Effect and drug sensitivity analysis of PRPF18 on HCC
2023-03-06ZHANGChaoshengXUZiyunFANYuchunNONGMinyuHUANGJiayiJIANGLihe
ZHANG Chao-sheng, XU Zi-yun, FAN Yu-chun, NONG Min-yu, HUANG Jia-yi, JIANG Li-he,,✉
1. Department of Pathophysiology, School of Basic Medicine, Youjiang Medical College for Nationalities, Baise 533000, China
2. Guangxi University Medical College, Nanning 530000, China
3. Youjiang Medical College for Nationalities, Baise 533000, China
Keywords:
ABSTRACT Objective: To analyze effects of PRPF18 on the proliferation, migration, invasion, and apoptosis of hepatocellular carcinoma cell line MHCC97H and drug sensitivity analysis.Methods: The expression, prognosis and drug sensitivity of PRPF18 in hepatocellular carcinoma have been analyzed by using bioinformatics.The expression of PRPF18 in HCC and normal hepatocytes was verified by qRT-PCR.PRPF18 was silenced by transient transfection in MHCC97H cells, and the silencing effect was verified by qRT-PCR.CCK-8 assay and colony formation assay were used to analyze the effect of silencing PRPF18 on the proliferation of MHCC97H cells.The effects of silencing PRPF18 on the migration and invasion of MHCC97H cells were analyzed by transwell assay and wound healing assay.Hoechst 33342 staining and mitochondrial membrane potential assay were used to analyze the effect of silencing PRPF18 on apoptosis of MHCC97H cells.Results: PRPF18 is highly expressed in HCC and is significantly associated with the prognosis of patients.Patients with high expression of PRPF18 have higher sensitivity to tipifarnib and sunitinib.After silenced PRPF18, the proliferation, migration, and invasion of MHCC97H cells were significantly lower than those of the control group.Silencing PRPF18 reduced mitochondrial membrane potential and promoted apoptosis.Conclusion: PRPF18 has a good prognostic value in HCC.Silencing PRPF18 may inhibit the proliferation, migration and invasion of MHCC97H cells and promote apoptosis.
1.Introduction
Globally, hepatocellular carcinoma (HCC) ranks fifth in cancer incidence and third in cancer-related mortality[1,2].Despite significant advances in treatment in recent years, including surgery,radiofrequency ablation, arterial chemoembolization, targeted therapies, and emerging immunotherapies, the survival rate of HCC patients still needs further improvement.Moreover, most patients are diagnosed at an advanced stage, resulting in a poor prognosis for most of them[3-5].Additionally, the availability of useful biomarkers for monitoring and diagnosing early-stage hepatocellular carcinoma remains limited.Therefore, researchers are continually seeking new prognostic markers and drug targets to enhance the prognosis of liver cancer patients.
Pre-mRNA processing factor 18 (PRPF18, also known as PRP18 or HPRP18) is widely expressed in various tissues, including adipose tissue and the brain.In budding yeast, PRP18 plays a role in intron-specific splicing and is associated with G1-S cell cycle progression[6].PRPF18 plays a crucial role in mRNA splicing.It is associated with U5 snRNP and plays a significant role in the catalytic step II of pre-mRNA splicing by stabilizing the interaction between the exon end and U5 snRNA loop 1[7,8].There are reports suggesting that the expression of PRPF18 is associated with disease-free survival in lung adenocarcinoma[9] and can be a potential target in endometrial cancer[10].However, the role of PRPF18 in liver cancer has not been documented in the literature so far.
In this study, we conducted a bioinformatics analysis of PRPF18 expression in liver cancer and analyzed its relevance to prognosis and drug sensitivity.We experimentally verified the impact of PRPF18 knockdown on the proliferation, migration, invasion, and apoptosis capacity of MHCC97H cells, aiming to provide new targets for clinical diagnosis and drug development in liver cancer.
2.Materials and Methods
2.1 Materials
2.1.1 Experimental Cells
The MHCC97H and HepG2 liver cancer cell lines were obtained from the Shanghai Institute of Cell Research, Chinese Academy of Sciences, and the LO2 human normal liver tissue cells were donated by Professor Wu Lichuan from Guangxi University.
2.1.2 Experimental Reagents
- DMEM culture medium (imported from the UK, batch number D6429)
- Fetal bovine serum (FBS) (from Gemini Corporation, USA, batch number 900-108)
- Penicillin-streptomycin antibiotic solution (from Solebao Technology, Beijing, batch number P1400)
- Phosphate-buffered saline (PBS) (from Solebao Technology,Beijing, batch number P1020)
- Lipofectamine 3000 (from Thermo Fisher Scientific, USA, batch number L3000015)
- Sterile, nuclease-free water (from Solebao Technology, Beijing,batch number R1600)
- RevertAid First Strand cDNA Synthesis Kit (from Thermo Fisher Scientific, USA, batch number K16225)
- Universal SYBR Green Master (from Roche, USA, batch number 04913914001)
- Cell proliferation assay kit (CCK-8) (from MCE Corporation,USA, batch number HY-K0301)
- 0.1% crystal violet staining solution (from Solebao Technology,Beijing, batch number G1063)
- 4% paraformaldehyde tissue fixative (from Solebao Technology,Beijing, batch number P1110)
- Hoechst 33342 reagent (from Biyun Tian, Shanghai, batch number C1025)
- Mitochondrial membrane potential assay kit (JC-1) (from Solebao Technology, Beijing, batch number M8650), and others.
2.1.3 Instruments
- Biological safety cabinet (from Thermo Fisher Scientific, USA)
- CO2incubator (from Shanghai Lixin Instruments)
- Pipettes (from Thermo Fisher Scientific, USA)
- Low-temperature refrigerator (from Haier, Qingdao)
- Fume hood (from Shenzhen Guanlan Equipment Factory)
- Microplate reader (from Beijing Tiangen)
- PCR thermal cycler (from Eppendorf, Germany)
- Real-time quantitative PCR instrument (from Roche, USA)
- High-speed centrifuge (from Beijing Dalong)
- Low-speed centrifuge (from Shanghai Anting Instruments)
- Cell counter (from Jiangsu Zhuohui Biological Technology)
- Multifunctional microplate reader (from BERTHOLD Technologies, Germany)
- Laboratory water purification system (from Shanghai Hetai)
- Inverted microscope and inverted fluorescence microscope (from Leica, Germany).
2.2 Methods
2.2.1 Data Collection
RNA sequencing (RNA-seq) data and corresponding clinical data for 374 liver cancer samples and 50 normal liver tissue samples were downloaded from The Cancer Genome Atlas (TCGA) database(https://portal.gdc.cancer.gov/)[11].
The TCGA data is publicly available and has been ethically approved for analysis.This study is based on open-access data and strictly follows the database’s publication guidelines and access policies, without being bound by additional ethical norms.
2.2.2 Differential Analysis of PRPF18 in Liver Cancer
Transcriptome data for 33 cancer types were downloaded from the TCGA database, and matrix transformation and gene expression extraction were performed using Perl software.Differential expression of PRPF18 across cancer types was analyzed using the“ggpubr” package in R.Differential expression of PRPF18 in the TCGA database was also analyzed and visualized using the “limma,”“ggplot2,” and “ggpubr” packages in R.
2.2.3 Analysis of the Relationship Between PRPF18 Expression and Prognosis in Liver Cancer Patients
The impact of PRPF18 on the survival of liver cancer patients in the TCGA database was evaluated using Kaplan-Meier survival curves based on the UALCAN database (https://ualcan.path.uab.edu/).Liver cancer patients were divided into high and low expression groups based on the median expression value of PRPF18.A p-value of less than 0.05 was considered statistically significant.
2.2.4 Cell Culture
MHCC97H, HepG2, and LO2 cells were cultured in high-glucose DMEM medium containing 10% fetal bovine serum (FBS) and 1%penicillin-streptomycin antibiotic solution.Cells were maintained in a 37 ℃ humidified incubator with 5% CO2[12].
2.2.5 PRPF18 Grouping
The experiment included two groups: the control group (nontargeting sequence, no PRPF18 knockdown), the siPRPF18 group (PRPF18 knockdown), and a positive control group (known positive).The experiment was repeated three times.
2.2.6 RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted according to the manufacturer’s instructions (Axygen, Suzhou, China), and cDNA was synthesized using the Thermo RevertAid First Strand cDNA Synthesis Kit.Quantitative real-time PCR (qRT-PCR) was performed using Roche FastStart Universal SYBR Green Master (Rox).The relative gene expression was calculated using the 2-ΔΔCtmethod, with GAPDH as the reference gene.Primers were sourced from Sangon Biotech(Shanghai).The primer sequences were as follows:
- GAPDH: Forward 5’-CAGGAGGCATTGCTGATGAT-3’; Reverse 5’-GAAGGCTGGGGCTCATTT-3’
- PRPF18: Forward 5’-TGACAAAAGGGTTTACTTGGGTG-3’;Reverse 5’-CTCATTGACGAAGGAAATGGCT-3’
2.2.7 Cell Transfection
qRT-PCR was used to validate the expression level of PRPF18 in liver cancer, and MHCC97H cells with the highest expression level were selected for transfection.siPRPF18 and control were transfected into MHCC97H cells at approximately 70% confluency according to the Lipofectamine 3000 reagent instructions.Transfection efficiency was assessed by qRT-PCR 48 hours after transfection.The siRNA sequence was as follows:
- Forward: 5’-GGCUACAAGCCUAUAAAUGTT-3’
- Reverse: 5’-CAUUUAUAGGCUUGUAGCCTT-3’
2.2.8 CCK-8 Cell Proliferation Assay
Transfected cells (4×103cells/well) were evenly seeded in a 96-well plate and incubated for 24, 48, 72, and 96 hours[13].CCK-8 reagent (10 μL/well) was added to the 96-well plate, and it was incubated in a cell culture incubator for 1 hour.The absorbance at 450 nm was measured using a microplate reader[14].
2.2.9 Colony Formation Assay (Continued)
After 48 hours of transfection, the cells were digested with trypsin,resuspended in complete culture medium, and plated at a density of 1.0×103cells per well in a 6-well plate.After 14 d of incubation, the cells were fixed with tissue cell fixative, methanol, and stained with 10% crystal violet.The stained colonies were counted and analyzed using ImageJ[14].
2.2.10 Wound Healing Assay
When MHCC97H cells reached 80% to 90% confluency 48 h after transfection, a straight scratch was made at the bottom of a 6-well plate using a 10 μL pipette tip.The wells were washed with PBS, and photographs were taken under a microscope.The culture medium was then replaced with serum-free DMEM, and photographs were taken again 24 h later[14].
2.2.11 Transwell Assay
Transfected MHCC97H cells were diluted to a concentration of 1×105cells/mL in serum-free DMEM medium.Two hundred microliters of cell suspension were added to the upper chamber of the Transwell migration assay (For Transwell invasion assay, the upper chamber was spread with Matrigel), and the lower chamber was filled with 600 μL of medium containing 10% FBS.After 24 h, cells were stained with crystal violet, and invasion was observed under a microscope.
2.2.12 Hoechst 33342 Staining
Cell apoptosis was detected using Hoechst 33342 staining(Beyotime Biotechnology, Shanghai, China).Transfected cells were seeded in a 6-well plate, and Hoechst 33342 dye was added to the culture medium.After 30 min of incubation in the dark, cells were washed twice with PBS and observed under a fluorescence microscope.
2.2.13 Measurement of Mitochondrial Membrane Potential
The mitochondrial membrane potential was assessed using the JC-1 assay kit (Beyotime Biotechnology, Shanghai, China).Transfected cells were seeded in a 6-well plate and stained with the JC-1 dye.After incubation in the dark, cells were observed under a fluorescence microscope.
2.2.14 Analysis of Drug Sensitivity of PRPF18
The drug sensitivity of PRPF18 in high and low expression groups was evaluated using the pRRophetic R package based on the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/).This analysis aimed to predict available drugs for liver cancer patients.
2.2.15 Statistical Analysis
Statistical analyses and graphing were conducted using R v4.0.4 software, SPSS 25.0 software, and GraphPad Prism 5 software.A p-value less than 0.05 was considered statistically significant, with significance levels denoted as follows:*P < 0.05,**P < 0.01,***P <0.001,****P < 0.0001.
3.Experimental Results
3.1 High Expression of PRPF18 in Liver Cancer
Using Perl and R software to evaluate PRPF18 expression across various cancer types in TCGA[15], the results showed that PRPF18 was highly expressed in tumor tissues such as CHOL,ESCA, STAD, while it was low in COAD, GBM, KICH, KIRC,PCPG, PRAD, READ, among others[16].Notably, in liver cancer,PRPF18 expression was significantly elevated[17] (Figure 1A, B).The expression levels of PRPF18 were further confirmed by qRTPCR, indicating that PRPF18 expression was higher in HepG2 and MHCC97H compared to the normal cell line LO2, and these differences were statistically significant (P=0.026; P=0.036) (Figure 1C).
3.2 Relationship Between PRPF18 Expression and the Prognosis of Liver Cancer Patients
Based on the median PRPF18 expression, liver cancer patients were divided into PRPF18 high-expression and low-expression groups.As shown in Figure 2, Kaplan-Meier survival analysis from the GEPIA 2 website[18] indicated that patients with high PRPF18 expression had a worse prognosis compared to those with low expression (Figure 2).
3.3 Cell Transfection Efficiency Assessment
After 48 h of transfection in MHCC97H cells, the transfection efficiency was assessed using real-time quantitative PCR.The results(Figure 3) showed that the mRNA expression in the siPRPF18 group was significantly reduced compared to the control group (P=0.016).

Fig 1 Expression of PRPF18
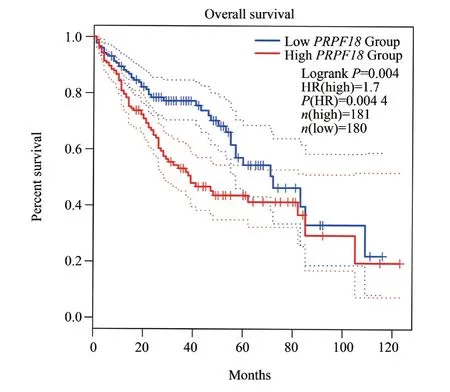
Fig 2 Analysis of the correlation between PRPF18 and the prognosis of patients with HCC
3.4 Silencing PRPF18 Suppresses the Proliferation of Liver Cancer Cells
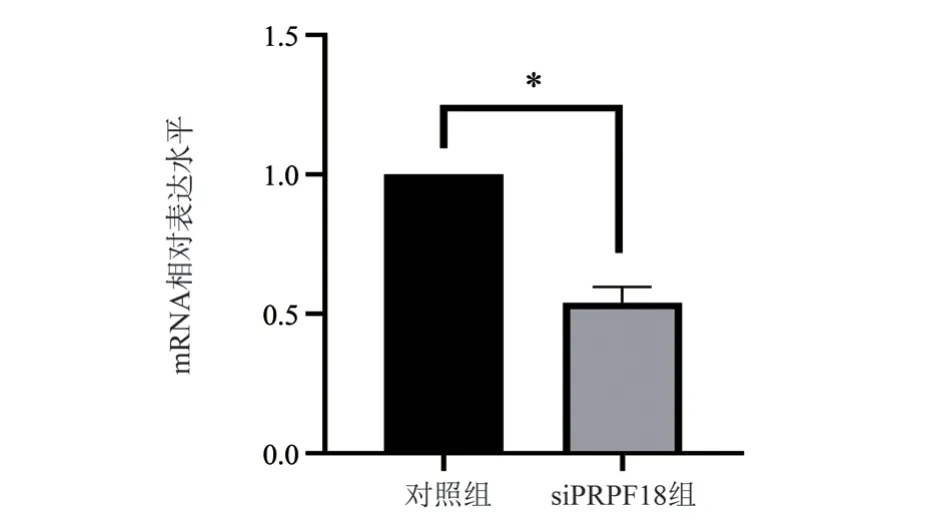
Fig 3 The transfection efficiency of PRPF18 in hepatocellular carcinoma cells was detected
The results of the CCK-8 assay indicated that as the time increased,the cell inhibition rate increased, and there was a statistically significant difference in cell OD values at 72 h and 96 h (P=0.005) (Figure 4A).In comparison to the control group, the siPRPF18 group showed a reduction in colony formation in the assay (P=0.006)(Figure 4B).These experimental results suggest that silencing PRPF18 can effectively inhibit the proliferation capacity of liver cancer cells.
3.5 Silencing PRPF18 Suppresses the Migration Ability of Liver Cancer Cells

Fig 4 Effect of PRPF18 on the proliferation ability of HCC cells
Based on the results of the wound healing assay, it was observed that the scratch closure rate in the siPRPF18 group was significantly lower compared to the control group (P=0.025) (Figure 5A).In the Transwell migration assay[19], the siPRPF18 group of liver cancer cells exhibited a significant reduction in vertical migration ability compared to the control group (P=0.000 2) (Figure 5B).These findings indicate that knocking down PRPF18 can inhibit the migration ability of liver cancer cells.

Fig 5 The Effect of PRPF18 on migration of HCC
3.6 Impact of PRPF18 on the Invasion Ability of Liver Cancer Cells
The number of invading cells in the siPRPF18 group was significantly reduced compared to the control group (P=0.005)(Figure 6).These results suggest that knocking down PRPF18 inhibits the invasion ability of liver cancer cells.
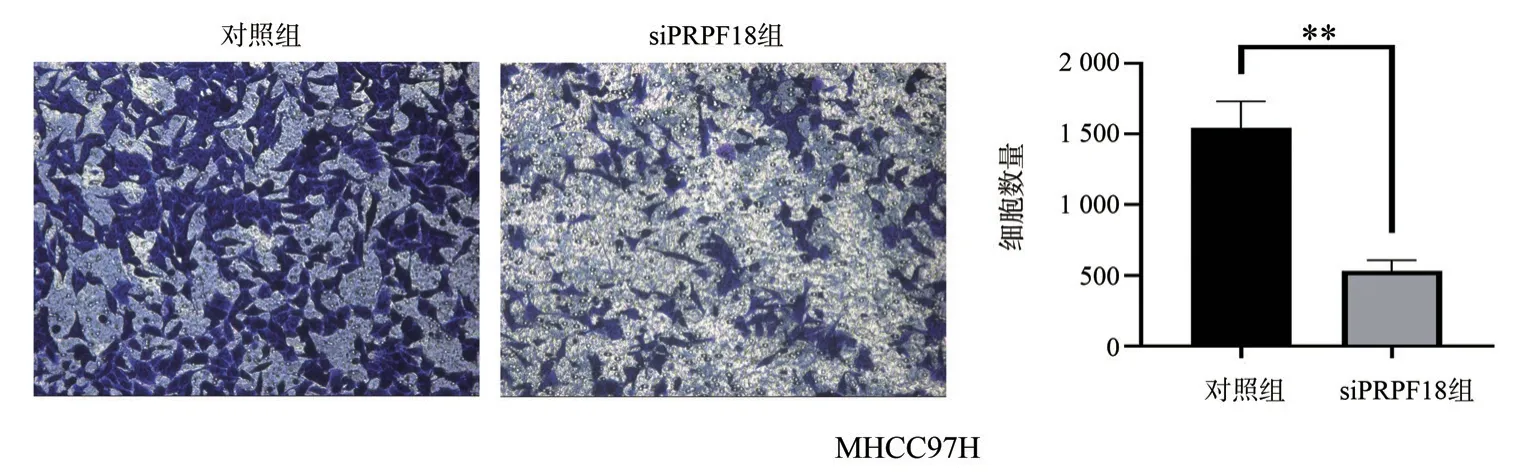
Fig 6 The Effect of PRPF18 on invasion ability of HCC
3.7 Knockdown of PRPF18 Reduces Mitochondrial Membrane Potential and Promotes Cell Apoptosis in Liver Cancer Cells
We further investigated the impact of PRPF18 on apoptosis in liver cancer cells.Hoechst 33342 staining was used to detect cell apoptosis, and the results showed that apoptosis in the siPRPF18 group was significantly higher compared to the control group (Figure 7A).In the measurement of mitochondrial membrane potential,the control group exhibited a significantly higher mitochondrial membrane potential compared to the siPRPF18 group (Figure 7B).These findings indicate that knocking down PRPF18 can reduce mitochondrial membrane potential and promote apoptosis in liver cancer cells.
3.8 Drug Sensitivity Analysis
A screening in the GDSC database identified the two drugs with the most significant differences in sensitivity related to PRPF18 in liver cancer.The results indicated that patients with high PRPF18 expression were more sensitive to tipifarnib and sunitinib (Figure 8).This information may contribute to sensitizing treatments with tipifarnib and sunitinib.

Fig 8 Drug senstivity analysis of PRPF18
4.Discussion
There has been limited research on the impact of PRPF18 on the malignant biological behavior of liver cancer and liver cancer cells.In this study, we first analyzed the differential expression and prognosis of PRPF18 in liver cancer using the TCGA database[20].The results revealed that PRPF18 was significantly overexpressed in liver hepatocellular carcinoma compared to normal tissue.Prognostic analysis showed that patients with high PRPF18 expression had poorer survival outcomes compared to those with low expression.Additionally, qRT-PCR confirmed that PRPF18 was overexpressed in MHCC97H and HepG2 cells compared to normal liver tissue cells(LO2).Therefore, PRPF18 is highly expressed in liver cancer and closely associated with the prognosis of liver cancer patients.
CCK-8 and colony formation experiments indicated that knocking down PRPF18 inhibited the proliferation of MHCC97H cells.Wound healing experiments demonstrated that knocking down PRPF18 suppressed the migration of MHCC97H cells.Transwell experiments showed that knocking down PRPF18 inhibited both migration and invasion of MHCC97H cells.Hoechst staining and mitochondrial membrane potential assays revealed that knocking down PRPF18 promoted apoptosis in MHCC97H cells.
The drug sensitivity analysis showed that patients with high PRPF18 expression were more sensitive to tipifarnib and sunitinib(P<0.001).Tipifarnib is a specific tyrosine kinase inhibitor that has been found to work well in combination with some other anti-tumor drugs.It can inhibit tyrosine kinases in multiple signaling pathways,such as Raf kinase[21,22], and can slow down the growth and spread of liver cancer cells.Sunitinib is an oral small-molecule inhibitor that targets tyrosine kinase activity in various receptors, including VEGFR1 and RET, among others[23].By reducing tumor angiogenesis,sunitinib can induce cancer cell apoptosis[24].Combining sunitinib with radiofrequency ablation significantly inhibits the growth of liver cancer[25].Therefore, analyzing the relationship between PRPF18 and drug sensitivity provides a reference for exploring the treatment of tipifarnib and sunitinib in liver cancer patients and reducing resistance.
In summary, PRPF18 is highly expressed in liver cancer and has prognostic value for liver cancer patients.Knocking down PRPF18 inhibits the proliferation, migration, and invasion of MHCC97H cells while promoting apoptosis.Our findings provide theoretical support for future targeted therapy in liver cancer.However, this study has some limitations as it did not conduct in vivo experiments,and further experiments are needed to confirm these results.
Author Contribution and Conflict of Interest Statement :
Zhang Chaosheng, Xu Ziyun and Fan Yuchun completed the research design and data analysis.Zhang Chaosheng and Xu Ziyun completed the article writing.Nong Minyu, Huang Jiayi and Wu Tong proofread the draft.Jiang Lihe guided the experiment and the paper.All the authors contributed to the further revision of this article.
Finally, all the authors have made contributions to the writing and revision of the article, and have some constructive suggestions and suggestions for improvement to ensure that the article can accurately express the complex research results.Each author has no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Advances in molecular mechanisms of oral submucosal fibrogenic carcinogenesis
- Research progress of ICOSL/ICOS pathway in maternal-fetal immune tolerance
- Research progress of sphingosine 1-phosphate and its signal transduction in central nervous system diseases
- Meta-analysis of the effects of high-intensity intermittent exercise on cardiopulmonary function rehabilitation in patients with stroke
- SPHK1 expression in gastric cancer and clinical value based on bioinformatics analysis
- Predictive value of controlling nutritional status score for progression to chronic critical illness in elderly patients with sepsis
