Characterization of a blaCTX-M-3,blaKPC-2 and blaTEM-1B co-producing lncN plasmid in Escherichia coli of chicken origin
2023-01-06WANGWenjingWANGYifuJlNYajieSONGWuqiangLlNJiamengZHANGYanTONGXinruTUJianLlRuichaoLlTao
WANG Wen-jing ,WANG Yi-fu ,JlN Ya-jie ,SONG Wu-qiang ,LlN Jia-meng,ZHANG Yan,TONG Xin-ru,TU Jian,Ll Rui-chao,Ll Tao
1 Hunan Provincial Key Laboratory of Tumor Microenvironment Responsive Drug Research,Institute of Pharmacy and Pharmacology,University of South China,Hengyang 421001,P.R.China
2 Shanghai Veterinary Research Institute,Chinese Academy of Agricultural Sciences,Shanghai 200241,P.R.China
3 Guilin Medical University,Guilin 541199,P.R.China
4 Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses,College of Veterinary Medicine,Yangzhou University,Yangzhou 225009,P.R.China
Abstract An extensively drug-resistant (XDR) Escherichia coli strain 258E was isolated from an anal swab sample of a chicken farm of Anhui Province in China.Genomic analyses indicated that the strain 258E harbors an incompatibility group N (IncN) plasmid pEC258-3,which co-produces blaCTX-M-3,blaKPC-2,blaTEM-1B,qnrS1,aac(6´)-Ib-cr,dfrA14,arr-3,and aac(6´)-Ib3.Multiple genome arrangement analyses indicated that pEC258-3 is highly homologous with pCRKP-1-KPC discovered in Klebsiella pneumoniae from a patient.Furthermore,conjugation experiments proved that plasmid pEC258-3 can be transferred horizontally and may pose a significant potential threat in animals,community and hospital settings.
Keywords: blaCTX-M-3,blaKPC-2,blaTEM-1B,IncN,plasmid,Escherichia coli
1.lntroduction
Avian breeding,one of the most important food industries worldwide,often gets involved in the same antibiotics used in human medicine to raise poultry.The undiscriminating use of such essential antibiotics in animal production would probably accelerate the development of antibiotic resistance in pathogens and commensal bacteria.This would result in therapy failures,economic losses and could serve as source of antimicrobial resistance gene pool for transmitting to humans (Tilmanet al.2002).Carbapenems,belonging to β-lactam antibiotics,possess broad spectrum of activity against bacteria and play a crucial role in treatment of bacterial infections,thus often act as “last-line agents” or “antibiotics of last resort”(Papp-Wallaceet al.2011;Karabayet al.2016;Davidet al.2019).Unfortunately,the overall increase in the use of carbapenems has led to the spread of carbapenem resistance in clinical and agricultural settings,which has been a worldwide phenomenon (Karabayet al.2016;Grundmannet al.2017).An essential feature that has facilitated widespread dissemination for resistance to carbapenems are frequently associated with mobile elements that allow bacteria to acquire new genetic materials from outside their clonal lineage (Partridgeet al.2018;Bonardiet al.2019).Gene transfer among bacteria is often mediated by self-transmissible or mobilizable plasmids (Eikmeyeret al.2012).In this scenario,we describe ablaCTX-M-3,blaKPC-2andblaTEM-1Bco-producing incompatibility group N (IncN) multidrug-resistant plasmid pEC258-3,which mediates carbapenem-resistance inEscherichia coli258E isolated from an anal swab sample of a chicken farm in China.
2.Materials and methods
2.1.Bacterial strains
A total of 671 anal swab samples of birds were collected between October 2019 and October 2020.The collected samples were enriched in Mueller-Hinton Broth (MHB) at 37°C for 24 h.The overnight culture was then streaked onto the MacConkey agar plates.The 302 colonies showingE.colimorphology were isolated and identified using the Phoenix™ 100 Automated Microbiology System(BD,Franklin Lakes,NJ,USA).
2.2.Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted by the Kirby Bauer disc diffusion method according to Clinical and Laboratory Standards Institute (CLSI) 2020 guidelines.Escherichia coliATCC 25922 was used as a quality control strain.
2.3.Bioinformatics analysis
Bacterial genomic DNA was extracted using a HiPure Bacterial DNA Kit (Magen,China) according to the manufacturer’s instructions.DNA quality and concentration were calculated using NanoDrop™ (Thermo Scientific,DE,USA).The genome sequencing was then performed using the Pacific Biosciences Platform (PacBio,USA).The PacBio long-read data were performed withde novoassembly by the Hierarchical Genome Assembly Process (HGAP),and complete genome sequences were further polished with Pilon to obtain accurate genome sequences.Genome function elements prediction included the prediction of coding genes,noncoding RNA and clustered regularly interspaced short palindromic repeats (CRISPRs).The Comprehensive Antibiotic Resistance (CARD) database was used to retrieve the pathogenicity genes and antibiotic resistance genes.Function annotation was completed by Blast search against different databases.Acquired antibiotic resistance genes,multilocus sequence typing (MLST),virulence genes,plasmid replicons,pathogenicity were identified using ResFinder 3.0,MLST 2.0,VirulenceFinder 1.5,PathogenFinder 1.1,respectively,available from the Center for Genomic Epidemiology(http://genomicepidemiology.org/).To construct the regional overview phylogeny,phylogenetic analysis incorporatingEscherichiaST602 isolates (https://pubmlst.org/escherichia/) was conducted and visualized by MicroReact.iTOL was used for additional visualizations.
2.4.Conjugation and transformation
Escherichia coliJ53 was used as the recipient in the conjugation assay,and mating was carried out on LB agar medium without selection.After 20 h,the mixed cultures were taken from the plates,suspended in saline,and plated onto MacConkey medium containing sodium azide(100 μg mL–1) and imipenem (32 μg mL–1).
2.5.Nucleotide sequence accession number
The complete nucleotide sequence ofE.coli258E was deposited as GenBank accession no.CP053855.The complete nucleotide sequences of plasmids were deposited as GenBank accession nos.CP097096–CP097098.
3.Results and discussion
MLST showed that the isolate 258E belonged to the epidemicE.coliST602.The results presented by Dayet al.(2019) indicated thatE.coliST602 dominated among food and veterinary isolates in UK.However,E.coliST602 of animal origin has not been reported in China.
Whole genome sequencing (WGS) analysis demonstrated that the chromosome ofE.coli258E was 4 715 664 bp in length and harboring ablaCTX-M-3,blaKPC-2andblaTEM-1Bco-producing IncN multidrug-resistant plasmid (pEC258-3) and two additional plasmids(pEC258-1 and pEC258-2) without antimicrobial resistance genes (ARGs).There was an antibiotic-related integrase Intl1 between chromosome 2 702 175–2 702 825 bp site ofE.coli258E.It is known that integrons are genetic elements that allow efficient capture and expression of exogenous genes and are widely known for their role in the dissemination of antibiotic resistance genes,particularly among Gram-negative bacterial pathogens(Golebiewskiet al.2007).The full length of plasmid pEC258-3 is 65 305 bp with GC content of 50.8%,and harbors 98 predicted open reading frames (ORFs).The resistance genes presented in the plasmid of pEC258-3 areqnrS1,dfrA14,arr-3,acc (6´)-Ib,blaCTX-M-3,blaKPC-2,andblaTEM-1B.
Except to fosfomycin and tetracycline drugs,E.coli258E displayed an extremely-drug resistant (XDR) profile including all tested β-lactams and aminoglycosides,quinolones,rifamycin,trimethoprim,macrolides and cephalosporins.Meanwhile,the transconjugant 258E-J53 showed the significantly different drug resistance profile compared with the parental strain (Table 1).
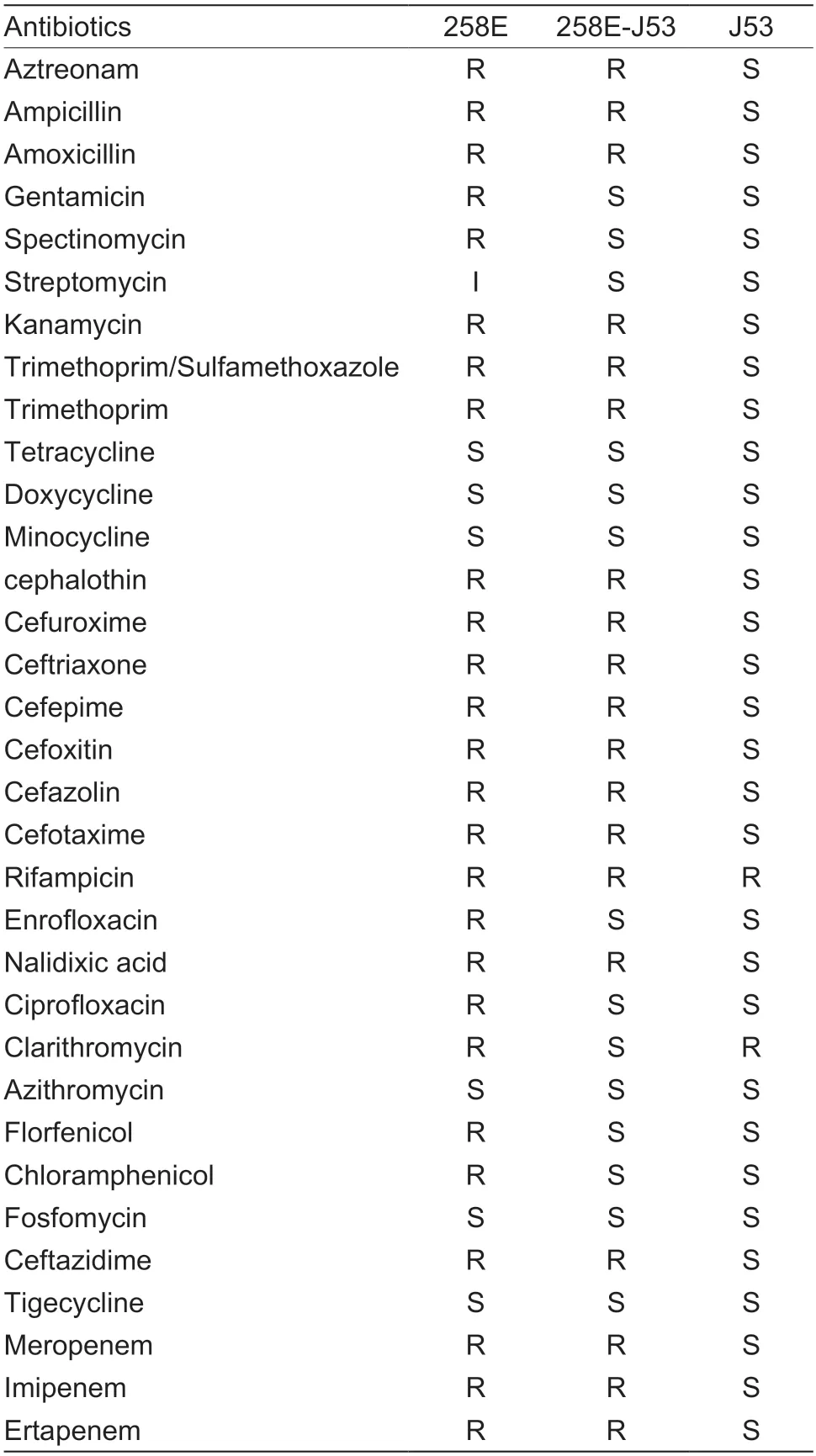
Table 1 Antibiotics susceptibility of 258E,258E-J53 and J53,respectively1)
When it comes to the molecular characterization and comparative analysis of plasmid pEC258-3,it is assigned to plasmid sequence type 7 (ST7) and belongs toincompatibility group N (IncN) plasmid.It was shown that plasmids belonging to the IncN incompatibility group often have been detected in human clinical samples,stables or slaughterhouses in animal associated environments,and is one of the most frequently encountered resistance plasmid types.Moreover,they are well described as a human health concern (Carattoli 2009;Eikmeyeret al.2012;Citterioet al.2020).Further analysis of the complete sequence indicated that pEC258-3 has a high similarity to theE.coliplasmid pECN580 (99.98% identity,100% coverage) andK.pneumoniaeplasmid pCRKP-1-KPC (99.96% identity,98% coverage) (Chenet al.2014;Zhanget al.2017).There are only few differences between these closely related plasmids.The core region of pEC258-3 and pECN580 both contain a replication module (repAandrepAbinding iterons),a stability operon(stbAB,ardBR,andumuCD),two transfer (tra) systems,and an anti-restriction system (EcoRII andEcoRII met).However,pECN580 is lack of a 371-bp fragment located between 930 and 1 300 site of pEC258-3 which is the DNA segment coding for a protein of 3-oxoacyl-[ACP] reductase(EC 1.1.1.100) FadG2.Whilst,there was a deletion in pCRKP-1-KPC in 31–32 kb site which codes an integrase TinR protein.Meanwhile,it is interesting that some homologous genes are placed in inverse sequences at particular separated sites (8 000 and 30 000 bp) of pCRKP-1-KPC compared with pEC258-3,and three elements,two IS6 family transpose and one class 1 integron integrase family Intl1,associated with gene inversion are found at the flank of this chromosome segments.These results suggest that pEC258-3 and pCRKP-1-KPC may evolve from a common plasmid and have independently spread to different species (E.coliandK.pneumoniae,respectively).Furthermore,gene environment analysis found that theblaKPC-2gene is linked to mobile componentISKpn6andISKpn27/IS1182.Transposon Tn3 (tnpR) was also found inblaKPC-2gene 5´ (Fig.1).Additionally,phylogenetic analysis showed that 258E located the cluster containing strains from the United Kingdom (Fig.2).
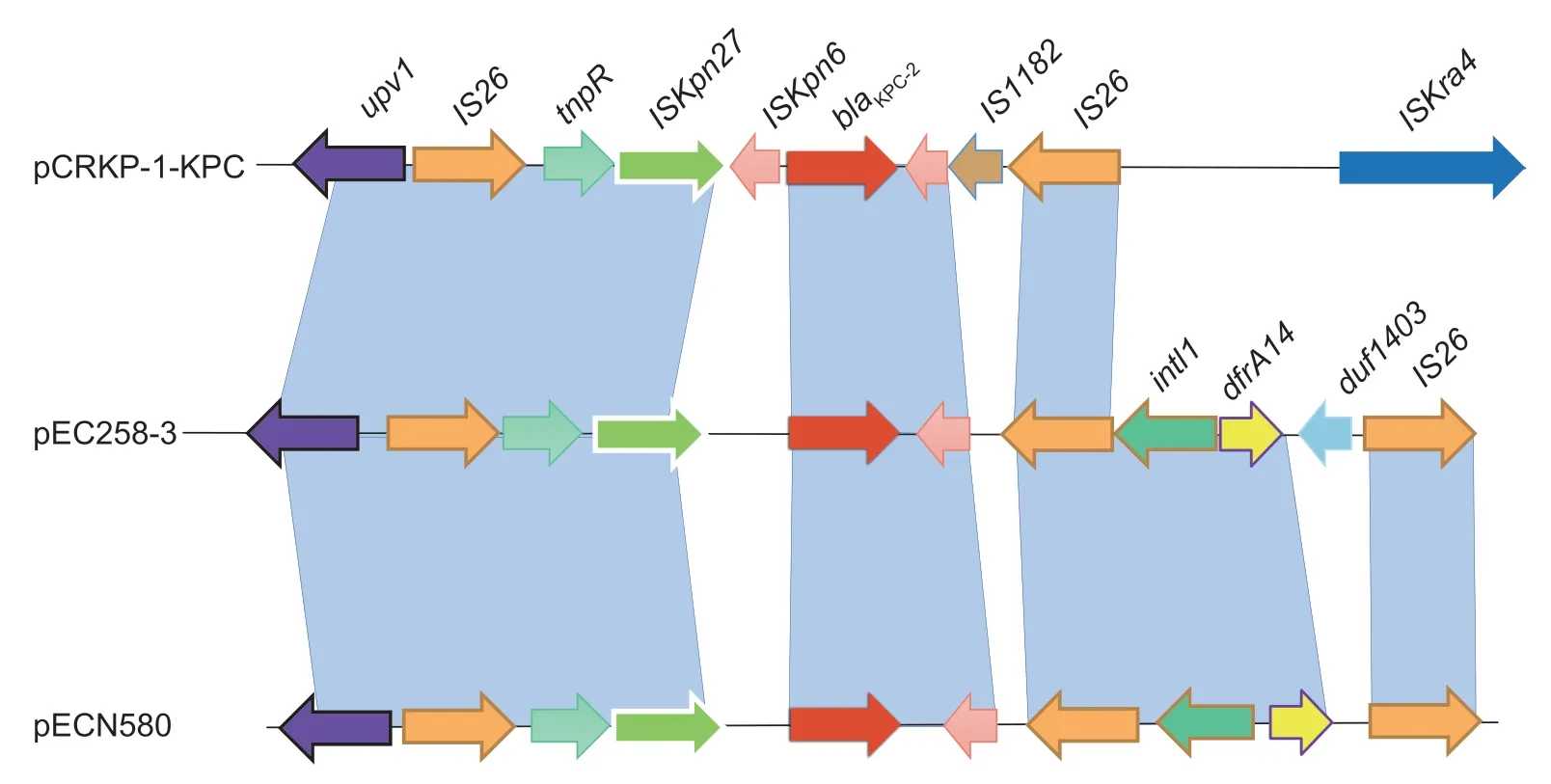
Fig.1 Comparison of genetic blaKPC-2 environments among pEC258-3 (GenBank accession no.CP097098),pECN580 (GenBank accession no.KF914891),and pCRKP-1-KPC (GenBank accession no.KX928750).Light-blue shading denotes shared regions of homology.Open reading frames (ORFs) are portrayed by arrows.
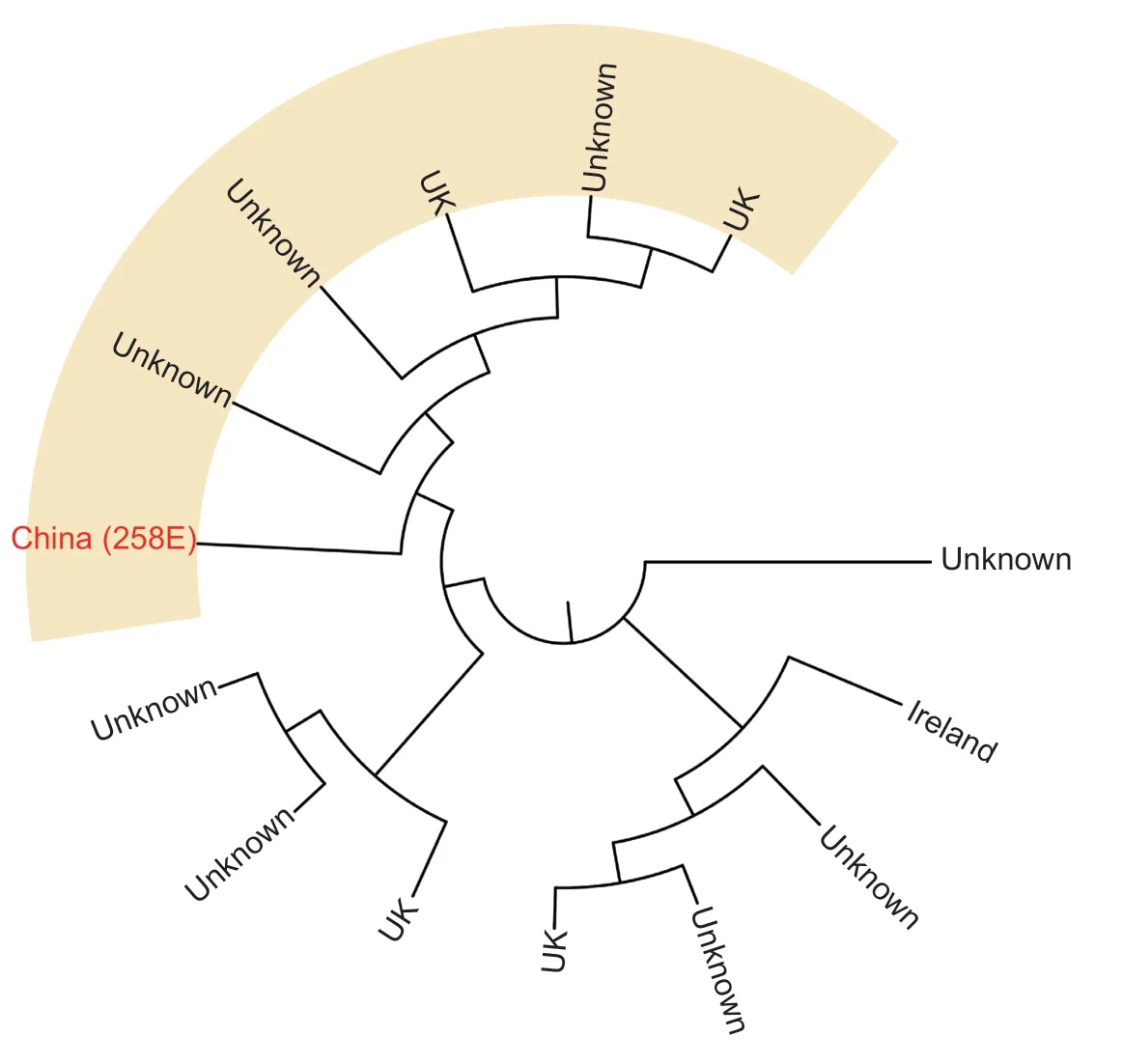
Fig.2 Phylogenetic analysis of 258E and global Escherichia coli ST602.Phylogenetic tree was built based on Escherichia ST602 isolates (https://pubmlst.org/escherichia/) and visualized by MicroReact.iTOL was used for additional visualizations.
4.Conclusion
To our knowledge,this is the first report of ablaKPC-2-harboring IncN plasmid from an XDRE.coliST602 of animal origin.Our findings highlight the plasticity and complexity ofblaKPC-2-harboring composite transposon-like element found in these promiscuous plasmids and also emphasize that the important role of bacteria of animal origin in the emergence of antibiotic resistance genes and facilitate resistance genes circulation within ecosystems,and the resistant strain of animal origin may serve as sources of antibiotic resistance genes to human health.
Acknowledgements
The work was funded by the National Key Research and Development Program of China (2018YFE0192600),the Shanghai Agriculture Applied Technology Development Program,China (T20200104),the Fundamental Research Funds for the Central Universities,China (2020JB05),and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences(CAAS-ZDRW202203).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable internaltional,national and institutional guidelines for the care and use of animals were followed.
杂志排行
Journal of Integrative Agriculture的其它文章
- Advances in studies on the physiological and molecular regulation of barley tillering
- Antioxidant lignans sesamin and sesamolin in sesame (Sesamum indicum L.): A comprehensive review and future prospects
- TaABI19 positively regulates grain development in wheat
- Genetic effects of Agropyron cristatum 2P chromosome translocation fragments in a wheat background
- Grain yield,nitrogen use efficiency and physiological performance of indica/japonica hybrid rice in response to various nitrogen rates
- Border effects of the main and ratoon crops in the rice ratooning system
