(34.2 Mb), which acts as a transcription factor and plays an important role in the shattering trait and grain weight (Wu et al, 2017).(4.3–4.8 Mb) was nearby(5.4 Mb), which has been identified as crucial for grain weight (Shomura et al, 2008; Duan et al, 2017; Liu et al, 2017).(5.6–7.1 Mb) impacting TD was found to nearby/(8.9 Mb). This gene is involved in encoding of gibberellin-regulated GASA/GAST/Snakin family protein precursor essential for the regulation of grain width and weight (Shi et al, 2020; Tang et al, 2021).(28.0 Mb) neared by(26.6 Mb), which encodes the GANT protein and plays an important role in grain weight and yield (Song et al, 2015).(23.7–25.0 Mb), associated with CD, CR, GL, GW and GLW, was found to nearby(19.1 Mb) (Zhou et al, 2015), which belongs to the SBP-box gene family member and plays a critical role in regulating grain length and weight.(26.3–27.9 Mb) overlapped with(26.5 Mb), which also belongs to the SBP-box gene family member and is crucial for the regulation of both grain weight and width (Wang et al, 2012).(21.2–23.7 Mb) was found to nearbyand(19.2 Mb).encodes the indole-3-acetic acid-amido synthetase and plays a vital role in determining grain weight and length (Wang et al, 2021).
,andshowed effects across three or more traits, thus, they were considered promising. The genes located in LD block region around the peak SNP (± 150 Kb) of each important QTL were excavated from the MSU Rice Genome Annotation Project (http://www.rice.uga.edu/). Then, all available SNPs located inside these genes were searched. Except for genes encoding hypothetical protein, transposon protein and retrotransposon protein, with SNP in coding region that leads to sense mutation, were considered as the candidate genes. Totally, five genes involved in the biological metabolism of plant hormone and starch synthesis were screened by this method (Tables S3 and S4), includingfor,andfor, andandfor.The expression levels of the five candidate genes in eight parents were detected using qRT-PCR (Fig. 1).showed no significant differences among the parental accessions, whereas,,andshowed 1.3–3.9-fold higher expression between the parental accessions.encodes a glucosidase II beta subunit-like domain containing protein. Glucosidase plays a crucial role in the metabolic pathway of glucose in organism. β-glucosidase is known to participate in cellulose metabolism, physiological and biochemical pathways (Ayaad et al, 2020).encodessucrose- phosphatase. Sucrose phosphate synthase is a rate-limiting enzyme that catalyzes the synthesis of sucrose 6-phosphate, whereas sucrose phosphatase hydrolyzes the phosphate radical on sucrose 6-phosphate to form sucrose. Sucrose phosphate synthase has high activity in photosynthetic tissues. It only uses uridine diphosphate glucose as glucose donor, hydrolyzes and removes phosphate groups through the action of phosphatase, and forms sucrose, which is the main pathway of sucrose biosynthesis (Barnaby et al, 2020).encodes an auxin efflux carrier component. Auxin, a crucial hormone composed of a simple small molecule based on the indole ring, plays an important role in various aspects of plant growth and development, including cell differentiation, division and elongation (Hou et al, 2021).encodesstarch synthase III, which is the second largest components of total soluble starch synthase activity in developingrice endosperm and plays an important role in starch biosynthesis in plants and the chalkiness and translucency of rice grains, as well as grain length and width (You et al, 2020).
This study contributes to our understanding of the genetic architecture underlying rice grain size. The QTLs and the significant SNPs identified in this study, particularly these exhibiting genetic background independency and environment stability, could be useful for breedingrice to improve grain size. Importantly, it is noteworthy that populations derived from multiparent crosses provide greater phenotypic and allelic diversity compared with conventional biparental populations, along with fewer confounding effects in terms of population structure and genetic relatedness than natural populations.
ACKNOWLEDGEMENTS
This study was supported by the earmarked fund for China Agriculture Research System (Grant No. CARS-01), Young Elite Scientists Sponsorship Program by China Association for Science and Technology (Grant No. 2020QNRC001), and Hunan Academy of Agricultural Sciences Scientific and Technological Innovation Project, China (Grant No. 2017JC10).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science;http://www.ricescience.org.
File S1. Methods.
Fig. S1. Distribution for grain appearance traits in Bandillomultiparent advanced generation intercross population.
Fig. S2. Principal component analysis for Bandillomultiparent advanced generation intercross population.
Fig. S3. Manhattan plot for grain appearance and shape related traits in Bandillomultiparent advanced generation intercross population using mixed linear model.
Fig. S4. Quantile-Quantile plot for grain appearance traits in Bandillomultiparent advanced generation intercross population using mixed linear model.
Table S1. Correlation analysis for milled grain appearance traits in Bandillomultiparent advanced generation intercross population.
Table S2. Analysis of variance for milled grain appearance traits in Bandillomultiparent advanced generation intercross population.
Table S3. Details of candidate genes related to grain appearance traits.
Table S4. Primers of five candidate genes used in qRT-PCR.
Ayaad M, Han Z M, Zheng K, Hu G, Abo-Yousef M, Sobeih S E S, Xing Y Z. 2020. Bin-based genome-wide association studies reveal superior alleles for improvement of appearance quality using a 4-way MAGIC population in rice., 28: 183–194.
Bai S, Yu H, Wang B, Li J. 2018. Retrospective and perspective of rice breeding in China., 45(11): 603–612.
Bandillo N, Raghavan C, Muyco P A, Sevilla M A L, Lobina I T, Dilla-Ermita C J, Tung C W, McCouch S, Thomson M, Mauleon R, Singh R K, Gregorio G, Redoña E, Leung H. 2013. Multi- parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding., 6(1): 11.
Barnaby J Y, Huggins T D, Lee H, McClung A M, Pinson S R M, Oh M, Bauchan G R, Tarpley L, Lee K J, Kim M, Edwards J D. 2020. Vis/NIR hyperspectral imaging distinguishes sub-population, production environment, and physicochemical grain properties in rice., 10(1): 9284.
Choi B S, Kim Y J, Markkandan K, Koo Y J, Song J T, Seo H S. 2018.functions as an E3 ubiquitin ligase for rice expansin-like 1., 19(7): 1904.
Descalsota G I L, Swamy B P M, Zaw H, Inabangan-Asilo M A, Amparado A, Mauleon R, Chadha-Mohanty P, Arocena E C, Raghavan C, Leung H, Hernandez J E, Lalusin A B, Mendioro M S, Diaz M G Q, Reinke R. 2018. Genome-wide association mapping in a rice MAGIC plus population detects QTLs and genes useful for biofortification., 9: 1347.
Duan P G, Xu J S, Zeng D L, Zhang B L, Geng M F, Zhang G Z, Huang K, Huang L J, Xu R, Ge S, Qian Q, Li Y H. 2017. Natural variation in the promoter ofcontributes to grain size diversity in rice., 10(5): 685–694.
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. 2006., a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrance protein., 112: 1164–1171.
Hou M M, Luo F F, Wu D X, Zhang X H, Lou M M, Shen D F, Yan M, Mao C Z, Fan X R, Xu G H, Zhang Y L. 2021. OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium., 229(2): 935–949.
Liu C, Song J L, Wang Y C, Huang X R, Zhang F, Wang W S, Xu J L, Zhang Y, Yu H X, Pang Y H, Bao J S. 2020. Rapid prediction of head rice yield and grain shape for genome-wide association study in indica rice., 96: 103091.
Liu J D, He Z H, Rasheed A, Wen W E, Yan J, Zhang P Z, Wan Y X, Zhang Y, Xie C J, Xia X C. 2017. Genome-wide association mapping of black point reaction in common wheat (L.)., 17(1): 220.
Ponce K S, Ye G Y, Zhao X Q. 2018. QTL identification for cooking and eating quality inrice using multi-parent advanced generation intercross (MAGIC) population., 9: 868.
Ponce K S, Meng L J, Guo L B, Leng Y J, Ye G Y. 2021. Advances in sensing, response and regulation mechanism of salt tolerance in rice., 22(5): 2254.
Ruan B P, Shang L G, Zhang B, Hu J, Wang Y X, Lin H, Zhang A P, Liu C L, Peng Y L, Zhu L, Ren D Y, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. 2020. Natural variation in the promoter ofdetermines grain width and weight in rice., 227(2): 629–640.
Shi C L, Dong N Q, Guo T, Ye W W, Shan J X, Lin H X. 2020. A quantitative trait locuscontrols rice grain size and yield through the gibberellin pathway., 103(3): 1174–1188.
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. 2008. Deletion in a gene associated with grain size increased yields during rice domestication., 40(8): 1023–1028.
Song X J, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J Z H, Kitano H, Sakakibara H, Jacobsen S E, Ashikari M. 2015. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice., 112(1): 76–81.
Tang Z B, Gao X Y, Zhan X Y, Fang N Y, Wang R Q, Zhan C F, Zhang J Q, Cai G, Cheng J P, Bao Y M, Zhang H S, Huang J. 2021. Natural variation inregulates grain length in rice., 19(1): 14–16.
Tatiana R, Julie D, Philippe L, Julien F, Isabelle R R, Noronirina V R, Tuong-Vi C, Kirsten V B, Alain R, Nourollah A, Louis-Marie R. 2021. Genome-wide association study of nitrogen use efficiency and agronomic traits in upland rice., 28(4): 379–390.
Wang D W, Sun W Q, Yuan Z Y, Sun Q, Fan K, Zhang C P, Yu S B. 2021. Identification of a novel QTL and candidate gene associated with grain size using chromosome segment substitution lines in rice., 11(1): 189.
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X. 2012. Control of grain size, shape and quality byin rice., 44(8): 950–954.
Wu W G, Liu X Y, Wang M H, Meyer R S, Luo X J, Ndjiondjop M N, Tan L B, Zhang J W, Wu J Z, Cai H W, Sun C Q, Wang X K, Wing R A, Zhu Z F. 2017. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication., 3: 17064.
Ying J Z, Ma M, Bai C, Huang X H, Liu J L, Fan Y Y, Song X J. 2018., a major QTL that negatively modulates grain length and weight in rice., 11(5): 750–753.
You H, Zhang O L, Xu L, Liang C, Xiang X C. 2020. Effects ofallelic variation on rice grain quality with differentbackgrounds., 100(15): 5344–5351.
Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q. 2018.acts as a transcriptional activator to regulate rice grain shape and appearance quality., 9(1): 1240.
Zhou H, Yun P, He Y Q. 2019. Rice appearance quality.: Bao J S. Rice: Chemistry and Technology 4 edn. AACC International Press: 371–383.
Zhou Y, Miao J, Gu H Y, Peng X R, Leburu M, Yuan F H, Gu H W, Gao Y, Tao Y J, Zhu J Y, Gong Z Y, Yi C D, Gu M H, Yang Z F, Liang G H. 2015. Natural variations inregulate grain shape in rice., 201(4): 1591–1599.
Copyright © 2023, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2023.04.001
Li Yongchao (yongchaoli66363@126.com)
4 November 2022;
28 April 2023
杂志排行
Rice Science的其它文章
- Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury
- Effect of GW8 Gene Editing on Appearance Quality of Erect-Panicle Type (dep1) Japonica Rice
- Transcriptome Analysis of oserf922 Mutants Reveals New Insights into Rice Blast Resistance
- ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice
- LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice
- Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa
