Integrated linkage mapping and genome-wide association study to dissect the genetic basis of zinc deficiency tolerance in maize at seedling stage
2022-12-02JinqinXuZhongfuNiFnjunChenXiuyiFuFutongYu
Jinqin Xu,Zhongfu Ni,Fnjun Chen,Xiuyi Fu,Futong Yu,*
a Key Laboratory of Plant-Soil Interaction(MOE),Centre for Resources,Environment and Food Security,College of Resources and Environmental Sciences,China Agricultural University,Beijing 100193,China
b State Key Laboratory for Agrobiotechnology,Key Laboratory of Crop Heterosis and Utilization(MOE),Beijing Key Laboratory of Crop Genetic Improvement,China Agricultural University,Beijing 100193,China
c National Maize Improvement Center of China,Beijing Key Laboratory of Crop Genetic Improvement,China Agricultural University,Beijing 100193,China
Keywords:Maize(Zea mays L.)Quantitative trait loci(QTL)Genome-wide association study(GWAS)Zinc(Zn)deficiency tolerance Candidate genes
ABSTRACT Zinc(Zn)deficiency is the most widespread micronutrient deficiency,affecting yield and quality of crops worldwide.Identifying genes associated with Zn-deficiency tolerance in maize is a basis for elucidating its genetic mechanism.A K22×CI7 recombinant inbred population consisting of 210 lines and an association panel of 508 lines were used to identify genetic loci influencing Zn-deficiency tolerance.Under-Zn and-Zn/CK conditions,15 quantitative trait loci(QTL)were detected,each explaining 5.7%-12.6%of phenotypic variation.Sixty-one significant single-nucleotide polymorphisms(SNPs)were identified at P<10-5 by genome-wide association study(GWAS),accounting for 5%-14% of phenotypic variation.Among respectively 198 and 183 candidate genes identified within the QTL regions and the 100-kb regions flanking these significant SNPs,12 were associated with Zn-deficiency tolerance.Among these candidate genes,four genes associated with hormone signaling in response to Zn-deficiency stress were co-localized with QTL or SNPs,including the genes involved in the auxin(ZmARF7),and ethylene(ZmETR5,ZmESR14,and ZmEIN2)signaling pathways.Three candidate genes were identified as being responsible for Zn transport,including ZmNAS3 detected by GWAS,ZmVIT and ZmYSL11 detected by QTL mapping.Expression of ZmYSL11 was up-regulated in Zn-deficient shoots.Four candidate genes that displayed different expression patterns in response to Zn deficiency were detected in the regions overlapping peak GWAS signals,and the haplotypes for each candidate gene were further analyzed.
1.Introduction
In prokaryotes,5%-6% of all proteins are Zn-binding proteins,and this proportion is estimated at about 10% in eukaryotes[1].Zinc(Zn)is the only trace element required for all six classes of enzymes and is a component of more than 1000 transcription factors and structural proteins[2].Zn deficiency is the most widespread micronutrient deficiency in crops.It affects more than 160 million ha of arable land worldwide and about 50% of cereal crop agricultural soils[3].Zn deficiency impairs crop yields while reducing the Zn content in grains and harming human health by dietary deficiency.More than 30% of the world population,especially in developing countries,are subject to dietary Zn deficiency[4].Most of these victims are women and children.Zn deficiency causes stunting and death in young children.In 2004,more than 450,000 children under the age of 5 died of Zn deficiency globally,accounting for 4.4% of deaths in this age class[5].Zn deficiency may lead to a high incidence of infectious diseases and complications during pregnancy or birth,including infant mortality[6].
Maize is the crop with the highest global annual yield,cultivated across a wide range of agroecologies and cropping systems[7].A record global maize grain production of 1054 million metric tonnes was achieved during 2016/2017[8].Maize not only is a nutrition source for humans,but is grown worldwide for multiple uses[9].It can be processed into industrial alcohol,beverages,ethanol fuel,starch,sweeteners,glue,and oil[10].Maize occupies dual roles as a model system for plant genetics and cytogenetics[11].Maize is more sensitive to Zn deficiency than other crops[12].Nearly half of Chinese farm soils are classified as critically Zn-deficient,especially in North China where most of the maize crop is planted[13].In maize,Zn deficiency causes stunted growth and morphological abnormalities,such as lower vascular bundle proportion and higher stomatal density,among changes in physiology,such as decreased net photosynthesis and lowered stomatal conductance[3,14].The biomass,yield,and quality of maize plants are impaired.
Few studies have focused on identifying genes that respond to Zn-deficiency stress in maize.The identification of Zn-sensitive genes would be helpful to improve the tolerance of maize coping with Zn deficiency,using either breeding by marker-assisted selection or transgenic technology.
Zn is taken up from soil by roots and reaches developing tissues through vascular tissues.There is exchange of Zn between tissues.For this purpose,a complex network for Zn homeostasis has developed that controls the uptake,distribution,and accumulation of Zn.Two major components responsible for Zn homeostasis in plants are chelators and transporters.Chelators for heavy metals are regulated by a few gene families,including NAS(nicotianamine synthase)and PC(phytochelatin).Transporters for metal ions are controlled by several gene families,including ZIP(ZRT/IRT-like protein),HMA(P1B-type metal ATPase),NRAMP(natural resistanceassociated macrophage protein),CDF(cation diffusion facilitator),and MTP(metal transport protein).Under Zn-deficient condition,Zn transporter genes from the ZIP gene family are up-regulated to increase Zn uptake[15,16].HMA9 is a metal efflux protein functioning in metal mobilization from mature leaves to new leaves in rice[17].AtMTP1 and AtMTP3 from the CDF gene family[18,19]and ZIF1 from the MFS gene family[20]are critical for sequestration,storage and distribution of Zn in Arabidopsis[18-20].NA(nicotianamine) is biosynthesized by trimerization of Sadenosylmethionine,catalyzed by NAS[21].Numerous studies[22-25]have confirmed that NA functions in chelation,transport,and homeostasis of various heavy metals,including Fe,Zn,and Cu.NA-deficient plants accumulate fewer ions of Fe,Cu,Zn,and Mn and alter their distribution in vivo[22,26].Overexpression of NAS in transgenic plants increases NA concentration and consequently metal concentration[27,28].Although these genes associated with Zn homeostasis have been identified and characterized in Arabidopsis and rice,information about the molecular mechanism of their action in maize is scarce.
Linkage analysis based on biparental population is an effective method to identify QTL for complex traits,but has limitations.First,it requires creation of a linkage population,a slow and resourceintensive process[29].QTL analysis usually produces large confidence intervals[30,31].It is expensive and time-consuming to narrow the confidence interval and finally to locate genes by fine mapping.Second,QTL mapping captures only a small proportion of the phenotypic variation between the two parents[32].Rare alleles can be missed owing to the lack of genetic diversity in biparental populations[33].
Recently,genome-wide association study(GWAS)has been developed as a powerful approach to identifying associated genomic regions at single-gene resolution using single-nucleotide polymorphism(SNP)data and rapid linkage-disequilibrium decay[34].Compared with conventional linkage analysis,GWAS is more timesaving and cost-effective[35,36],owing to its higher resolution[37-39]and more diverse genetic germplasm with different genetic backgrounds[40].However,population substructure and genotyping errors in GWAS can lead to high false-positive rates[41-43].In contrast,QTL mapping based on a biparental population gives a lower false-discovery rate,resulting from its advantages in comparing pairs of alleles[44].Linkage mapping based on biparental populations,together with GWAS relying on linkage disequilibrium,constitutes a powerful approach to dissect the genetic architecture of complex traits using natural variation in crops.
Complex quantitative traits in maize have been investigated by linkage analysis and GWAS[45-47].But the genetic basis of the mechanisms of Zn deficiency tolerance in maize is poorly understood because of a lack of reports investigating the performance under Zn-deficient condition.Loci controlling traits associated with Zn-deficiency tolerance have been detected by linkage analysis in rice and wheat[48,49].Moreover,Lee et al.[50]combined biparental QTL mapping and GWAS to detect QTL controlling root development,biomass accumulation,and grain yield in rice under Zn deficiency and to identify candidate genes conferring tolerance to Zn-deficiency stress.However,to our knowledge,no studies have combined linkage analysis and GWAS to dissect the genetic architecture of Zn-deficiency tolerance in maize.The goals of the present study were to identify loci associated with Zn-deficiency tolerance using QTL mapping and GWAS,and to further identify candidate genes for these loci.
2.Materials and methods
2.1.Plant materials
A recombinant inbred population containing 210 lines derived from the cross K22×CI7 was used in a previous study[51].The parent line K22 is a Zn-inefficient inbred line,and CI7 is highly Zn-deficiency tolerant,as revealed by our previous study[52]comparing Zn efficiency among 20 maize inbred lines.An association panel composed of 508 diverse inbred lines(AM508)was used for GWAS.Based on population structure,all lines were classified into four subgroups:stiff stalk(SS),non-stiff stalk(NSS),tropical-subtropical(TST)and admixed group(MIX).Detailed information for this panel has been previously presented[53,54].
2.2.Plant culture in hydroponics
Maize seeds were sterilized for 30 minutes in a 10% solution of H2O2and washed with distilled water.After being soaked in saturated CaSO4for 10 hours,the seeds were then germinated on moist filter paper in the dark at room temperature.Two days later,the germinated seeds were wrapped in moist filter paper roll and grown.At the stage of two visible leaves,seedlings were selected and transferred into a 40-L black container.Plants were cultured in hydroponics under Zn-deficient(3×10-4mmol L-1Zn-EDTA for QTL analysis,2×10-4and 5×10-4mmol L-1Zn-EDTA for GWAS)and Zn-sufficient conditions(1×10-2mmol L-1Zn-EDTA).Besides,the adjusted Hoagland nutrient solution contained(in mmol L-1):0.5 NH4NO3,0.5 CaCl2,1.5 Ca(NO3)2,0.75 K2SO4,0.65 MgSO4,0.1 KCl,0.25 KH2PO4,1.0×10-3H3BO3,0.35 Fe(II)-EDTA,8.0×10-3CuSO4,1.2×10-2MnSO4,4.0×10-5(NH4)Mo7O24,4.0×10-3NiCl.Two experiments were performed for association mapping.Each experiment contained Zn-deficient(-Zn)and Zn-sufficient(CK)treatments,and each treatment with three replicates.Solution pH was set at 5.5-6.0.Nutrient solution was renewed every 3 days and aerated with a pump.The growth chamber conditions were as follows:a 14-hour light period from 8:00 to 22:00 at 28 °C,and a 10-hour dark period at 22 °C.The mean light intensity measured at the canopy was 350μmol m-2s-1and the relative humidity was 60%.
2.3.Phenotyping
Phenotype data for Zn score(ZnSc),plant height(PH),shoot(SDW)and root(RDW)dry weight,root-to-shoot ratio(R/S ratio)were collected for two plants for each replicate.A Zn score for each plant under Zn deficiency was visually recorded three times following day 15 after transplanting.A scale of 0-5 was used to assess the Zn-deficiency tolerance of maize at seedling stage(Fig.S1A).Under Zn-deficient condition,tolerance scoring reflected the degree of inhibition in growth and development for the whole plant,as well as the areas of chlorosis and necrotic patches distributed on middle and young leaves.Score 0:plants develop about three leaves with one shoot,and show stunted growth and reduced plant height,wrinkled leaf margins,and small,malformed leaves.More than 50%of areas on middle and young leaves appear chlorotic and turn pale yellow.Score 1:plants have developed 4 leaves and 1 shoot,and plant height and leaf elongation are strongly suppressed as with score-0 plants.Shortened and malformed leaves are prominent symptoms,and 30%-50% of areas on middle and young leaves show Zn-deficiency chlorosis with necrotic patches distributed on margins and tips of leaves.Score 2:plants develop 5 leaves and 1 shoot,showing malformations and distorted leaf growth like those of score-1 plants.All leaves are shortened and small,and 20%-30% of area of middle and young leaves show brown patches.Zn-deficiency chlorosis is lower than in score-0 and score-1 plants.Score 3:compared with score-2 plants,reduction in plant height and internode length are decreased.But extension of all leaves is strongly inhibited,and necrotic spots are shown on 10%-20%of the area of middle and young leaves.Score 4:plants grow well but still display banded chlorosis on leaf margins of middle and young leaves.Score 5:plants are green and healthy.
The experiment was terminated on day 21 after transplanting.Plant height(PH)was measured first,and the shoot and root of each plant were stored in envelopes separately before drying.After all samples were dried at 105 °C for 30 min and dried at 75 °C till constant weight,shoot(SDW)and root(RDW)dry weights were measured,and R/S ratio(R/S)was calculated as root to shoot dry weight for each plant.
2.4.Genotyping
For linkage mapping,the high-density bin map generated using 2386 SNPs covered 1720 cM over 10 chromosomes.For GWAS,this panel consisted of 1.03 million high-quality SNPs,and 56,110 SNPs that were genotyped with the MaizeSNP50 BeadChip[54].A total of 559,285 high-quality SNPs with minor allele frequency(MAF)>0.05 were used.
2.5.Data analysis
Comparison of trait means of the two parents in the K22×CI7 RIL population was compared by two sample t-test.The linear mixed effect function lmer in the lme4 package of R version 3.1.1 was fitted to each line in the population to obtain the best linear unbiased prediction(BLUP)value for each trait:yijk=μ+Gi+Ej+Gi×Ej+R(E)jk+εi,where yijkis the phenotypic value of the ith inbred line in the jth environment and kth replication,μis the grand mean,Giis the genetic effect of the ith inbred line,Eiis the environmental effect of the jth environment,Gi×Ejis the interaction between genotype and environment for the ith inbred line and jth environment,R(E)jkis the kth replication in the jth environment,andεijkis the residual error.These variance components were used to calculate the broad-sense heritability as H2=,whereis genetic variance,is the interaction of genotype and environment,is residual error,and e and r are the numbers of environments and replications,respectively.The ratio of the values in the-Zn treatment to the values in the CK treatment(-Zn/CK)for each trait was calculated from the BLUP of each line.
2.6.QTL mapping
QTL were identified by composite interval mapping(CIM)in Windows QTL Cartographer version 2.5.The scanning interval between markers was set at 0.5 cM,and the window size was set at 10 cM.Model 6 was selected for detecting QTL and estimating their effects.The threshold logarithm of odds(LOD)values in this study was estimated by permutation tests with a minimum of 1000 replicates at a significance level of P<0.05.The confidence interval of the QTL position was determined using the 1-LOD interval method.
2.7.Genome-wide association analysis
A genome-wide association analysis of traits associated with Zn deficiency tolerance was performed using a mixed linear model(MLM)incorporating the population structure and relative kinship matrix[42].GWAS for Zn score under Zn deficiency,plant height,shoot and root dry weight,R/S ratio under Zn deficiency and ratios of-Zn to CK was performed using TASSEL 3.0.Using the‘‘no compression”and‘‘population parameters previously determined”(P3D)options,MLM was fitted to detect associations between phenotype and genotype.The threshold P=1×10-5was used to identify significant SNPs and was determined based on quantilequantile(Q-Q)plots and distribution of P-values for all traits.
2.8.Identification and annotation of candidate genes
For GWAS,as in our previous study[54],the linkage disequilibrium(LD)of this association panel was 50 kb.Accordingly,the 100-kb interval flanking each significant SNP(50 kb upstream and downstream of the significant SNP)was selected for identifying candidate genes[54,55].Their functional descriptions were identified using the maize B73 reference genome assembly version 2 available in the MaizeGDB(http://www.maizeGDB.org/)and Gramene(https://www.gramene.org/)databases.
2.9.Haplotype and linkage disequilibrium(LD)analysis
The SNPs within the 1.5-kb promoter region of candidate gene and the nonsynonymous SNPs in the coding sequence(CDS)were used for haplotype analysis.An LD heat map of each candidate gene was generated with the LD heatmaps package in R.
2.10.RNA extraction and gene expression quantification
Total RNA was extracted from shoots and roots of plants with a Total RNA Extraction Kit(TIANGEN,Beijing,China).The cDNA was synthesized following Fast Quant RT Super Mix Reverse Transcription Kit instructions(TransGene,Beijing,China).Quantitative realtime PCR was performed using SYBR Green Real-time RT-PCR(Takara,Kusatsu,Shiga,Japan)and an ABI7500 Fast Real-Time PCR System(Applied Biosystems).The relative gene expression level was calculated using the 2-ΔΔCT method[56].Each realtime PCR experiment contained three technical replicates.
3.Results
3.1.Phenotypic variation in linkage population and GWAS panel
Symptoms of Zn deficiency in plants were consistent with those in our previous studies,including chlorosis in young leaves and reduced leaf size.Under Zn-deficient condition,K22 showed stunted growth and developed five leaves only on day 21 after transplanting,whereas plants of CI7 developed at least seven leaves(Fig.S1B).Compared with control plants(Fig.S1C),K22 showed severe Zn-deficiency symptoms,including short,curled leaves and shortened internodes and petioles(Fig.S1B),whereas CI7 showed few symptoms(Fig.S1B,C).
The parents of the QTL mapping population differed in their responses to Zn deficiency(Fig.S2).Zn deficiency reduced the plant height and shoot and root dry weight of the sensitive parent K22 by 59%,56% and 45%,respectively.In contrast,there were no significant differences in plant height and shoot dry weight of the tolerant parent CI7 between the CK and the-Zn treatments.Under Zn-deficient condition,shoot and root dry weights of CI7 were respectively 2.5-fold and 1.4-fold higher than those of K22.R/S ratios were significantly elevated under Zn deficiency.The R/S ratio of K22 was higher than that of CI7,indicating that K22 is more sensitive to Zn-deficiency status.However,no significant differences between CI7 and K22 in shoot and root dry weight or R/S ratio were observed under Zn-sufficient condition.
All the traits of the lines in the RIL population and GWAS panel varied widely under Zn-deficient and Zn-sufficient conditions(Fig.1).Traits showed differential responses to Zn deficiency stress(Fig.S3).On average in the QTL mapping population and GWAS panel,Zn deficiency reduced plant height and shoot and root dry weight by 18.9% and 31.9%,35.9% and 40.0%,33.3% and 25.8%,respectively.Low-Zn stress increased the R/S ratio in the linkage and association experiments by 15.4% and 25.0%,respectively.The coefficients of variation in the linkage population and GWAS panel ranged from 19.3%to 45.4%and from 25.2%to 64.6%,respectively(Table S1).Under Zn deficiency,plant height,shoot and root dry weight,and R/S ratio in the QTL mapping population and GWAS panel followed normal distributions(Fig.S4).The broadsense heritability of each trait under Zn deficiency in the GWAS panel was higher than 80%(Table S1).Spearman correlation analysis in the RIL and association populations showed that ZnSc was significantly correlated with PH,SDW and RDW,R/S(Table S2).
3.2.QTL detection for traits associated with Zn-deficiency tolerance
Based on the high-density genetic map,15 QTL were identified under Zn deficiency at an empirical threshold logarithm of odds(LOD)value of 3.2 estimated from 1000 permutation tests.These 15 QTL were distributed on chromosome 3,5,6,7,8 and 10,as follows:2 on chromosome 3,4 on chromosome 5,1 on chromosome 6,1 on chromosome 8,4 on chromosome 7,and 3 on chromosome 10(Fig.2;Table S3).The physical lengths of the intervals of these QTL ranged from 0.2 to 45.5 Mb,with a mean of 6.5 Mb.
3.2.1.QTL for Zn score
The locus qKC-ZnSc5-1 with the second-largest effect,mapped on chromosome 5,showed a positive effect,indicating that this allele from the tolerant parent CI7 increased the Zn score by 0.31.A locus qKC-ZnSc6-1 with a small fraction(6.8%)of explained variation detected on chromosome 6 showed a negative effect,suggesting that alleles from the sensitive parent K22 contributed to increased Zn score.
3.2.2.QTL for plant height
qKC-PH3-1 was identified on chromosome 3 explaining 6.2% of phenotypic variation.qKC-PH3-1 displayed a negative effect,indicating that the alleles increasing plant height were derived from K22.The third largest effect locus qKC-PH7-1,detected on chromosome 7,accounted for 10.9% of phenotypic variation,and CI7 alleles at this locus accounted for an increase of 3.97 cm in plant height.
3.2.3.QTL for shoot and root dry weight,and R/S ratio
Three small-effect QTL(qKC-SDW3-1,qKC-SDW5-1,qKC-SDW5-2)controlling shoot dry weight and two loci(qKC-RDW5-1,qKC-RDW8-1)controlling root dry weight were mapped on chromosome 3,5,and 8,explaining 6.9%-8.2%of phenotypic variation.The estimated additive effect of each locus ranged from-0.08 to 0.08,and the additive effect of two QTL(qKC-SDW5-1,qKCSDW5-2)for SDW and the loci qKC-RDW5-1 for RDW originated from the tolerant parent CI7,whereas the alleles of two QTL(qKC-SDW3-1,qKC-RDW8-1)came from the sensitive parent K22.Four minor QTL(qKC-R/S7-2,qKC-R/S10-1,qKC-R/S10-2,and qKCR/S10-3)controlling R/S ratio,together with the two large-effect loci qKC-R/S7-1 and qKC-R/S7-3 were identified on chromosomes 7 and 10,accounting for 5.7%-12.6% of phenotypic variation.
3.3.Genome-wide association analysis of Zn-deficiency tolerance
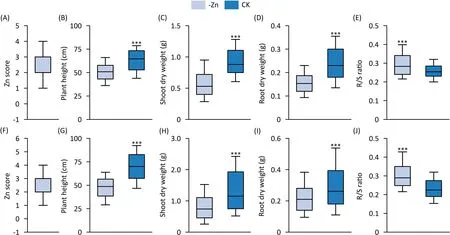
Fig.1.Differences in traits in response to Zn deficiency in linkage(A-E)and association(F-J)populations.(A,F)Zn score.(B,G)Plant height.(C,H)Shoot dry weight.(D,I)Root dry weight.(E,J)R/S ratio.The upper and lower lines represent the 90th and 10th percentiles,respectively;The solid line in each box represents the median value.The upper and lower edge of each box represent the 75th and 25th percentiles,respectively.***,indicate difference between the-Zn and CK treatment at P<0.001.
The Q-Q plots for GWAS based on the BLUP values of the traits implied that the associations were well controlled for population structure(Fig.S5).At a significance level of P<1×10-5,61 significant SNPs were mapped on 10 chromosomes(Fig.3;Table 1).The phenotypic variance explained by these SNPs ranged from 5% to 14%.Among the 61 SNPs,2,8,5,14 and 28 were associated with the single-trait Zn score,plant height,shoot and root dry weight,and R/S ratio,respectively.The SNP chr2.S_20527298 was found significant for both Zn score and plant height,and another three(chr9.S_59587835,PZE-109040807,and PZE-100000518)were associated with both shoot and root dry weights.

Fig.2.Fifteen quantitative trait loci(QTL)associated with Zn score(ZnSc),plant height(PH),shoot(SDW)and root dry weight(RDW),root-to-shoot ratio(R/S)were detected in the K22×CI7 population.The positions of candidate genes are painted in gray columns,including ZmARF7(GRMZM2G475263),ZmEIN2(GRMZM2G102754),ZmVIT(GRMZM2G107306)and ZmYSL11(GRMZM5G812538).
3.4.Candidate genes identification
Linkage analysis and GWAS are complementary methods for QTL detection.To reduce the effects of genetic background,we combined QTL mapping and GWAS to detect loci associated with Zn deficiency tolerance.Not only overlapping regions,but regions detected either by linkage analysis or by GWAS were considered for candidate gene identification.
For QTL mapping,four QTL co-localization regions were identified on chromosomes 3,5,7,and 10,and 198 genes were identified within these regions(Chr.3,1,467,202-1,686,795 bp;Chr.5,206,563,089-208,072,333 bp;Chr.7,153,175,123-154,297,456 bp;Chr.10,123,956,269-132,034,269 bp)in the K22×CI7 population(Table S4).
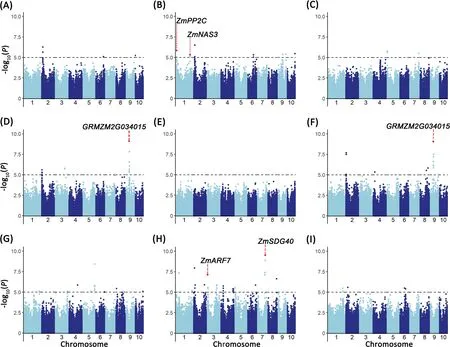
Fig.3.Genome-wide association results for traits associated with Zn deficiency tolerance in a maize association panel under Zn-deficient condition.Manhattan plots for Zn score(A),plant height(-Zn,(B);-Zn/CK,(C)),shoot dry weight(-Zn,(D);-Zn/CK,(E)),root dry weight(-Zn,(F);-Zn/CK,(G)),and R/S ratio(-Zn,(H);-Zn/CK,(I))are shown.Gray dashed lines in Manhattan plots correspond to thresholds of P<1×10-5.Red dots mark significant SNPs corresponding to the candidate genes associated with Zn deficiency tolerance,including ZmPP2C(GRMZM5G833774),ZmNAS3(GRMZM2G478568),ZmARF7(GRMZM2G475263),ZmSDG40(GRMZM2G092131)and GRMZM2G034015.
Candidate gene ZmARF7(GRMZM2G475263)which is involved in auxin regulation in plants,was not only localized between two adjacent peak SNPs for qKC-PH3-1 and qKC-SDW3-1 on the QTL co-localization on chromosome 3(Fig.S6A),but also identified in the 57-kb region of the significant SNP chr3.S_1613904(Fig.3H).The expression of ZmARF7 was significantly down-regulated in Zn-deficient shoots and roots(Fig.S7A,B).The R/S ratio of inbred lines containing haplotype Hap 1 was the lowest and significantly lower than those of Hap 2 and Hap 3(Fig.S8).This finding indicated that Hap 1 was the favorable allele for stable R/S ratio in response to Zn deficiency.ZmTERF18(GRMZM2G017355)and ZmTERF19(GRMZM2G017429)which encode mitochondrial transcription termination factor family proteins,were identified in the interval between two adjacent peak SNPs for each locus(qKC-ZnSc5-1 and qKC-RDW5-1)on the QTL co-localization regions on chromosome 5(Fig.S6B;Table S4).

Table 1Significant SNPs for traits associated with Zn deficiency tolerance detected by GWAS.
Candidate gene GRMZM5G812538,encoding the yellow stripelike transporter ZmYSL11,was located within the co-localization of qKC-R/S10-1 and qKC-R/S10-3(Fig.2;Table 2).ZmYSL11 was found to be Zn deficiency-inducible in the shoots of maize inbred lines(Fig.S7C).These findings suggested that ZmYSL11 might be involved in the mechanism underlying Zn deficiency tolerance.
As previously reported[54,57],the SNPs detected by GWAS and the QTL associated with the same target traits can be compared to further identify priority candidate genes.In the present study,significant SNPs were compared with the QTL detected in the K22×CI7 population in this study and the Zn efficiency-associated loci detected in the Ye478×Wu312 population in our previous study[52].As a result,on chromosome 1,the significant SNP chr1.S_8274996 which was located in the reported minor-effect locus qZEAL-R/S1-1 in the Ye478×Wu312 population,was associated with the candidate gene GRMZM5G833774 encoding a phosphatase 2C family protein in maize(Fig.3B;Table 2).The expression of GRMZM5G833774(ZmPP2C)was significantly upregulated under Zn deficiency in shoots and not in roots(Figs.S9A,B,and S10A,B).Six major haplotypes were identified in the 1.5-kb promoter region and CDS(Fig.4A-C).The plant height of the inbred lines containing Hap 2 was the highest(mean:52.6 cm),indicating that Hap 2 is a favorable allele for increased plant height in response to Zn deficiency in maize.
For GWAS,a 100-kb region(50 kb upstream and downstream)flanking each significant SNP was selected for identifying candidate genes.A total of 183 candidate genes was identified in the 100-kb regions flanking the 61 significant SNPs(Table S5).Among them,the gene GRMZM2G478568(ZmNAS3)encoding nicotianamine synthase 3 in maize[58],was localized in the 39-kb region flanking the significant SNP chr1.S_259817863.ZmNAS3 has been proposed[59,60]to be involved in Zn-deficiency tolerance in maize.Nine major haplotypes were identified and the plant height of the lines containing Hap 8 was the highest(mean:54.5 cm)(Fig.4D-F),indicating that Hap 8 is a favorable allele for increased plant height under low-Zn stress.
Candidate gene GRMZM2G092131 encoding the SET domain group protein in maize was found to contain the SNP chr7.S_144602513(P=3.11E-10),which was the most significant SNP detected by GWAS in this study(Fig.3H;Table 2).Expression of GRMZM2G092131(ZmSDG40)was significantly up-regulated in Zn-deficient shoots and roots(Figs.S9C,D and S10C,D).Seven major haplotypes were identified in the 1.5-kb promoter region and CDS(Fig.5A).An LD heat map showed that GRMZM2G092131 was located in a region with relatively high LD(Fig.5B).The R/S ratio of Hap 4 was significantly lower than the R/S ratios of the other five haplotypes,indicating that Hap 4 is the favorable allele for stable root-to-shoot ratio in response to low-Zn stress(Fig.5C).
Candidate gene GRMZM2G034015 was identified within the peak GWAS signal chr9.S_59587835 controlling both shoot and root dry weights under Zn deficiency(Fig.3D,F;Table 2).GRMZM2G034015 was markedly induced under Zn deficiency in both shoots and roots(Figs.S9E,F,and S10E,F).Six major haplotypes were identified in the promoter and coding region of GRMZM2G034015(Fig.5D,G).The LD heat map indicated a relatively high LD level around GRMZM2G034015(Fig.5E,H).Hap 1 was found to have the highest shoot(mean:1.40 g)and root(mean:0.44 g)dry weights,significantly higher than those of the other five haplotypes(Fig.5D,F,G,and I),indicating that Hap 1 is the favorable allele for increased shoot and root biomass accumulation under Zn deficiency.
4.Discussion
4.1.Comparison of loci with previous studies
To date,there have been few reports on QTL mapping under Zn deficiency in maize[52,61].However,there were several previous studies[62-66]of the traits investigated in the present study.Based on the physical position,8 QTL detected by linkage mapping together with 17 significant SNPs identified by GWAS were colocalized with QTL identified in previous studies(Table S6),suggesting that the loci detected in this study are valuable for candidate gene identification.On chromosome 2,the significant SNP chr2.S_20527298 associated with Zn score and plant height,and chr2.S_20959597 associated with R/S ratio,were co-localized with loci controlling three traits related to plant height detected by Zhang et al.[66].The genomic region overlapped by qKC-PH3-1,qKC-SDW3-1 and chr3.S_1613904 on chromosome 3 was also identified within the intervals of two QTL controlling ear height and yield in previous studies[64,66].The significant SNP chr4.S_235396188 on chromosome 4 and the co-localization of a major-effect QTL(qKC-ZnSc5-1)and a minor-effect QTL(qKCRDW5-1)on chromosome 5 were co-localized with MQTL52 and MQTL62 which both controlled plant height,leaf angle and yield[64].The loci qKC-SDW5-1 controlling shoot biomass and the chr5.S_1525638 associated with R/S ratio were both co-localized with the QTL for total root length previously identified[63].
Apart from the physiological traits investigated in this study,QTL mapping of grain Zn concentration has been performed in maize[67-69].The significant SNP chr3.S_173585799 on chromosome 3 and the locus qKC-R/S7-2 on chromosome 7 were colocalized with loci controlling Zn and Mn concentration in grain[68].Eleven significant SNPs together with two minor-effect QTL distributed on chromosome 2,4,7,8,9,and 10 were colocalized with 16 QTL associated with various traits identified in previous studies[62-66,68,70].These findings suggest that these genomic regions harbor several genes with pleiotropic effects not only on root traits at seedling stage,but also on plant height under normal condition and on salt tolerance,yield,and mineral concentration.
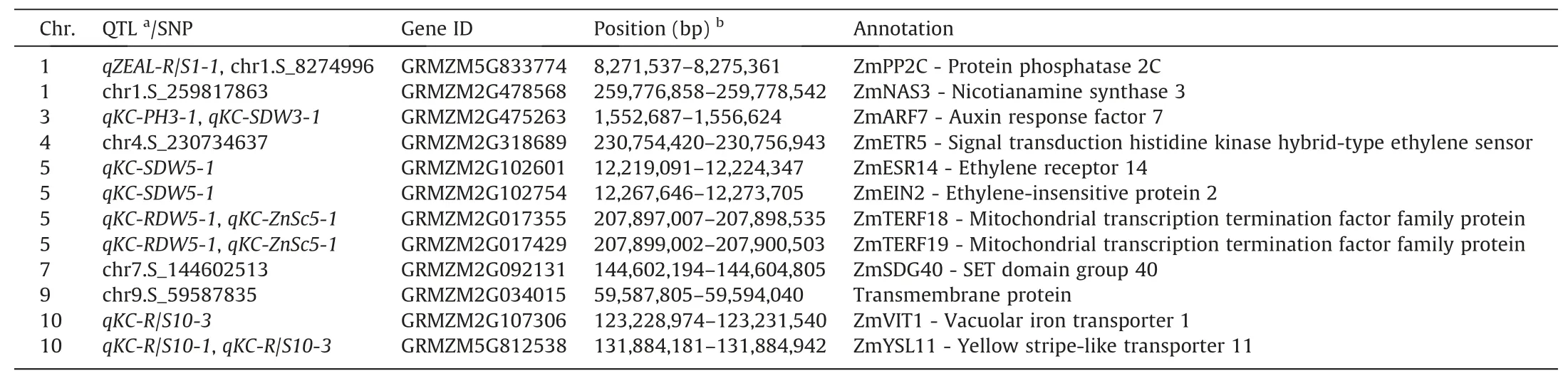
Table 2Candidate genes associated with Zn deficiency tolerance identified by linkage mapping and GWAS in maize.
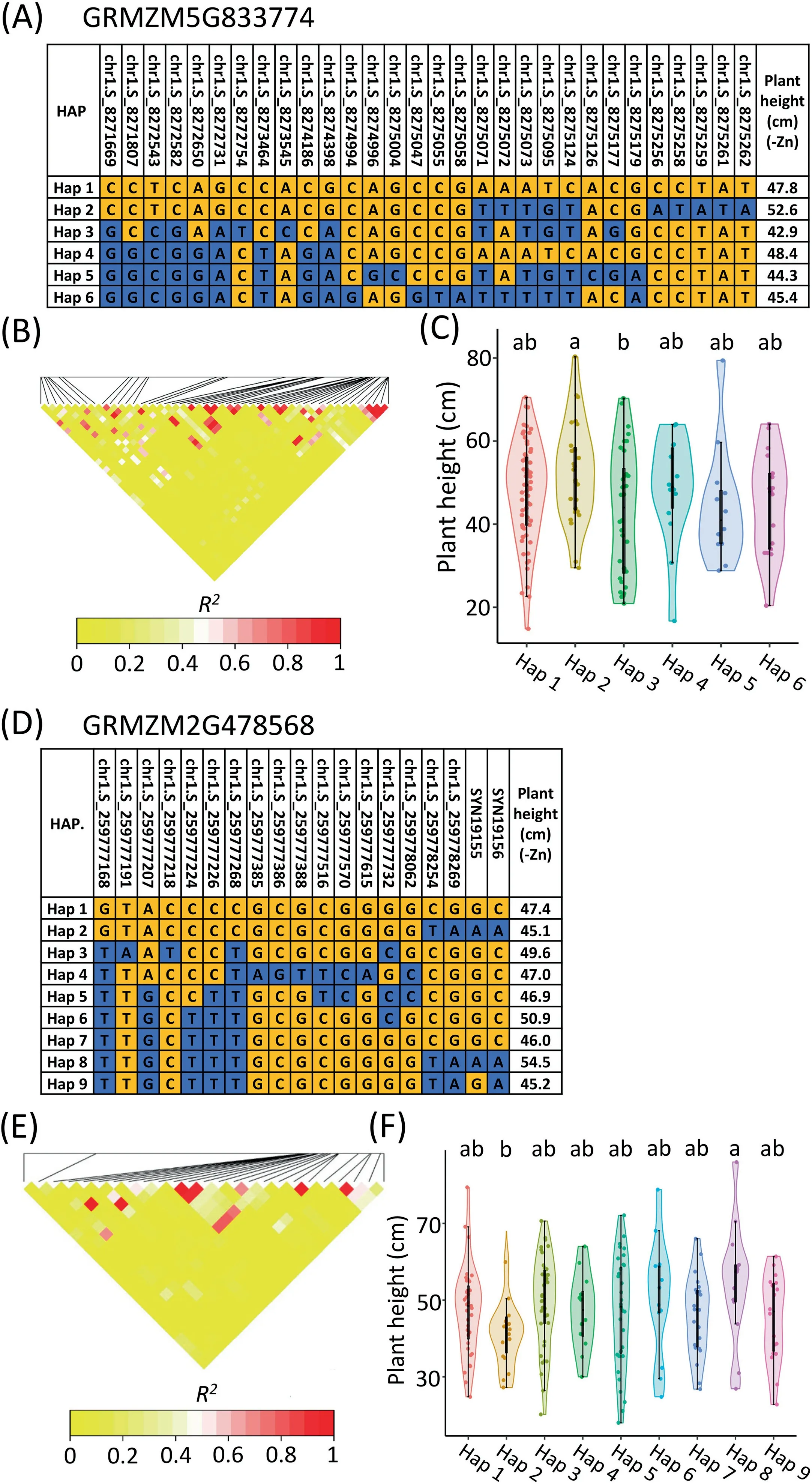
Fig.4.Associated regions and haplotype analysis of GRMZM5G833774(ZmPP2C)and GRMZM2G478568(ZmNAS3)containing chr1.S_8274996 controlling plant height and chr1.S_259817863 controlling R/S ratio,respectively.(A,D)LD heat map of GRMZM5G833774(A)and GRMZM2G478568(D).The triangle represents the genomic region of the candidate gene.(B,C,E and F)Marker information and haplotype analysis of GRMZM5G833774(B,C)and GRMZM2G478568(E,F).Violin plot describes the distribution of a given trait for each haplotype.Different letters represent difference at P<0.05.
4.2.Co-localization with genes involved in phytohormone regulation
Zn deficiency signal may be linked with hormone signaling pathway that indirectly regulates downstream Zn-responsive genes,including genes responsible for Zn acquisition,uptake and transport and genes involved in Zn chelator biosynthesis and release[71].It is reported[72]that differentially expressed genes in Zn-deficient leaves or roots are up-regulated by Zn deficiency,including genes likely involved in auxin and ethylene signaling.In the present study,several genes known to be involved in ethylene regulation were detected by QTL mapping and GWAS.

Fig.5.Associated regions and haplotype analysis of GRMZM2G092131(ZmSDG40)and GRMZM2G034015 containing chr7.S_144602513 controlling R/S ratio and chr9.S_59587835 controlling shoot and root dry weights,respectively.(A,D,and G)LD heat map of GRMZM2G092131(A)and GRMZM2G034015(D,G).The triangle represents the genomic region of the candidate gene.(B,C,E,F,H,and I)Marker information and haplotype analysis of GRMZM2G092131(B,C)and GRMZM2G034015(E-F,H-I).Violin plot describes the distribution of a given trait for each haplotype.Different letters represent difference at P<0.05.
ZmARF7,encoding auxin response factor 7 in maize,was colocalized with the overlapping region of qKC-PH3-1 and qKCSDW3-1 on chromosome 3,and also located in the 57-kb region flanking the significant SNP chr3.S_1613904(Figs.3H,S6A;Table 2).AtARF18 and OsARF1 are orthologs of ZmARF7 in Arabidopsis and rice,respectively[73].Both AtARF18 and OsARF1 are proposed to be repressors[74,75].It is reported[76]that transgenic rice carrying an antisense of OsARF1 showed stunted growth,poor vigor,short curled leaves and sterility,suggesting that OsARF1 may influence fertility and regulate genes controlling growth and reproduction.In agreement with these findings,expression of ZmARF7 was down-regulated in both shoots and roots.A greater decrease in down-regulation was found in the Zn-efficient parent CI7,not in Zn-inefficient parent K22(Fig.S7A,B).These findings suggest that ZmARF7 may be associated with differences in the ability to tolerate Zn deficiency in maize.
The significant SNP chr4.S_230734637 was associated with the candidate gene GRMZM2G318689 encoding ethylene sensor ETR5 in maize(Table 2).The locus qKC-SDW5-1 was co-localized with GRMZM2G102601 and GRMZM2G102754,which encode ethylene receptor 14(ZmESR14)and ethylene-insensitive protein 2(ZmEIN2),respectively(Fig.2).Ethylene receptors in Arabidopsis act as negative regulators of the ethylene-signaling pathway[77].The homolog of ZmETR5 in Arabidopsis encodes AtEIN4 proteins which activate CTR1,leading to a suppression of the downstream ethylene signaling component EIN2[78].The C-terminal end of EIN2 allows the ethylene signal to reach the downstream components EIN3 and EILs which activate the expression of ethylene target genes[79,80].Thus,ZmESR14,ZmETR5,and ZmEIN2 may be involved in regulation of ethylene signaling in maize.
4.3.Co-localization with genes responsible for Zn uptake and transport
In addition to the genes involved in ethylene and auxin regulation pathways,genes encoding Zn chelators and Zn transporters were identified.GRMZM2G478568,which was characterized as ZmNAS3 in maize,was located in the 50-kb region flanking the significant SNP chr1.S_259817863(Fig.3).A previous study[21]has shown that NAS catalyzes the synthesis of NA.On one hand,NA acts as a metal chelator and participates in internal transport via stable complexes Zn-NA distributed in the cytosol and phloem as well as in the xylem[81].On the other hand,NA can be a precursor of MAs(mugineic acid family phytosiderophores)which include deoxymugineic acid(DMA),and be involved in long-distance transport of DMA-Zn in plants[82-84].The expression of ZmNAS3 and its paralog ZmNAS5 are consistently up-regulated under Zn deficiency[3].Zn deficiency greatly increases the expression of OsNAS3 in the shoot and root[59].Activated OsNAS3 expression also leads to increased tolerance to both Fe and Zn deficiencies[60].
Two QTL(qKC-R/S10-1,qKC-R/S10-3)detected on chromosome 10,were co-localized with ZmYSL11(GRMZM5G812538)(Fig.2).YS-like proteins,such as AtYSL1 and OsYSL2,are reported[85-88]to transport metals via NA complexes,including Zn(II)-NA,Cu(II)-NA and Fe(II)-NA,Mn(II)-NA.The locus qKC-R/S10-3 was co-localized with candidate gene ZmVIT1(GRMZM2G107306)(Fig.2).The VIT family proteins in rice have been implicated in Zn transport.OsVIT1 and OsVIT2 function to transport Fe(II),Zn(II)and Mn(II)into vacuoles in yeast[89].OsVIT1 and OsVIT2 are suggested[89]to function in Fe/Zn translocation between flag leaves(source organ)and grain(sink organ).ZmYSL11 was expressed and significantly induced under Zn deficiency only in shoots(Fig.S7C),and greater up-regulation was found in the Zn-efficient parent CI7,suggesting that ZmYSL11 may be involved in Zn transport in response to Zn deficiency in maize.
4.4.Candidate genes associated with tolerance to abiotic stress
ZmPP2C(GRMZM5G833774)contained the significant SNP that was co-localized with the QTL reported to be associated with Zn deficiency in maize(Fig.3B;Table 2).The serine/threonine protein phosphatase 2Cs(PP2Cs)have been implicated[90]as negative modulators of protein kinase pathways.Abscisic acid(ABA)is a phytohormone involved in adaption to environmental stress and regulation of plant development.ZmPP2C overexpression attenuated ABA inhibition of seed germination and root growth of transgenic plants[91].ZmPP2C-A10 interacts with ZmPYL ABA receptors and ZmSnRK2 kinases,suggesting that ZmPP2C-A10 is involved in mediating ABA signaling and functions as a negative regulator of drought tolerance in maize.Our results showed that ZmPP2C was significantly up-regulated in shoots(Figs.S9,S10),further suggesting that ZmPP2C is associated with Zn deficiency tolerance.
The most significant SNP associated with R/S under Zn deficiency corresponded to the gene GRMZM2G092131 which encodes a SET domain group protein in maize(Fig.3H;Table 2).Histone lysine modification is catalyzed by histone lysine methyltransferase SET DOMAIN GROUP(SDG)proteins which harbor an evolutionarily conserved SET domain as catalytic module[92].SDG725 encodes a H3K36 methyltransferase,and its down-regulation causes wideranging defects in rice,including dwarfism,shortened internodes,erect leaves,and small seeds[93].These phenotypes are very similar to those of Zn-deficient plants.Chromatin immunoprecipitation analyses[93]showed that levels of H3K36me2/3 were reduced in chromatin in some regions of these brassinosteroid-related genes in SDG725 knockdown plants,and that SDG725 protein is able to directly bind to these target genes.In the present study,ZmSDG40 was highly Zn deficiency-inducible in both shoots and roots(Figs.S9,S10),suggesting that ZmSDG40 may be involved in the mechanism underlying tolerance to low-Zn stress.
Two candidate genes,characterized as ZmTERF18(GRMZM2G017355)and ZmTERF19(GRMZM2G017429)in maize[94],were located in the overlapping region of qKC-RDW5-1 and qKC-ZnSc5-1.Recently[94],stress-related cis-element analysis in maize suggested that ZmTERF19 might regulate 11 drought-,high-salt-and cold responsive elements.ZmTERF8,which was identified[94]as a paralog of ZmTERF18 and ZmTERF19,was down-regulated in darkness by more than twofold,suggesting that mTERF genes are generally required for chloroplast development and photosynthesis in maize.These findings indicate that mTERF genes in maize can be influenced by environmental signals and respond to abiotic stress.These candidate genes participating in hormone signaling pathways and/or abiotic stress responses may also be involved in the disorder of maize plant hormone regulation under low-Zn stress.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Jianqin Xu:Formal analysis,Visualization,Writing-original draft,Writing-review&editing.Zhongfu Ni:Resources,Supervision,Writing-review & editing.Fanjun Chen:Formal analysis,Writing-review & editing.Xiuyi Fu:Formal analysis,Visualization.Futong Yu:Conceptualization,Funding acquisition,Supervision,Writing-review & editing.
Acknowledgments
This study was supported by the National Key Research and Development Program of China(2016YFD0200405).The authors thank Professor Xiaohong Yang(China Agricultural University)for providing the RIL population and the association mapping population.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2022.05.004.
杂志排行
The Crop Journal的其它文章
- Profiling seed soluble sugar compositions in 1164 Chinese soybean accessions from major growing ecoregions
- The mitochondria-localized protein OsNDB2 negatively regulates grain size and weight in rice
- Delaying application time of slow-release fertilizer increases soil rhizosphere nitrogen content,root activity,and grain yield of spring maize
- Effects of increasing panicle-stage N on yield and N use efficiency of indica rice and its relationship with soil fertility
- Yield sustainability of winter wheat under three limited-irrigation schemes based on a 28-year field experiment
- Identification of a stable major QTL for fresh-seed germination on chromosome Arahy.04 in cultivated peanut(Arachis hypogaea L.)
