Cloing and Bioinformatics Analysis of ndk Gene from Vibrio alginolyticus
2023-08-02YujiaZHANGShiWANGJianZHONGWeijieZHANGXingXIAOZhiqingWEIHuanyingPANGNaWANG
Yujia ZHANG, Shi WANG, Jian ZHONG, Weijie ZHANG, Xing XIAO, Zhiqing WEI, Huanying PANG*, Na WANG
1. College of Fisheries, Guangdong Ocean University, Zhanjiang 524025, China; 2. Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture / Key Laboratory of Control for Diseases of Aquatic Economic Animals of Guangdong Higher Education Institutes, Zhanjiang 524025, China; 3. Zhanjiang Customs, Zhanjiang 524022, China; 4. Chinese Academy of Inspection and Quarantine, Beijing 100176, China
Abstract [Objectives] The paper was to clone and analyze bioinformatics of ndk gene from Vibrio alginolyticus. [Methods] A pair of specific primers was designed based on the ndk gene sequence of V. alginolyticus HY9901. The full length of ndk gene was amplified by PCR and bioinformatics analysis was performed. MEGA 5.0 software was used to construct NDK phylogenetic tree by neighbor-joining method. SWISS-MODEL program was used to obtain the three-dimensional structural model of single subunit from NDK protein. [Results] The ndk gene, molecular structural formula C702H1094N192O214S7, was 426 bp in total, encoding 141 amino acids, with the theoretical molecular weight of 15.871 99 kD and the theoretical pI value of 5.13. The prediction results of protein subcellular localization, SignalP 5.0, TMHMM Server 2.0 and SoftBerry-Psite showed that NDK mainly existed in the cytoplasm, and the protein was unstable and hydrophobic. There was neither signal peptide cleavage site, nor transmembrane region and KEGG metabolic pathway. The amino acid sequence had two protein kinase C phosphorylation sites, a casein kinase II phosphorylation site, a N-myristoylation site, three microbody C-terminal target signal sites, and a nucleoside diphosphate kinase active site. Homology analysis showed that the NDK of V. alginolyticus had high homology with that of V. diabolicus, with a similarity of 98.58%. Analysis of the structural functional domain revealed that the protein had one NDK structural functional domain. The prediction results of secondary structure showed that the α-helix, random coil, β-sheet and extended strand accounted for 53.19%, 28.37%, 7.09% and 11.35%, respectively. Analysis of NDK protein via STRING database demonstrated that the proteins interacting with NDK protein were NrdA, NrdB, GmK, CmK, TmK, PyrG, PyrH, RelA, FolE and SpoT. [Conclusions] The study plays a positive role in the prevention and control of vibriosis and the improvement of the current aquaculture environment.
Key words Vibrio alginolyticus, Gene cloning, ndk, Bioinformatics analysis
1 Introduction
In recent years, frequent outbreaks of vibriosis have seriously affected the development of aquaculture industry in China[1]. As one of the most common bacterial diseases[2], vibrio is widely distributed in the ocean, estuary and other water environment[3], and has a great impact on marine organisms.Vibrioalginolyticusis a kind of halophilic, thermophilic, facultative anaerobic gram-negative brevibacterium, without capsule or spore, which is one of the main dominant species in the marine environment[4]. When the water temperature is 25-35 ℃,V.alginolyticuseasily causes serious vibriosis of fish, shrimp, shellfish and other aquatic economic animals[5-6]. This bacterium is also a zoonotic pathogen, and human ingestion of vibrio-contaminated food can cause diarrhea, otitis media, enteritis and other diseases[7-8]. The pathogenicity ofV.alginolyticusdepends on the interaction with the host, and its invasion and proliferation of the host cause tissue damage and disturb the metabolism of the host cells[9]. The main virulence factors ofV.alginolyticusinclude extracellular products[10], lipopolysaccharides[11], iron carriers[12], and attachment factors. In addition, the pathogenicity ofV.alginolyticusis also regulated by the type III secretion system[13], which is a hot spot of pathogenic bacteria research in recent years.
The type III secretion system (T3SS) is needle-like, and is a strictly controlled virulence mechanism. WhenV.alginolyticusinfects the host, T3SS can directly inject virulence protein into the host cells, and interfere with the normal metabolism of the host cells, resulting in the death of the host cells[14]. Nucleoside diphosphate kinase (NDK), as an important enzyme of nucleoside triphosphate (NTPs) or its deoxidized derivatives, plays an important role not only in nucleotide metabolism, but also in bacterial growth, regulation of bacterial virulence and regulation of gene transcription[15]. Studies have shown that NDK is involved in the regulation of T3SS. ExsA is an important transcriptional regulatory protein ofPseudomonasaeruginosa, and plays an important leading role in regulating the activation of T3SS transcription; knocking out thendkgene ofP.aeruginosacan up-regulateexsA, thereby regulating the transcription of T3SS genes[16]. Additionally,ndkis involved in adhesion to host cells. After the deletion ofndkgene, the biofilm formation ability ofAeromonasveroniiand its invasion and adhesion ability in EPC cells were significantly reduced[17]. Previous studies indicate thatndkmay regulate the expression of virulence related genes ofP.aeruginosaat the transcriptional level. Transcriptomic sequencing showed that the deletion ofndkgene significantly inhibited the expression oflasA,lasB,aprA,aprDEF,plcBandrhlABCgenes ofP.aeruginosa[16]. In addition to playing an important regulatory role in bacterial virulence,ndkis also critical in regulating bacterial adaptability in the host. Studies have revealed that the NDK ofMycobacteriumtuberculosisdisrupts macrophage phagocytosis by hindering EEA1 recruitment to phagosomes and preventing the formation of mature phagosomes, or by inhibiting RILP recruitment to late phagosomes and preventing the formation of phagolysosomes[18]. Chopraetal.[19]have shown that NDK is an important cytotoxic factor secreted byM.tuberculosis, and can induce ATP-dependent P2Z receptor to mediate macrophage death.
In recent years, there have been many researches on NDK ofP.aeruginosa,M.tuberculosisandA.veroniiat home and abroad, but related researches ofndkinV.alginolyticushave not been reported yet. Therefore, in order to explore the function ofndkgene, thendkgene ofV.alginolyticuswas cloned in this study, and its sequence was analyzed bioinformatically, which would lay a foundation for further research on the regulation mechanism of protein virulence toV.alginolyticus.
2 Materials and methods
2.1 Materials
2.1.1Strains and vectors.V.alginolyticusHY9901 was a virulent strain preserved in Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, andEscherichiacoliDH5α competence was also preserved by the laboratory. The clone vector pMD18-T was purchased from Takara.
2.1.2Main reagents. ExTaqDNA polymerase was derived from Takara. Bacterial genome DNA extraction kit and DNA glue recovery kit were purchased from Tiangen Biotech Co., Ltd., and other reagents were imported or domestic analytical pure. PCR primer synthesis and sequencing were completed by Sangon Biotech (Shanghai) Co., Ltd. The antibiotic ampicillin (Amp+) was used at a concentration of 100 μg/mL.
2.2 Methods
2.2.1Extraction of total DNA fromV.alginolyticusHY9901.V.alginolysticuswas spread on TSA plate. Single colonies were picked and inoculated on TSB medium, and oscillated at 28 ℃ for more than 12 h. Appropriately 1 mL of bacterial solution was loaded into a centrifuge tube and centrifuged at 5 000 rpm for 3 min to collect thalli. Genomic DNA was extracted according to the instructions of the kit, and stored at -20 ℃ for later use.
2.2.2Cloning ofndkgene. According to thendkgene sequence ofV.alginolyticus(accession No.:CP098034) on Genbank, it was aligned with the full genome sequence ofV.alginolyticusHY9901. A pair of primers was designed according to thendkgene sequence ofV.alginolyticusin Genbank. Forward primer P1:5’-ATGGCTCTAGAAAGAACATTTTCA-3’; reverse primer P2:5’-TTAGCGAGAGAAAACTTCTGATTCA-3’. Using the total DNA extracted fromV.alginolyticusas a template, PCR was performed in the following procedures:pre-denaturating at 95 ℃ for 3 min; denaturating at 95 ℃ for 30 s, annealing at 59 ℃ for 30 s, extension at 72 ℃ for 40 s, 33 cycles; extension at 72 ℃ for 5 min. PCR products were detected by electrophoresis on 1% agarose gel and purified by DNA gel cutting kit.
2.2.3Sequencing of PCR products. According to the instructions, PCR products were connected to pMD18-T vector, then transformed intoE.coliDH5α competent cells, and screened on the Amp+ resistant LB plate. Finally, positive clones were sent to Sango Biotech for sequencing.
2.2.4Bioinformatics analysis ofndkgene fromV.alginolyticusHY9901. Sequence homology alignment and similarity analysis were performed using NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi); nucleic acid homology was aligned via DNAMAN Version 6.0 (Lynnon Biosoft); ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and ExPASy Proteomics Server (http://ca.expasy.org) was used to derive amino acid sequence, determine open reading frame (ORF), calculate molecular weight (Mw), and predict theoretical isoelectric point (pI),etc.; signal peptide sequence was predicted by online analysis software SignalP 5.0 Server (http://www.cbs.dtu.dk/services/SignalP); transmembrane domain structure was predicted using TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM); the distribution of functional sites in amino acid sequences was predicted using SoftBerry-Psite(http://linux1.softberry.com/berry.phtml?topic=psite&group=programs&subgroup=proloc); the functional domain of protein structure was analyzed via SMART website (http://smart.embl-heideberg.de/); subcellular localization was predicted using PSORT II Prediction (http://psort.hgc.jp/form2.html); Clastal 2.0 and MEGA 5.0 software was used to construct the phylogenetic tree by neighbor-joining method; SWISS-MODEL program of ExPASy server (http://www.swissmodel.expasy.org/) was used for modeling; KEGG website (https://www.genome.jp/kegg/) was used to search for metabolic pathways; STRING database was used to search protein network interaction (http://string.embl.de/).
3 Results and analysis
3.1 Full-length gene amplificationThendkgene was amplified by PCR assay. The amplified products were analyzed by agarose gel electrophoresis, and specific bands of about 426 bp were amplified (Fig.1). Sequencing of the amplified product and the cloned vector pMD18-T showed that thendkgene contained an open reading frame of 426 bp, encoding 141 amino acids. The amplified product of the gene was submitted to GenBank (accession No.:QQ620230).

Note:M. DL 2000 DNA Marker; 1-2. PCR products of ndk.
3.2 Physicochemical propertiesThe physicochemical properties of NDK protein ofV.alginolyticuswas analyzed by ExPASy software. The results showed that the total number of atoms was 2 209 and the molecular structure formula was C702H1094N192O214S7. The theoretical molecular weight was 15.871 99 kD and the theoreticalpIwas 5.13. The instability coefficient was 48.94 (threshold value 40), so the protein was unstable. The fat coefficient was 775.53, the total mean hydrophilicity was -0.335, and the total hydrophobicity of the protein was low. The protein was free of tryptophan (Trp), pyrrolysine (Pyl), and selenocysteine (Sec). The total number of acid amino acid residues (Asp+Glu) was 23, and that of basic amino acid residues (Arg+Lys) was 16, and Met was at the N terminus. The half-lifeinvivowas longer than 20 h in yeast and longer than 10 h inE.coli, and the half-lifeinvitrowas 30 h in mammalian reticular cells.
3.3 Sequence analysisThe N-terminal signal peptide structure of the amino acid sequence ofndkgene was predicted by SignalP 5.0 Server program, and no signal peptide cleavage site was found in the gene. Prediction made by TMHMM Server 2.0 program showed that the protein had no transmembrane region. Prediction by SoftBerry-Psite demonstrated that the amino acid sequence contained two protein kinase C phosphorylation sites (102-104 aa, 110-112 aa), a casein kinase II phosphorylation site (93-96 aa), a N-myristoylation site (101-106 aa), three microbody C-terminal target signal sites (25-27 aa, 37-39 aa, 84-86 aa), and a nucleoside diphosphate kinase active site (114-122 aa) (Fig.2). The prediction results of protein subcellular localization suggested that the NDK protein was most likely to be located in the cytoplasm (78.3%), followed by the nucleus (13.0%), and the probability of being located in mitochondria and peroxisome was 4.3%.
3.4 Homology and evolutionary analysisHomology analysis was performed using DNAMAN software. The NDK amino acid sequences ofV.anguillarum,V.campbellii,V.cholerae,V.diabolicus,V.fluvialis,V.harveyi,V.mimicus,V.parahaemolyticusandV.vulnificuswere aligned with that ofV.alginolyticus. The results showed that the NDK ofV.alginolyticushad high homology with those of otherVibriostrains, and the similarity withV.diabolicuswas as high as 98.58% (Fig.3). Multiple sequence similarity alignment indicated that thendkgene inVibriostrains was highly conserved.
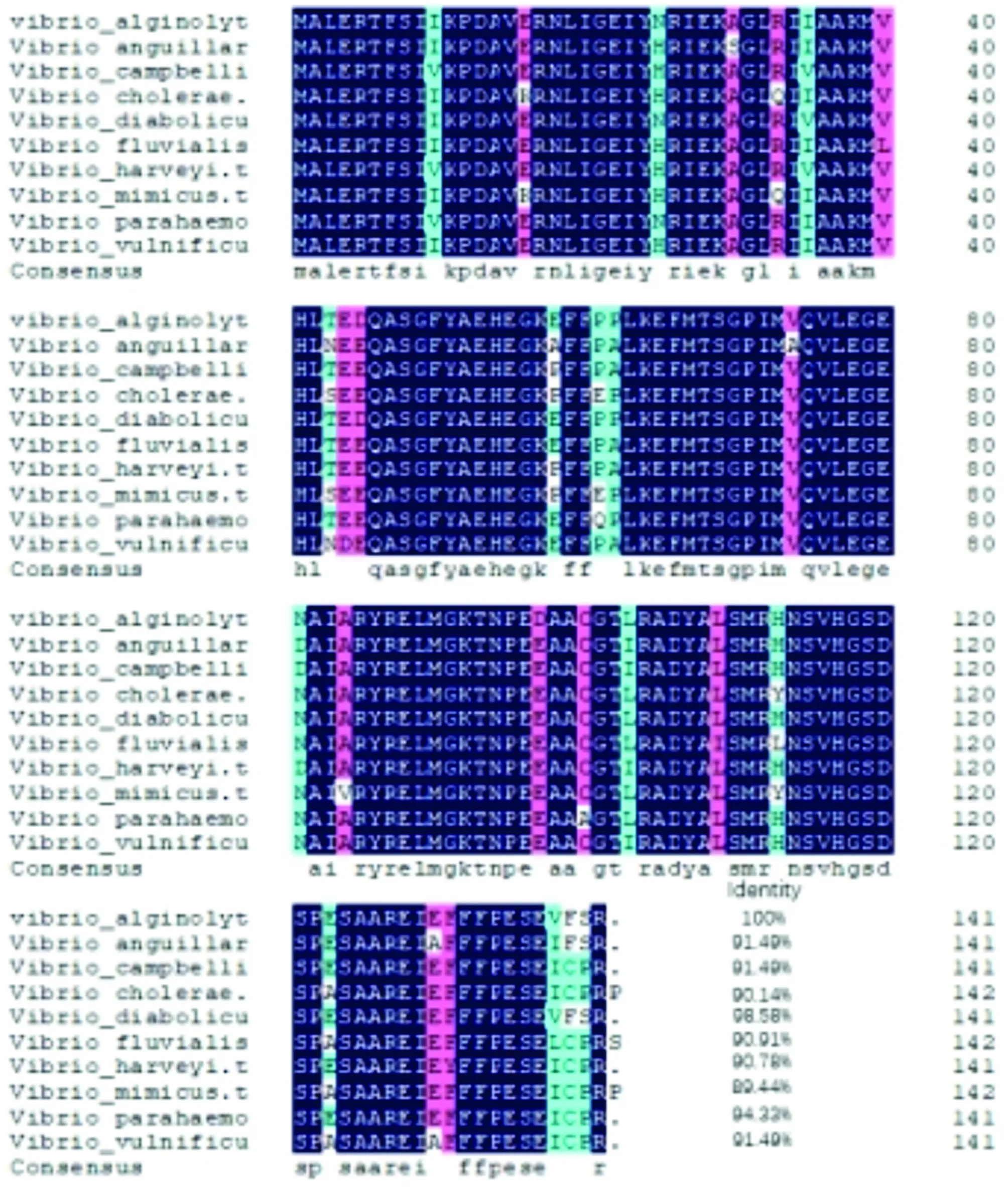
Fig.3 Homology comparison of amino acid sequences of NDK protein from Vibrio alginolyticus and other Vibrio strains
The phylogenetic tree was constructed by combining the NDK amino acid sequence ofV.alginolyticusand otherVibriostrains using the neighbor-joining method of MEGA 5.0. The results showed thatV.alginolyticusandV.diabolicusclustered into the same subfamily, indicating that they were closely related, which was also consistent with the results of morphological and biochemical characteristics classification (Fig.4).
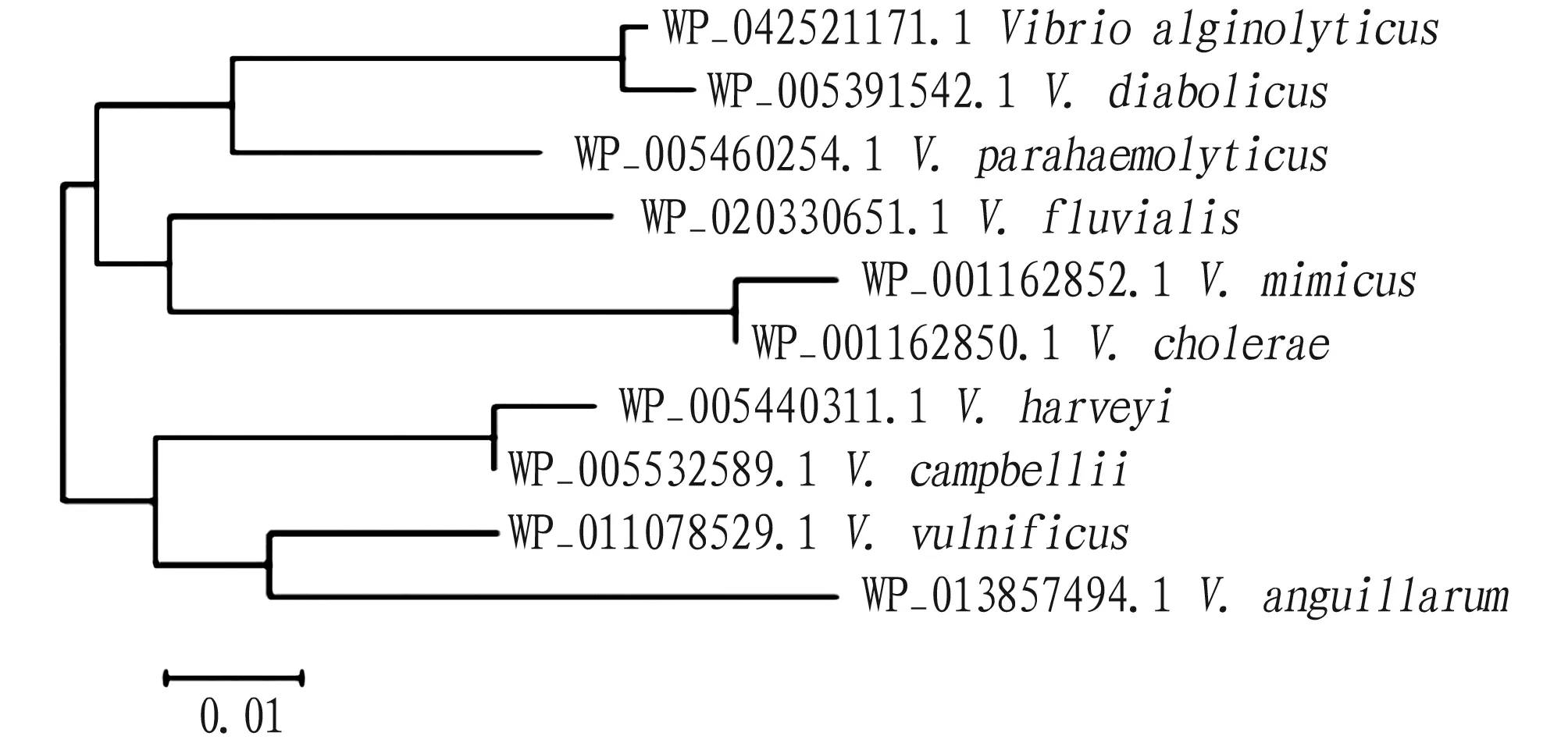
Fig.4 Phylogenetic tree of NDK protein constructed by neighbor-joining method
3.5 Subunit structureThe amino acid sequence ofndkgene was submitted to SWISS-MODEL program, and homologous proteins were automatically searched as templates to obtain the tertiary structure model of single subunit of NDK protein (Fig.5).
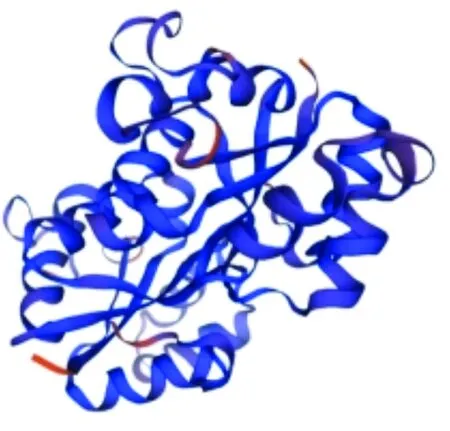
Fig.5 Three-dimensional structural model of NDK protein subunit of Vibrio alginolyticus
3.6 Prediction of functional domain and secondary structure
The functional domains of amino acid structure were analyzed using the SMART website program, and the prediction results showed that NDK protein had a functional domain, namely NDK (Fig.6), which started at 3 aa and terminated at 140 aa, without hidden domains or functions.
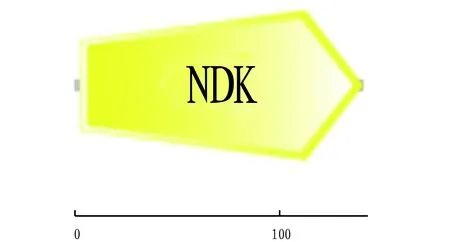
Fig.6 Functional domain of NDK protein
The secondary structure of NDK protein was analyzed by SOPMA software. The results showed that the α-helix, random coil, β-sheet and extended strand accounted for 53.19%, 28.37%, 7.09% and 11.35%, respectively (Fig.7).
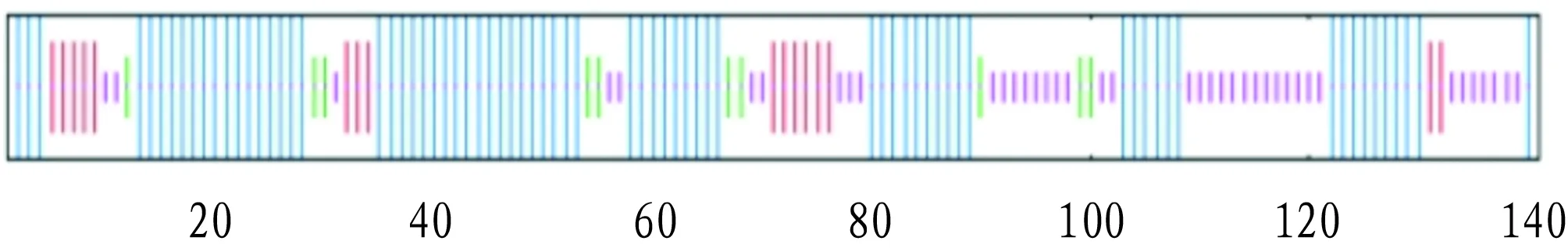
Note:The line in blue represents α-helix; the line in purple represents random coil; the line in red represents extended strand; and the line in green represents β-sheet.
3.7 Network interaction of NDK proteinThe cross-linking analysis of NDK protein was performed using STRING database, and the results displayed that the main proteins interacting with NDK protein were NrdA, NrdB, GmK, CmK, TmK, PyrG, PyrH, RelA, FolE, and SpoT (Fig.8).
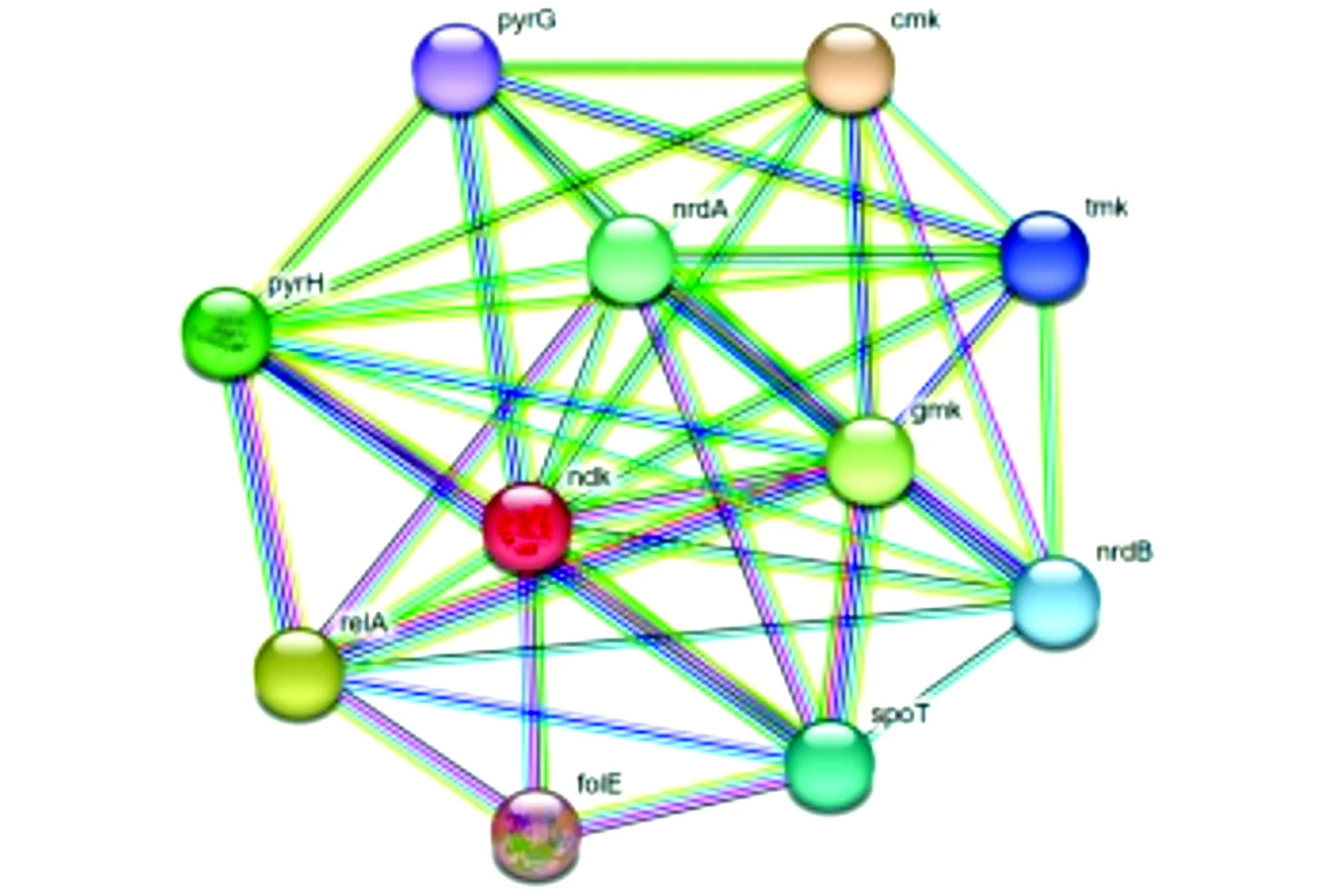
Fig.8 Network interaction of NDK protein
4 Discussion
At present, bioinformatics analysis is an important method to analyze the structure and function of proteins, and can accurately predict the evolutionary relationship, physical and chemical properties and advanced structure of proteins[20-21]. Wang Sen[22]analyzed the NDK ofAspergillusflavusbioinformatically, and the results showed that there was no signal peptide or transmembrane region in NDK protein, which was similar to that in this study. In this study, thendkgene ofV.alginolyticuswas cloned successfully and bioinformatic analysis was performed. Thendkgene was 426 bp in total, encoding 141 amino acids, with the theoretical molecular weight of 15.871 99 kD and the theoretical pI value of 5.13. This protein was unstable and hydrophobic. Through online prediction, it was found that there was no signal peptide, transmembrane region or KEGG metabolic pathway inndkgene. The secondary structure was mainly composed of α-helix and a small amount of random coil, extended strand and β-sheet. Homology analysis indicated that NDK proteins were highly similar invibrio, and the affinity withV.diabolicuswas the closest. The amino acid sequence had two protein kinase C phosphorylation sites, a casein kinase II phosphorylation site, a N-myristoylation site, three microbody C-terminal target signal sites, and a nucleoside diphosphate kinase active site. Phosphorylation of protein is a post-translational modification, and phosphorylation of histidine plays a key role in the regulation of cells in prokaryotes. The NDK that produces NTP transfers phosphoric acid from the active site to the histidine residue of the substrate protein, thereby playing the role of histidine kinase[23].V.alginolyticuslacks endoplasmic reticulum and Golgi apparatus, and can not amidate or glycosylate the translated protein, but this does not affect phosphorylation modification[24].
In recent years, the prevalence of vibriosis has brought huge economic losses to aquaculture industry. The use of antibiotics has caused increasing problems such as bacterial resistance and drug residues[25]. NDK is a multifunctional protein that plays a crucial role in nucleotide metabolism, regulation of bacterial virulence, transcription and expression of virulence genes, regulation of apoptosis, regulation of inflammation,etc.[23]. At present, thendkgene ofV.alginolyticushas not been studied, so the in-depth exploration ofndkgene will play a positive role in the prevention and control of vibriosis and the improvement of the current aquaculture environment.
杂志排行
Asian Agricultural Research的其它文章
- Analysis of Application Status and Recommendations for Shade-Tolerant Lawns and Ground Cover Plants in the West Lake Scenic Area
- Design of Diversified Intelligent Control System for Energy-saving Optimization of Solar Greenhouse in North China
- Determination of Lead in Lime-preserved Egg by Microwave Digestion and Flame Atomic Absorption Spectroscopy
- Analysis of Land Use Change and Driving Factors in Mojiang County Based on PLUS Model
- Intellectual Property Protection and New Development Pattern Construction of Xinjiang Jujube
- Current Situation, Reasons and Suggestions of Income and Consumption of Rural Residents
