CRISPR/Cas: Promising Technology for Virus Detection and Antiviral Therapy
2023-02-04,,
,,
(Hunan Provincial Key Laboratory of Medical Virology,Institute of Pathogen Biology and Immunology,College of Biology,Hunan University,Changsha 410082,China)
Abstract: Frequent viral epidemics in recent years urge the development of efficient and convenient techniques for virus detection and antiviral drugs development.As the most popular gene-editing tools,engineered systems based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) possess high specificity and efficiency in nucleic acid targeting and cleavage,which have also been widely used in virological research and medical practice.This review briefly introduces the features of three most commonly used CRISPR/Cas systems,including Cas9,Cas12 and Cas13,and comprehensively concludes their application in virus detection and antiviral therapeutics.For virus detection,Cas9 enhances the sensitivity and accuracy of biosensor detection by combining it with biosensors such as fluorescent sensors,electrochemical sensors,and lateral flow chromatography.As for Cas12 and Cas13,multiple technologies have been developed to detect DNA and RNA viruses based on their trans-cleavage activity,such as SHERLOCK and DETECTR.As CRISPR/Cas-based antiviral techniques,Cas9 has been employed to cleave viral DNA,including genomes of DNA viruses and intermediate DNA of retroviruses,while Cas13 has been used to target viral RNA,including genomes of RNA viruses and viral mRNA.Although CRISPR/Cas systems have shown multiple advantages in sensitivity,efficiency and convenience,they still bear some limitations,such as offtarget effect,immunogenicity and carcinogenicity.In summary,this manuscript provides an overview of current progress in the application of different CRISPR/Cas systems as promising tools in virus detection and antiviral therapy.
Key words: CRISPR/Cas system;virus detection;antiviral therapy;biosensor
Infectious diseases caused by viruses constantly pose great concerns to public health with frequent occurrence of viral epidemics or even global pandemics in recent years,such as the worldwide pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the widespread chronic disease due to persistent infection of hepatitis B virus (HBV).Therefore,it is essential to develop effective and efficient tools to facilitate the treatment and diagnosis of pathogens.
The nucleic acid of viruses is the major target for virus detection and antiviral drugs.Different gene editing techniques are capable of targeting and remolding aimed nucleic acid sequences,which can also serve as tools for viral nucleic acid detection and modulation.Engineered systems based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are innovative genome editing techniques with significant advantages in sensitivity,efficiency and convenience,compared with other gene editing tools,e.g.zinc finger nucleases and transcription activator-like effector nucleases[1].Recently,the application of CRISPR/Cas systems in virus detection and antiviral development has been being rapidly explored.In this review,we briefly introduce the principle and types of CRISPR/Cas systems,and then focus on the application of the three most frequently used Cas proteins,Cas9/12/13,in therapeutics and detection for a variety of human pathogenic viruses,aiming to providing a comprehensive view of the current progress in the field and to promote the treatment of viral infectious diseases.
1 CRISPR/Cas system
CRISPR sequences were first observed by Ishinoet al.[2]as a series of DNA array in the genome ofEscherichia coli,and CRISPR/Cas systems were identified as adaptive immunity components of archaea and bacteria that act against viruses or foreign plasmids[3].The main constituents of functional CRISPR/Cas complexes include CRISPR RNAs (crRNAs) and Cas proteins.The crRNAs are capable of binding to their target DNA or RNA by complementary base pairing,which activates Cas endonucleases and results in the cleavage of the target DNA or RNA[4].Bacteria employ such strategy to eradicate invading genetic elements.
Currently reported CRISPR/Cas systems can be classified into 2 classes,6 types,and 33 subtypes.Class I CRISPR/Cas systems utilize multiple-effector-protein Cas complexes to cleave their targets,further divided into 3 types,Type I,Type II and Type IV,based on the effector proteins[3].Class II systems harbor single-effector Cas proteins with multiple domains to perform their function,based on the Cas proteins,it can be divided into three types,including Type II (Cas9),Type V (Cas12) and Type VI (Cas13) (Tab.1)[5].Among the three types of Class II Cas proteins,Cas9 and Cas12 are RNA-guided DNA endonucleases that recognizes and cleaves DNA strands complementary to their crRNAs,while Cas13 is a group of RNA-guided ribonuclease that recognize and cleave RNA strands complementary to their crRNAs (Fig.1)[6-8].Re-markably,Cas12 and Cas13 exhibit trans-cleavage activity on single-strand DNA and RNA,respectively.These two Cas proteins are capable of cleaving non-target nucleic acid sequences upon activation,which is quite useful for amplifying biosensor signals[6-7].Since Class II CRISPR/Cas systems are relatively simple in their structure and action,they have been widely engineered and applied to gene editing,nucleic acid detection and antiviral therapy.

Fig.1 Nucleic acid targeting and cleavage activity of Cas9,Cas12 and Cas13
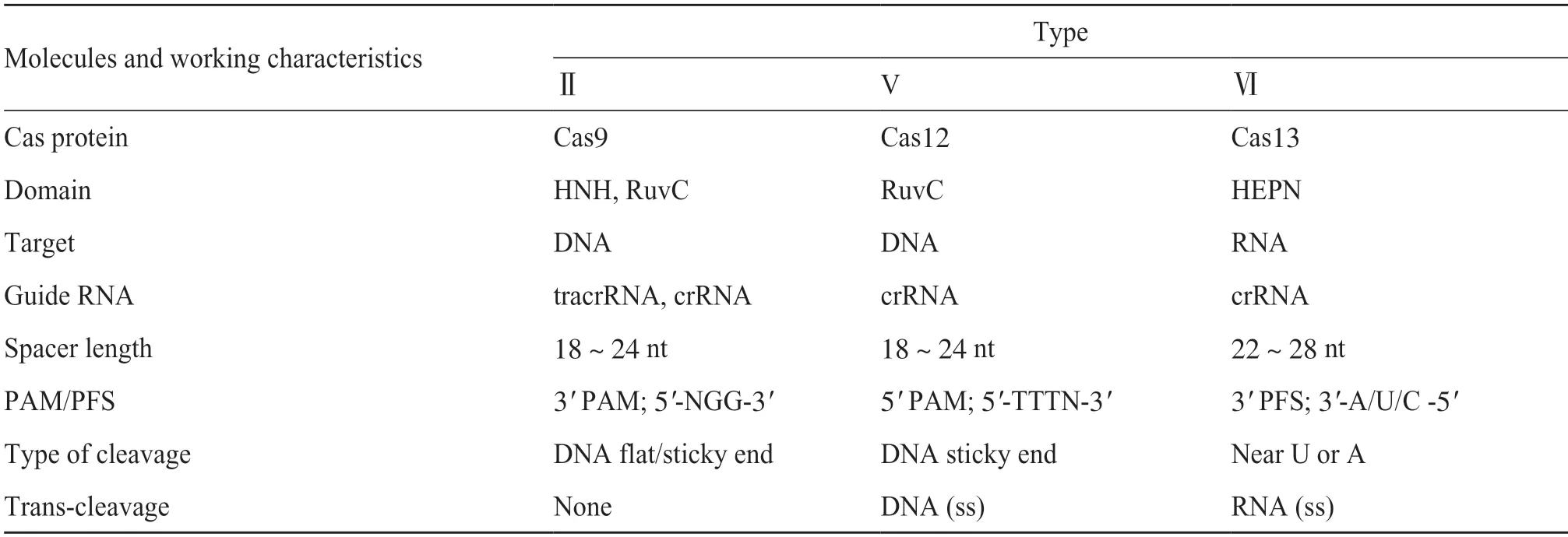
Tab.1 Class II CRISPR/Cas systems
2 CRISPR/Cas based techniques for detection of viruses
All the three types of Class II CRISPR/Cas systems have been applied to detecting viruses.The two major virus-detection approaches utilizing the CRISPR/Cas system include the RNA-guided target recognition system(Cas9) and the target-recognition-triggered accidental cleavage systems (Cas12 and Cas13)[9].There are four primary categories for detecting signals,namely: realtime detection utilizing fluorescent signals,colorimetric detection relying on visual observation,electrochemical detection,and detection based on alternative signals(Tab.2).
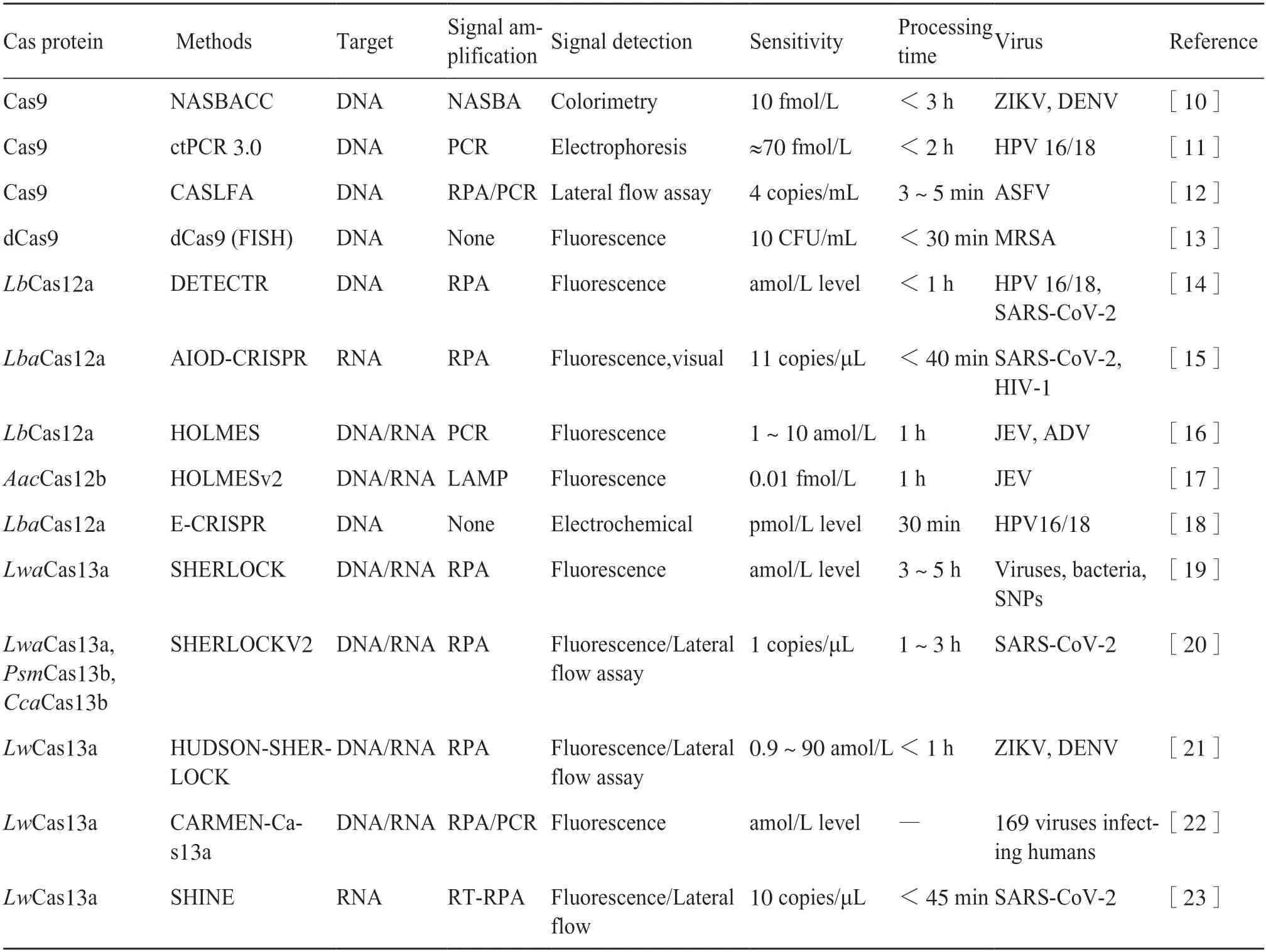
Tab.2 Application of CRISPR/Cas systems in virus detection
Fluorescent probes have been widely used due to their ease of operation and high sensitivity.One of the frequently employed methods for detecting fluorescence signals involves utilizing a reaction system that incorporates singlestranded DNA or RNA reporter genes labeled with fluorescent groups at their ends.In this system,when the Cas effector protein and crRNA complex specifically recognize and cleave the target genes,the reporter gene may undergo non-specific cleavage,resulting in the release of fluorescein and subsequent display of the obtained outcomes.
2.1 Application of CRISPR/Cas9 system in virus detection
Cas9 is a prominent protein within the type II CRISPR/Cas system,which has garnered significant attention and extensive utilization in research.For the CRISPR/Cas9 system to work,single-guide RNA (sgRNA),which is a combination of crRNA and trans-activating RNA(tracrRNA),must be present.This sgRNA serves as a guide for the Cas9 enzyme,enabling it to accurately identify and cleave certain double-stranded DNA sequences that include protospacer adjacent motif (PAM) sites[24].Complementary sgRNAs can be strategically generated based on various gene fragments in order to enable precise recognition and cleavage of distinct target sequences.
Pardeeet al.[10]were the first to introduce the application of the CRISPR/Cas system in conjunction with nucleic acid-dependent amplification technology (NASBA) for the purpose of detecting the Zika virus (ZIKV).They combined Cas9 and NASBA to establish a platform,NASBACC (NASBA-CRISPR Cleavage),for visual interpretation of ZIKV test results with the help of a Toehold biosensor.The platform can identify and genotype the ZIKV within 3 h by taking advantage of NASBA’s efficient amplification of target RNA and CRISPR/Cas9’s ability to specifically recognize targets.
In 2017,Wang and his colleagues devised a multiplex approach for the identification of human papillomavirus (HPV).This method utilizes the Cas9 protein to target the viral genesL1andE6/E7amplified by PCR(ctPCR),enhancing the sensitivity of the diagnostic assay and facilitating to distinguish HPV16 and HPV18 variants[25].The test has undergone significant enhancements to enable the quantitative identification of HPV16/18 through the utilization of qPCR (ctPCR 3.0).This assay allows standard detection of target DNA in 2 h by adding Cas9 and sgRNA directly to the qPCR reaction and performing additional isothermal incubation prior to the qPCR program.It also avoids the operation of opening the test tube throughout the analysis process,thus simplifying the experimental process[11].
Wanget al.[12]combined lateral flow nucleic acid assays with CRISPR/Cas9 tools.The technique,called CRISPR/Cas9-mediated lateral flow nucleic acid assay(CASLFA),integrates the CRISPR/Cas9 system with a lateral flow device to detect target dsDNA.CASLFA is a simple method consisting of a PCR or recombinase polymerase amplification (RPA) step with biotinylated primers and a short (3~5 min) lateral flow assay to detect African swine fever virus (ASFV) with high specificity and sensitivity.
In addition to detection using the targeted cleavage function of Cas9,nuclease-deactivated Cas9 (dCas9),which has lost its ability to shear but retains its ability to bind,is also well suited for pathogen detection.ctPCR detects nucleic acids using a CRISPR/CasII-type system.Zhanget al.[26]created a diagnostic assay based on two dCas9 proteins fused to luciferase split structural domains as a diagnostic test.Binding of the two dCas9 to adjacent target DNA stimulates the reorganization of the luciferase,leading to the emission of a luminescent signal that can be easily detected by a luminometer.In 2017,Guket al.[13]developed a method for the detection of methicillin-resistantStaphylococcusaureus(MRSA) called DNA-FISH by incorporating the CRISPR/dCas9 system into the fluorescencein situhybridization (FISH) technological process.The method can detect MRSA as low as 10 CFUs and can differentiate with high specificity between MRSA and methicillin-sensitiveStaphylococcusaureus(MSSA).
The development of the CRISPR chip represents a significant advancement in CRISPR diagnostics[27].The CRISPR-Chip integrates the CRISPR concept with a graphene-based electrical transistor[28].The CRISPR/Cas9 chip utilizes dCas9 protein specifically connecting to the target DNA without inducing any cleavage.For example,the dCas9 protein is attached to a graphene transistor,and when the DNA extracted from a biological specimen is introduced,dCas9 binds to the target DNA,thereby altering the electroconductivity of the graphene and the electrical properties of the crystalline tube.The CRISPR device exhibits a remarkable level of sensitivity,enabling the detection of target DNA at an impressively low concentration of 1.7 fmol.Additionally,the procedure itself demonstrates exceptional speed,with results obtained in less than 15 min.The utilization of CRISPR-Chip has been employed for the identification of clinical samples obtained from individuals exhibiting Duchenne muscular dystrophy mutations[27].
These studies demonstrate the potential of dCas9 as a powerful nucleic acid detection platformin vitro,whereas its detection sensitivity still needs to be improved[13].
2.2 Application of CRISPR/Cas12 system in virus detection
Cas12 is an RNA-guided endonuclease containing RuvC and NuC structural domains.Among the three Cas12a proteins (LbCas12a,sCas12a,andFnCas12a) widely used for nucleic acid detection,LbCas12a shows the strongest trans-cutting activity.
Chenet al.[14]proposed the DNA recycling endonuclease inhibiting CRISPR trans gene reporter (DETECTR) as a CRISPR/Cas diagnostic platform in 2018.This method relies on the trans-cleavege activity of the Cas12a protein,which will be activated after Cas12a recognizes the targeted DNA (Fig.2).By combining RPA,it not only improves the respective detection and analysis levels but also lower the requirement of equipment.This technology has been successfully applied to the detection of HPV[14]and SARS-CoV-2[29].
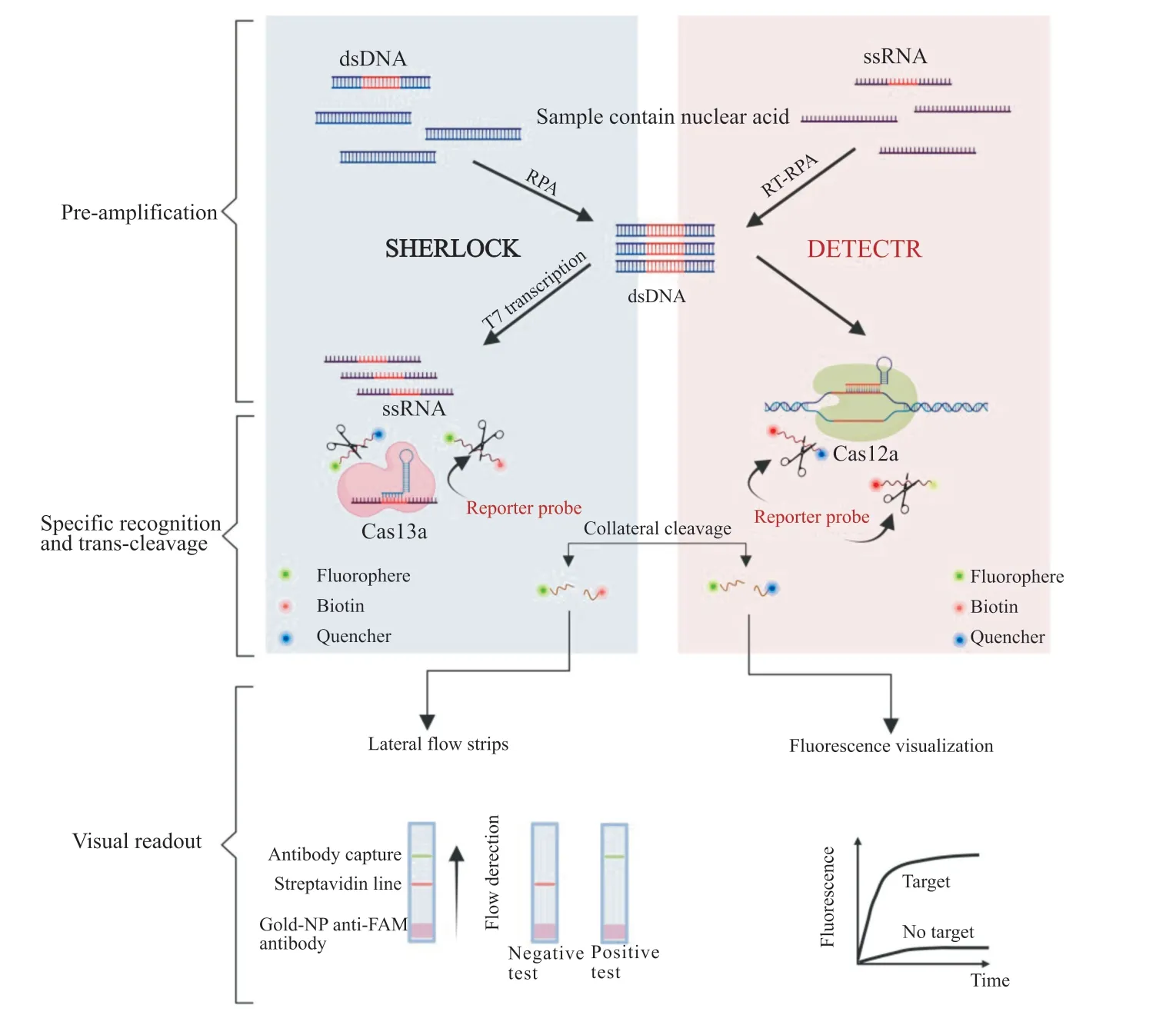
Fig.2 Mechanism and workflow of SHERLOCK and DETECTR assays
HPV16 and HPV18 are the causes of most HPVrelated cancers.DETECTR systems were first created for HPV diagnosis.Hepatitis B virus (HBV) and hepatitis D virus (HDV) co-infection,and hepatitis C virus (HCV)infection are among the leading causes of death from infectious viral diseases[30].DETECTR is able to detect the virus with sensitivity,specificity,and robustness at an amolar concentration and can differentiate between viral subtypes.It is worth mentioning that the DETECTR analysis can be completed within 1 h[14].In addition,the DETECTR system,by combining with RT-LAMP,can detect theNandEgenes of SARS-CoV-2,with a detection time of only 40 min at an amolar concentration[31].
However,the DETECTR assay is performed in two steps and is prone to cross-contamination.To avoid this possibility,Liet al.[16]used a physical method to place the isothermal amplification system and the CRISPR/Cas12a system in the same tube,separated them,and then mixed the two systems after the isothermal amplification reaction,thus realizing a one-step detection that avoided cross-contamination and simplified the operation steps.In recent years,a research team has established a light-activated,one-step RPA-CRISPR/Cas method that has been successfully used for the rapid detection of the ASFV,with a detection time of only 40 min and a detection sensitivity of 2.5 copies[32].Compared with the step-by-step conventional assay,in which isothermal amplification is performed first and then the CRISPR/Cas system is added,this assay greatly simplifies the operation process under the premise of consistent sensitivity.
In the one-step test,Dinget al.[15]developed an allin-one dual CRISPR/Cas12a (AIOD-CRISPR) system in which all components are mixed in the one-pot reaction system and incubated at the same temperature,thus eliminating the need for separate amplification and transfer of amplification products.In the meantime,the system utilizes dual crRNAs to achieve the detection of SARSCoV-2 and HPV1,which makes the operation simpler with higher sensitivity and specificity.All reactions were conducted at a temperature of 37℃ for a duration of 40 min.The limit of detection (LOD) for AIOD-CRISPR is determined to be 1.2 copies for DNA targets and 4.6 copies for RNA targets.
In 2018,LIet al.[16]combinedLbCas12a with PCR amplification to develop a technique called the one-hour low-cost multipurpose highly efficient system (HOLMES).This technique can detect DNA viruses such as Japanese encephalitis virus (JEV) and Aujeszky’s disease virus (ADV) with a sensitivity of 110 amol/L.It can also accurately distinguish viral genotypes as well as single nucleotide polymorphisms (SNPs) in humans.Nevertheless,the utilization of this methodology in conjunction with PCR amplification to enhance stability also results in prolonged detection times and heightened reliance on specialized equipment.
With the discovery of Cas12b,the team made the modified HOLMES V2,which combines the LAMP amplification and the CRISPR/Cas12b system.This updated system can be integrated into a one-step system with a constant temperature,which therefore brings much convenience to nucleic acid detection[17].The lowest concentration of target single-or double-stranded DNA detected by Cas12b is about 1 nmol.The combination of Cas12b and LAMP technology can be combined to specifically detect DNA as low as 0.01 fmol DNA,which is equivalent to the sensitivity of Cas12a.HOMLESv2 can also detect trace amounts of DNA and RNA if isolates are reverse transcribed during sample preparation.
In 2019,Dai and his colleagues established the E-CRISPR system.It has an electrochemical signal detection module that uses Cas12a’s trans-cutting activity and a modified ssDNA reporter gene with a methylene blue electrochemical tag attached to the surface of the sensor[18].When the target sequence is present,the Cas12a protein cuts the ssDNA-MB reporter gene and lowers the amount of the electrochemical signal that is being sent.This technique achieved picomolar sensitivity in the detection of HPV16 nucleic acids within 1 h.
2.3 Application of CRISPR/Cas13 system in virus detection
Cas13 is an RNA-guided and RNA-targeted nuclease with two HEPN structural domains[6].It acts specifically on RNA targets to perform specific recognition and non-specific cleavage functions[33].This property has quickly shown its unique advantages in nucleic acid detection and disease diagnosis and has become one of the research hotspots for developing rapid detection tools.Among a series of Cas13 proteins,Cas13a is the first isoform to be specified and the first effector protein to be applied in nucleic acid detection.
In 2017,Zhang Feng and colleagues introduced specific high-sensitivity enzymatic reporter unlockin (SHERLOCK),a diagnostic platform based on the CRISPR/Cas type VI system[23].This technology is based on the same principle as DETECTR but relies on the activity of theLwaCas13a (Leptotirichia wadei) nuclease,which recognizes and cleaves the target RNA while trans-cutting the ssDNA reporter molecule to generate a fluorescent signal[34](Fig.2).
The SHERLOCK system has been employed for the detection of the SARS-CoV-2 virus[34].The sensitivity and specificity of the fluorescence reading within the detection limit of 42 RNA copies were 100%,and the flow reads were 97% and 100%,respectively.In addition to the diagnosis of SARS-CoV-2,SHERLOCK has been successfully used for the diagnosis of ZIKV,dengue virus(DENV),and even some bacteria such asPlasmodium falciparum[35],which can be accomplished in a short amount time with high sensitivity.Moreover,all components of the SHERLOCK reaction system can be lyophilized and stored,which facilitates transportation without affecting the sensitivity and specificity of the assay.However,the SHERLOCK assay has one major drawback:it is qualitative,not quantitative.The upgraded SHERLOCKV2 uses a combination of four types of Cas proteins (LwaCas13a,PsmCas13b,CcaCas13b,andAsCasl2a) to cleave reporter molecules labeled with different fluorescent motifs and performs the assay in FAM,Cy5,HEX,and TEX channels,which realizes quantitative and simultaneous detection of the four nucleic acid sequences.However,it still cannot realize single-tube multiplex detection[36].In addition,this improved system further amplified the detection signal by using the CRISPR-associated enzyme Csm6,which increased the detection sensitivity[20].This method enables direct detection of nucleic acids in pathogenic samples at very low concentrations (as low as 1 copies/μL) in biological fluids.
In order to simplify the workflow,Zhang Feng’s team also introduced the Heating Unextracted Samples to Eliminate Nucleases (HUDSON) technology,which successfully simplified the nucleic acid extraction step in the pathogen detection process.HUDSON can efficiently extract viral nucleic acids from a wide variety of clinical samples including urine,whole blood,plasma,serum,saliva,etc.without affecting the sensitivity of the lateral flow strips[21].The Hudson-Sherlock pipeline can detect multiple targets in a single clinical sample.Myhrvoldet al.[21]showed that the test had a high sensitivity for detecting DENV using pre-defined biological samples with a reported turnaround time of less than 1 h.HUDSONSHERLOCK successfully differentiated the four serotypes of DENV and ZIKV in biological samples from different regions of the world,with 100% specificity,100%sensitivity,and 100% concordance in most samples.
To improve CRISPR-based assays,in 2020,Ackermanet al.[22]combined CRISPR diagnostics and microfluidics to develop the combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) assay platform.CARMEN is based on microfluidics,which uses a collection of droplet emulsions with PCR-or RPAamplified inputs and CRISPR/Cas assay mixtures (Cas13 protein,specific crRNA,and reporter genes).Each experimental sample or assay mixture is then combined with a solution-based fluorescent color code that is used as an optical identifier.CARMEN-Cas13 can simultaneously detect 169 viruses infecting humans,thus representing a highly multiplexed platform for the rapid and sensitive detection of human pathogens.Ackermanet al.[22]have also shown that CARMEN-Cas13 can find clinically important viral mutations in a multiplexed way,subtype all human influenza A strains,and find dozens of HIV mutations that make the virus resistant to drugs.CARMENCas13 has also successfully detected ZIKA sequences.Many CRISPR tests with different readouts have been developed to provide rapid and accurate detection of pathogenic nucleic acids.
To better enable on-site detection,the Arizti-Sanz team integrated amplification and CRISPR/Casl3 system assays into a one-step diagnostic platform,streamlined highlight of infection to navigate pandemic (SHINE),which improves the sensitivity of nucleic acid detection by adding RNase H.And the effective nucleic acid detection of SARS-CoV-2 was achieved by using in-tube fluorescence and a companion smartphone app[23].The SHINE diagnostic platform minimizes the requirement for equipment and also reduces the result reading bias.
3 CRISPR/Cas based antiviral strategy
Viral genomes are DNA or RNA carrying genetic information of viruses whose replication and transcription are critical for viral life cycle.Meanwhile,viral life cycle is highly dependent on many host cellular proteins.Both viral genome and virus-related host proteins are targets of antiviral drugs,but the development of classical antiviral therapy is usually hindered by several problems such as low specificity and limited effectiveness.Recently,CRISPR/Cas systems targeting viral genomes or host genes critical for viral infection are considered as promising antiviral strategy,due to their high specificity and programmability for different viruses.Moreover,CRISPR/Cas systems can be easily configured to target different viruses by switching the guide RNA.
3.1 Antiviral strategy targeting viral DNA using Cas9
Among different CRISPR/Cas systems,CRISPR/Cas9 is the most mature tool for the development of therapy against a variety of viruses (Tab.3).

Tab.3 Application of CRISPR/Cas9 systems as antivirals
3.1.1 Hepatitis B virus
HBV is one of the major causes of chronic liver disease.According to WHO reports,more than 250 million people worldwide suffer from chronic HBV infection,which kills nearly 900 000 people every year.In hepatocytes,the HBV genome migrates into the nucleus where it is converted into covalently closed circular DNA(cccDNA).cccDNA is the template for the synthesis of viral proteins and pregenomic RNA[37].Recent anti-HBV therapies,such as interferons and nucleotide analogs,can hardly eliminate the replicative template of HBV cccDNA.However,removing cccDNA is still a critical challenge for treatment of persistent HBV infection.
A variety of Cas9-based anti-HBV trials have been reported.Linet al.[38]edited theP1,S1,PS2,andPS3regions of HBV genome with Cas9 and specific gRNAs,leading to significantly reduced expression of HBV protein.In another study,combined use of multiple gRNAs targeting HBV genome enhancer I and sgRNA targetingpreC/Ccould not only efficiently inhibit the expression of HBsAg and HBeAg but also effectively eliminate cccDNA in HepAD38 cells[39].Moreover,Seegeret al.[40]successfully suppressed HBV infection and effectively reduced the expression of HBV proteins by targeting the overlap regions of thePandSregions of the HBV genome.All the above studies show the effectiveness of the CRPISPR/Cas9 system in blocking HBV replication.
3.1.2 Human herpes virus
The gene expression of herpes simplex virus 1(HSV-1),a typical pathogenic human herpes virus,is al-tered at different infectious stages,and no viral protein production occurs during viral latency until viral reactivation.Chemical antiviral medications,such as acyclovir,can inhibit viral replication only in the active state but not in the latent phase of infection[55].In 2014,Biet al.[41]reported that CRPISPR/Cas9 technology can be used to edit HSV-1 genome.Subsequently,Roehmet al.[42]used this technique to target and delete theRL2gene from the HSV-1 genome and successfully inhibit the replication and reactivation of HSV-1.
Epstein-Barr virus (EBV) is another pathogenic herpes virus detected ubiquitously in population.Wanget al.[43]used CRPISPR/Cas9 technology to target different regions of the EBV genome,resulting in arrested cell proliferation and reduced virus titer.Subsequently,Yuenet al.[44]used two sgRNA to delete 558 bases of theBARTpromoter region encoding viral microRNA,effectively suppressing the expression of the microRNA of the virus.This provides a new and effective method for EBV genome editing.In addition,van Diemenet al.[45]used a single sgRNA to targetEBNA1(EBV nuclear antigen 1),which resulted in a decrease of EBV titer in Akata-Bx1 cells with an efficiency of 40%~60%.The efficiency of the combination of the two kinds of EBV sgRNA is as high as 95%,which indicates that targeting the different regions of the EBV genome by CRISPR/Cas9 can effectively reduce the virus titer in the infected cells.
3.1.3 Papillomavirus
HPV is the most common sexually transmitted virus in the USA[56].High-risk HPV types (Type 16 or Type 18) are considered as the major cause of cancer in women,while infection with low-risk types (Type 6 or Type 11) leads to genital warts,laryngeal papillomatosis,and recurrent respiratory papillomatosis[46].The viral oncoproteins produced by theE6andE7genes of HBV are important effectors of tumor formation,inhibiting the tumor suppressorsp53and retinoblastoma proteins,respectively.Therefore,targetingE6andE7with CRISPR/Cas9 is a very promising approach leading to cell cycle arrest and cell death induction in HPV-positive cervical cancer cells.
Kennedyet al.[46]targeted the ORFs ofE6andE7genes in the HPV-18 and HPV-16 genomes of SiHa cells and HeLa cells,but Zhenet al.[47]targeted the promoters ofE6andE7genes.The results showed both methods effectively blocked the expression ofE6andE7genes,thus accelerating the apoptosis of infected cells.In addition,Huet al.[48]targeted only theE7gene of HPV-16 in SiHa and Caski cells and also induced apoptosis in human papillomavirus-positive cells.Therefore,CRPISP/Cas9 can be used as an effective tool to target and remove papillomavirus infected cells.
3.1.4 Human immunodeficiency virus
According to the statistics from the World Health Organization (WHO) (http://www.who.int/hiv/en/),about 37.7 million people were infected with human immunodeficiency virus (HIV) by the end of 2020,with about 1.5 million newly infected individuals.Highly active antiretroviral therapy (HAART) is still the main therapeutic strategy for HIV-1 patients.However,HAART cannot efficiently eliminate latent viral reservoirs,making HIV/AIDS a chronic and incurable disease[57].The CRISPR/Cas9 system has the potential to treat HIV/AIDS by targeting co-factors or the HIV-1 genome to reduce HIV-1 infection and eliminate provirus[58].
In 2013,CRISPR/Cas9 was first applied to HIV/AIDS treatment.Ebinaet al.[49]successfully used CRISPR/Cas9 to inhibit HIV-1 previral expression in Jurkat cell lines by targeting TAR and NF-κB binding cassettes located in theU3region orRregion,respectively.Liaoet al.[50]also demonstrated that targeting multiple sites in the HIV-1 genome could improve the efficiency of excision and destroy the non-integrated previral genome.In addition,combining two effective sgRNAs targeting different regions of the HIV genome prevents the virus from replicating and escaping[51].Excision of HIV-1 previral DNA in animal models was reported in 2017[52].The researchers demonstrated the feasibility and efficiency of destroying HIV-1 provirus in three different animal models using an all-in-one adeno-associated virus (AAV)combined with multiple sgRNAs and SaCas9,laying the foundation for the design of human clinical trials.
In addition to targeting the HIV-1 genome,the CRISPR/Cas9 technology can also be used to resist HIV-1 entry into cells by editing theCCR5orCXCR4co-receptor.Wanget al.[53]used CRISPR/Cas9 to knock out theCCR5gene in CD4 cells and showed anti-HIV-1 resistance.Even a few days after transcription,there were no potential off-target site mutations.Liuet al.[54]designed sgRNA targetingCXCR4andCCR5genes,respectively,and edited the target genome using CRISPR/Cas9 technology.The results showed thatCXCR4andCCR5-edited CD4+T cells resist HIV-1 infection without obvious cytotoxic side effects.
The strategy of reactivating dormant viruses in host cells and inducing cell killing and antiviral immune responses via HAART is called “shock and kill” .Nevertheless,drugs used to reactivate viral gene expression,such as histone deacetylase (HDAC) inhibitors[59],have been observed to have serious side effects in patients.CRISPR/Cas9-mediated viral transcriptional activation provides an alternative method for targeting and activating viral gene expression in latent HIV-1 reservoirs.The virus gene transcript is activated by binding dCas9 proteins to targeted DNA sequences through sgRNAs,and HIV is then cured by a “shock and kill” strategy[60].
3.2 Antiviral strategy targeting viral RNA using Cas13
Compared with Cas9,Cas13 does not modify DNA sequence,hence,the cell may have the opportunity to tolerate and recover from the off-target effect of Cas13 without causing permanent damage to the cell DNA.A majority of pathogenic viruses have ssRNA genomes,making Cas13 a powerful tool for disrupting RNA virus genomes and consequently inhibiting viral replication.Earlier studies illustrated that Cas13a[61]and Cas13b[62]could be used to specifically reduce the expression of host mRNA in mammalian cells.Building upon this work,Freijeet al.[63]developed a system called Cas13-assisted restriction of viral expression (CARVER),which can inhibit RNA viruses by using Cas13 to cleave viral genomes(vRNA) or mRNA.CARVER reduced the RNA levels of several RNA viruses: IAV in MDCK (Madin-Darby Canine Kidney) or A549 (adenocarcinomic human alveolar basal epithelial cell);LCMV (lymphocytic choriomeningitis virus) in HEK 293FT cells;and VSV (Indiana vesiculovirus) in HEK 293FT cells.This work demonstrated the ability of Cas13 to efficiently reduce RNA levels even when RNA levels were quickly increasing.
Liet al.[64]demonstrated that Rev-targeting crRNA inhibits HIV-1 infection most efficiently.Cas13a not only diminishes the level of newly synthesized viral RNA,but it also targets and destroys the viral RNA that enters cells within the viral capsid.However,Cas13a cannot cut and eliminate HIV DNA;in theory,it cannot fundamentally cure HIV.The use of Cas13a,driven by crRNAs targeted atNS3,reduced the level and contagion of DENV2 virus RNA.The above results show that CRISPR/Cas13a can be used as a powerful antiviral technology to inhibit a wide variety of RNA viruses.
A few months after the start of the COVID-19 pandemic,PAC-MAN (prophylactic antiviral CRISPR in human cells) was developed[65].This method harnesses the potent activity of Cas13d (CasRx),an ortholog that is small enough to be packaged into AAVs[66].Abbottet al.[65]compared the genome sequences of 47 patients with COVID-19 and Middle East respiratory syndrome (MERS).The crRNA library for the two highly conserved genes of SARS-CoV-2,RNA-dependent RNA polymerase (RdRp) and nucleocapsid (N),were screened insilicousing bioinformatic methods.By targeting these two conservative genes,the PAC-MAC can inhibit the replication of the virus in human epidermal cell lines,with 85% and 70% inhibition rates,respectively.The PAC-MAN system was able to target more than 90% of the coronavirus genes,including the coronavirus associated with severe acute respiratory syndrome (SARS),MERS and novel coronavirus disease-2019 (COVID-19),with just six crRNAs.The above experiments show that PAC-MAN proved to be a potential anti-SARS-CoV-2 strategy.
4 Challenges in CRISPR/Cas system application
Although CRISPR/Cas systems exhibit promising advantages in virus detection and antiviral therapy,their application,especially in clinical practice,is still limited by several challenges,
4.1 Off-target effects
The nucleic acid detection technology based on the CRISPR/Cas system has high sensitivity,specificity,costeffectiveness,and efficiency and has important clinical value in the rapid detection of infectious diseases,early diagnosis of cancer,and genotyping.However,the existence of off-target effects shows that these newly developed technologies still need to be continuously studied and improved.Off-target effects are the main limiting factor affecting the clinical application of CRISPR technology.In recent years,researchers have conducted in-depth research on the factors affecting the off-target effect and found that the current strategies to solve the off-target effect are mainly “predicting off-target sites” and “optimizing the design strategy of sgRNA and the structure of the Cas enzyme”.Tools such as CRISPR and CHOPCHOP can be used to design gRNAs online and analyze potential off-target sites,allowing researchers to select gRNAs with low off-target effects as much as possible.Tru-RFNs are created by combining the orthogonal strategies of truncating the 5 end of gRNAs with dimerization-dependent RFNs,which exhibit reduced residual undesired monomeric cleavage behavior[67-68].Vakulskaset al.[69]demonstrated that a RNP complex with the R691A SpCas9 mutant introduces highly efficient gene editing in human HSPCs with reduced off-target editing.
4.2 Immunogenicity of the Cas proteins
CRISPR/Cas-based antivirals could be a promising platform for counteracting the risks of antiviral resistance,whereas their effectiveness and safety require further investigation before realizing their clinical application.Kimet al.[70]demonstrated that chemical synthesis and phosphatase treatment of sgRNA to remove its 5'-ppp can inhibit innate immune responses of cell death and reduce cell death.
4.3 Delivery of CRISPR/Cas components
Another major challenge in CRISPR/Cas applications is how to effectively deliver this large complex into virusinfected cells.According to previous reports,the main delivery vectors include adenoviral,lentiviral,and adenoassociated viral vectors[71].Adeno-associated viral vectors have the characteristics of safety,low toxicity,and efficient delivery[72].However,due to their small packaging size,they can only accommodate a small exogenous gene.
In addition to viral delivery systems,numerous nonviral delivery systems have been developed to facilitate CRISPR/Cas delivery,including inorganic nanoparticles,lipid nanoparticles,polymer nanoparticles,electroporation,hydrodynamic injection,and microinjection[73].Among them,the lipid nanoparticles stand out for their non-toxicity,low immunogenicity,and increased Cas enzyme expression timein vitroandin vivo.
4.4 Viral escapes from CRISPR/Cas systems
Van Diemenet al.[45]found that Cas9/gRNA can inhibit HIV-1 replication,but due to NHEJ repair,mutations can occur around the cutting site,which is likely to cause the virus to escape repression by the CRISPR system.To cope with this escape mechanism,Wanget al.[74]proposed alternative solutions such as modifying sgRNA,reprogramming Cas9 nuclease,and inhibiting NHEJ activity.In addition,multiple cleavage sites can be created by using multiple CRISPR sgRNAs to avoid virus escape.
4.5 Carcinogenicity of CRISPR/Cas systems
Recent studies have reported that CRISPR/Cas9 gene editing techniques may induce mutations of thep53gene,triggering apoptosis in human pluripotent stem cells.p53mutations are a key driver of tumorigenesis,Mutant p53proteins not only lose their tumor suppressive activities but often gain additional oncogenic functions that endow cells with growth and survival advantages.Therefore,p53should be closely monitored when using the CRISPR/Cas9 system to treat diseases[75].
5 Conclusion and perspectives
Currently,a variety of techniques based on different CRISPR/Cas systems are being rapidly developed and applied to virus detection and antiviral therapy,showing promising advantages surpassing classical technqiues.
Cas9 is the first Cas protein engineered and applied to virus detection and antiviral therapy.Cas9-based techniques,such as NASBACC,ctPCR and CASLFA,have been applied to the detection of several viruses with a high sensitivity of fmol/L or even amol/L.However,lacking trans-cleavage activity,Cas9-based detection is usually coupled with DNA amplification to generate detectable signals,which requires additional reagents or facilities.Meanwhile,Cas9 cannot be used to detect RNA.Thus,the application of Cas9 in virus detection is relatively limited.Nevertheless,the high endonuclease activity of Cas9 makes it an ideal tool for antiviral therapy,which has been widely used to specifically and efficiently remove viral DNA,especially persistent viral DNA such as cccDNA of HBV and provirus integrated in host genome.
The trans-cleavage activity of Cas12 and Cas13 enable easy signal amplification using fluorophorequencher DNA or RNA probes,which shows great advantage in virus detection.Based on this feature,a variety of Cas12/13-derived techniques for viral detection have been developed.Among these techniques,DETECTR,SHERLOCK/SHERLOCKv2,and CRISPR-Chip are considered as breakthroughs in the field of molecular diagnostics,which allow rapid,sensitive,accurate and inexpensive diagnostic tests under fully isothermal conditions with minimal equipment and hands-on training of personnel.Introducing these CRISPR/Cas derived techniques into clinical diagnosis is a great favor for epidemic control,especially in remote areas.Cas13 has also been applied to antiviral therapy to destruct viral RNA,generating several unique techniques such as CARVER and PAC-MAN.However,Cas13-derived techniques are still relatively immature and more optimization is required for their application in clinical practice.
In summary,though facing many challenges,CRSIPR/Cas systems provide promising approaches for virus detection and antiviral therapy.Fortunately,the barriers for the application of CRSIPR/Cas systems are being overcome with the rapid improvement of related techniques,and clinical application of CRSIPR/Cas systems in virus detection and antiviral therapy can be with great promise.
