Identification of a stable major QTL for fresh-seed germination on chromosome Arahy.04 in cultivated peanut(Arachis hypogaea L.)
2022-12-02MoningZhngQingZngHuLiuFiynQiZiqiSunLijunMioXionLiChnyuLiDbingLiuJunjiGuoMngyunZhngJingXuLiShiMngiTinWnzhoDongBingynHungXinyouZhng
Moning Zhng,Qing Zng,Hu Liu,Fiyn Qi,Ziqi Sun,Lijun Mio,Xion Li,Chnyu Li,Dbing Liu,Junji Guo,Mngyun Zhng,Jing Xu,Li Shi,Mngi Tin,Wnzho Dong,Bingyn Hung,*,Xinyou Zhng,*
a Innovation Base of Zhengzhou University,Henan Academy of Agricultural Sciences/Henan Academy of Crop Molecular Breeding/State Industrial Innovation Center of Biological Breeding/Key Laboratory of Oil Crops in Huang-Huai-Hai Plains,Ministry of Agriculture/Henan Provincial Key Laboratory for Oil Crops Improvement,Zhengzhou 450002,Henan,China
b School of Agricultural Sciences,Zhengzhou University,Zhengzhou 450002,Henan,China
c State Key Laboratory of Cotton Biology,Institute of Cotton Research,Chinese Academy of Agricultural Sciences,Anyang 455000,Henan,China
d College of Applied Science and Technology of Hainan University,Danzhou 571737,Hainan,China
e College of Agriculture,Henan University of Science and Technology,Luoyang 471023,Henan,China
Keywords:Peanut QTL mapping Fresh-seed germination(FSG)Dynamic germination KASP
ABSTRACT Fresh-seed germination(FSG)impairs peanut production,especially in areas where the peanut harvest season coincides with rainy weather.Developing FSG-resistant cultivars by molecular breeding is expected to mitigate yield loss and quality impairment caused by FSG.However,the genetic control of FSG awaits elucidation.In this study,FSG at 1,3,5,7,and 9 days post-imbibition in three environments were tested,and quantitative-trait loci(QTL)associated with FSG were mapped in a peanut recombinant inbred line population by leveraging existing high-density peanut genetic maps.Of 24 QTL identified in 13 linkage groups,qFSGA04 was a stable major QTL on linkage group 04(LG04).It was consistently detected in five germination stages and three environments.By designing and validating DNA markers in the confidence interval of qFSGA04,we identified one single-nucleotide polymorphism and one InDel closely associated with FSG that could be used as linked markers for FSG resistance in peanut breeding.
1.Introduction
Peanut(Arachis hypogaea L.),a source of edible oil and protein,is native to South America and naturalized throughout the temperate region of the world[1-4].Owing to extensive selection for high yield and improvement in quality traits during the domestication of wild peanut and the development of modern commercial cultivars,an undesirable trait of fresh seed germination(FSG)prior to harvest has become a growing problem,especially in regions where the harvest season coincides with wet weather[5,6].It is estimated[5]that FSG can cause up to 50% yield reduction and severely impair nut quality.FSG-affected peanut seeds are susceptible to pathogen infection,which renders them vulnerable for seed contamination by aflatoxin or other mycotoxins detrimental to human health[7].
Studies of traits associated with FSG,including seed dormancy,preharvest sprouting(PHS),and in situ germination,have been conducted but have focused mainly on grain crops.Construction of recombinant inbred line(RIL)populations using parents with contrasting values for agronomic traits,coupled with map-based cloning,has been commonly used to study the molecular mechanisms underlying PHS in cereal crops,as exemplified by the successful cloning of several causal genes of PHS,such as AlaAT of Qsd1[8,9]and MKK3 of Qsd2-AK[10]in barley,TaPHS1 of Qphs.pseru-3AS[11-13],and MKK3 of Phs1[14,15]in wheat.Despite its economic significance to the peanut industry,FSG research on peanuts has lagged,owing mainly to a lack of genetic variation and suitable DNA markers,in addition to the peculiarity of underground peanut fruit maturation,which makes it difficult to observe and characterize FSG.
The rapid development of molecular breeding methods and recent advent of deep-sequencing technologies have greatly facilitated precision breeding and trait improvement in peanut.Wholegenome sequences of multiple peanut types,including cultivated tetraploid species and their progenitor species,are now publicly available[1,3,4],enabling genome-wide molecular breeding for peanut traits[16-18].Exploiting genome sequence data,singlenucleotide polymorphism(SNP)markers have been widely used in the development of high-density genetic maps owing to their abundance,rich polymorphism,and wide distribution in the genome,facilitating quantitative-trait loci(QTL)mapping for complex peanut traits such as yield[19,20],quality[21,22],and disease resistance[23,24].
Two peanut QTL,qfsd-1 and qfsd-2,associated with FSG have been reported[5]from QTL-mapping studies in an F2population derived from two Spanish-type parents with contrasting FSG.The first was located on chromosome A05,and the second on chromosome B06.Advancing the F2to a RIL population led to the discovery of two additional QTL located on B05 and A09 with marker intervals spanning 2.4 Mb and 0.74 Mb,respectively[6].The parents in these studies were all Spanish types,and the genetic influence of botanic type on FSG remains unknown.FSG is a dynamic process best observed over a period of time,and environmental factors strongly influence the severity of FSG.
In the present study,QTL for FSG were identified in a RIL population of 521 lines derived from a cross between a Spanish-type cultivar and a runner-type cultivar in five germination stages of three growth environments.The objectives of this study were(1)to identify the differences of peanut fresh seed germination between parents and within RIL population;(2)to conduct QTL mapping of peanut fresh seed germination;(3)to develop KASP(kompetitive allele-specific PCR)markers for QTL with stable major genetic effects.
2.Materials and methods
2.1.Plant materials
A RIL population of 521 lines was developed by single-seed descent from a cross between female parent Yuanza 9102(YZ9102)and male parent wt09-0023.YZ9102 is a Spanish-type cultivar released in 2002 by Henan Academy of Agricultural Science,and is high-yielding but susceptible to FSG,with FSG up to 85%[25].wt09-0023 is a runner-type high-oleic and FSG-resistant cultivar with FSG less than 10%,provided by Dr.Kim Moore(AgResearch Consultants Inc.,Tifton,GA,USA).F2:9-F2:11plants of the RIL were used for phenotyping.To validate diagnostic markers,a panel of 96 cultivars(Table S1)randomly selected from four botanical types was combined into a verification group[26].
2.2.Field trials and phenotyping
The RIL population and its parents were planted in Zhengzhou(Henan province),Ledong(Hainan province),and Xinxiang(Henan province),in May 2020,November 2020,and May 2021,respectively.These trials are referred to here as Zhengzhou 2020,Ledong 2020,and Xinxiang 2021.All field experiments were conducted in a randomized complete block design with two replicates.Each line was planted in a plot with two rows,each row with 20 seeds 20 cm apart,and a row spacing of 30 cm.Field-trial management followed standard agricultural practice[27].
At harvest,mature pods from each line were immediately shelled and 90 mature kernels with intact seed coats and uniform size were selected,washed with sterile water,and placed on filter papers in a Petri dish.Each treatment consisted of three dishes containing 30 seeds with three replications.Approximately 50 mL of sterile water was added to each dish to maintain a moist germination environment.The dishes were maintained in a chamber with constant temperature of 28±2°C in the dark.The number of germinated seeds was recorded at 1,3,5,7,and 9 days postimbibition(DPI).
Fresh peanut pods harvested in Ledong 2020 were also immediately collected for whole-shell germination testing.One hundred mature pods with intact shells were harvested from each line and immediately immersed in water.Germinated seeds were counted at 3 DPI.
2.3.Statistical analysis of phenotypic data
Statistical parameters,distribution of phenotypic data,and error distributions were estimated with QTL IciMapping software[28].Broad-sense heritability across three environments for FSG in each investigation stage was calculated as.
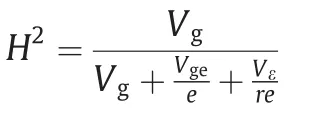
where e represents the number of environments and r the number of replicates.FSG rate was recorded as:number of fresh seeds with radicle protruding through the seed coat/total number of fresh seeds×100%.
2.4.QTL mapping
Based on our previous study[29],a linkage map containing 5120 SNPs in 20 linkage groups was used for QTL mapping.Genotyping was conducted by digesting genomic DNA with EcoRI and DNA sequencing on an Illumina HiSeq 4000(Illumina,Inc.,San Diego,CA,USA)platform.The sequencing depths for the parents and RILs were approximately 25×and 5×,respectively.The genetic map covered 3179 cM with a mean marker interval of 0.6 cM.QTL for FSG in each environment and stage were detected based on the replication mean using QTL IciMapping[28,30],setting the mapping step size as 1 cM and the logarithm of odds(LOD)threshold as 3.0.The QTL region of linkage group 04(LG04)was drawn with MapChart 2.3[31].QTL were named as:q+the abbreviated trait name+linkage group number,or named as q+the abbreviated trait name+linkage group number.+a number designating one of multiple QTL in a single linkage group,following the International Rules of Genetic Nomenclature[11].
2.5.KASP marker development and validation
Using the sequences of the flanking markers,detected QTL were mapped to the reference genomes of A.hypogaea cv.Tifrunner(https://www.peanutbase.org/).SNPs polymorphic between the parents in the region of the putative QTL were converted to KASP markers[32].One hundred RILs with lowest and 100 RILs with highest FSG scores were selected for marker validation.The other verification group comprised 96 cultivars.The 200 RILs and 96 cultivars were then genotyped with the KASP markers and classified by their genotype similarity to YZ9102 or wt09-0023.
3.Results
3.1.Phenotypic analysis of FSG in RIL population
FSG did not follow a normal distribution in the RILs,but errors appeared to be normally distributed(Table 1;Fig.S1).The FSG rate of YZ9102 was higher than that of wt09-0023 in all three environments(Table 1).The FSG rate ranged from 0 to 100%.The coefficient of variation(CV)of FSG ranged from 0.51% to 1.14%among the three environments,and the broad-sense heritability(H2)from 0.83 to 0.84.Significant environmental effects on FSG were observed:the mean FSG was significantly higher in Ledong 2020 than in the other two environments(Table 1).

Table 1Phenotypic variation of FSG in parents and RIL population across five germination periods in three environments.
3.2.Identification of QTL for FSG
A total of 24 QTL for FSG with LOD score above 3.0 were mapped during five germination periods under three growth environments(Table S2).These QTL had phenotypic variation explained(PVE)of 0.82%-15.83%,and LOD values of 3.0-51.4.They were located on 13 LGs,i.e.,LG01 and LG11,LG02 and LG12,LG04 and LG14,LG05,LG06 and LG16,LG07 and LG17,LG10 and LG20.
Among the 24 QTL,qFSGA04 on LG04,with PVE of 8.0%-14.9%and LOD of 16.2-30.1,was consistently detected(Fig.1;Table S2). Flanked by markers A04.100806014 and A04.101505919,qFSGA04 was assigned to a 0.70-Mb(100.806-10 1.506 Mb)genomic region of pseudomolecule A04 of Tifrunner.The confidence interval of qFSGA04 was 50.5-51.5 cM with the left flanking marker A04.100070836 and right marker A04.102865415(Fig.2).
3.3.Gene annotation in the qFSGA04 confidence interval
The confidence interval of qFSGA04 contained 54 genes(Table S3).In this interval 415 SNPs and 41 InDels were found,of which one SNP and one InDel were located in the exons of Arahy.N8MLZ0 and Arahy.ISSF38,respectively(Tables 2,S4).The SNP in Arahy.04:101089930 was located in an exon of an unknown gene(Arahy.ISSF38).The InDel in Arahy.04:100184540 was located in an exon of a gene(Arahy.N8MLZ0)encoding ribosomal RNA small subunit methyltransferase,NEP1(Tables 2,S3).
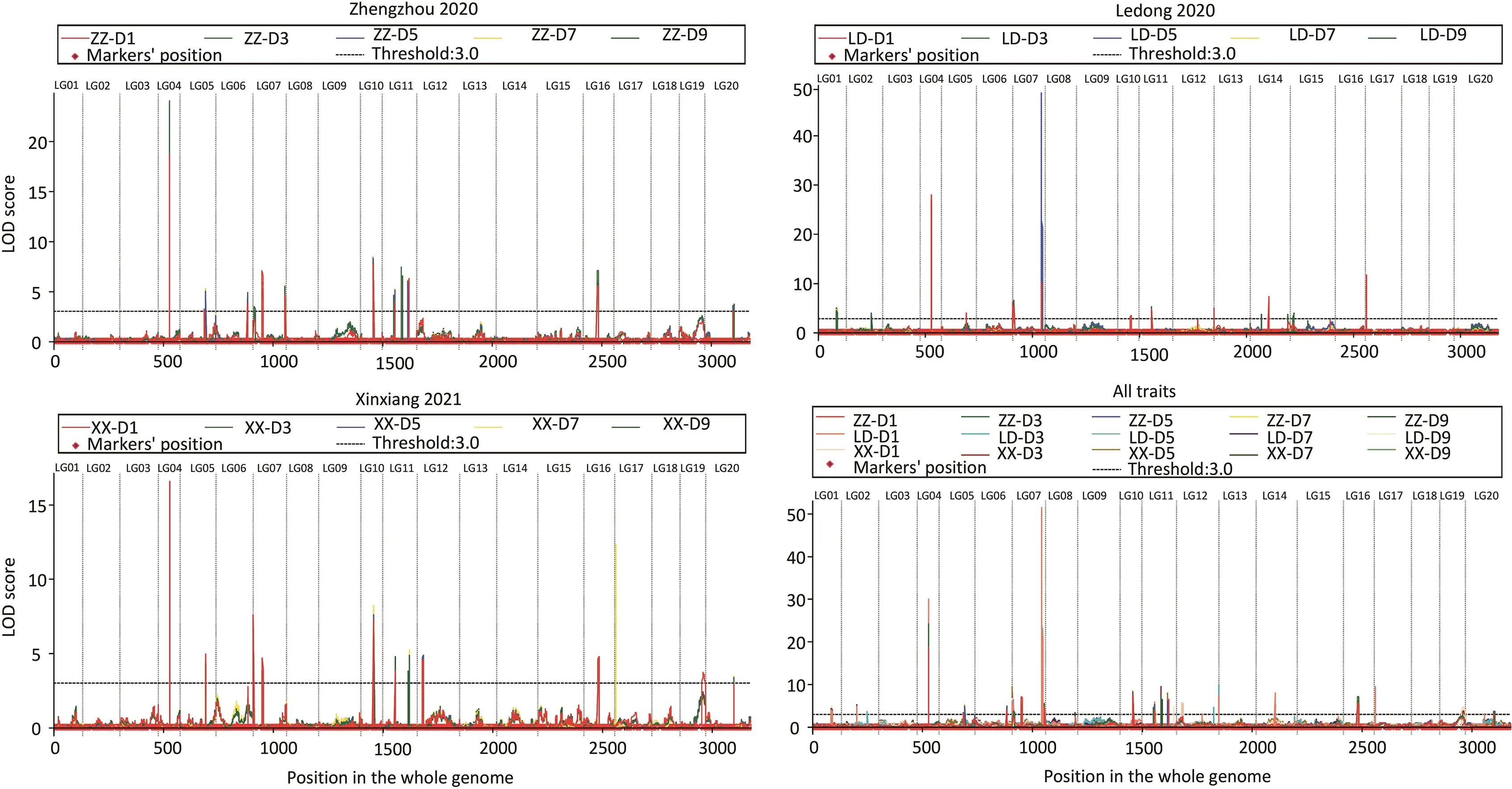
Fig.1.QTL mapped on LG04.LOD curves of FSG across the whole genome under five germination periods in three environments.ZZ,LD,and XX represent respectively Zhengzhou 2020,Ledong 2020,and Xinxiang 2021.D1,D3,D5,D7,and D9 represent the recording date at 1,3,5,7,and 9 days post-imbibition.
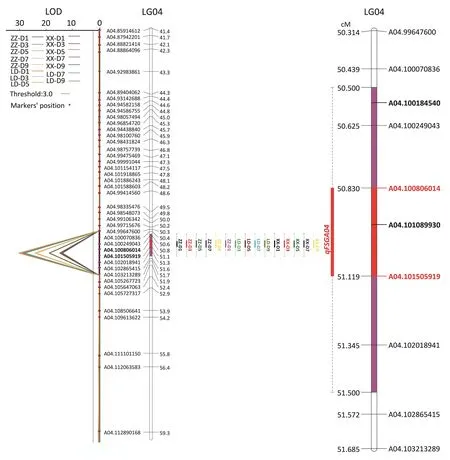

Table 2Nucleotide types of SNPs located in candidate genes.
3.4.Marker validation for FSG
KASP markers were designed to identify missense mutations associated with the identified SNP and InDel,and validated in the verification groups(Fig.3).Among all the validation materials,one SNP in Arahy.04:101089930 and one InDel in Arahy.04:100184540 were found to be associated with FSG.T:T(YZ9102 type)and G:G(wt09-0023 type)represented the genotype at Arahy.04:101089930,while T:T(YZ9102 type)and/:/(wt09-0023 type)represent the InDel at Arahy.04:100184540.In both indoor and field germination tests,the mean FSGs of T:T were significantly higher than those of G:G at Arahy.04:100184540,and the mean FSGs of T insertion were significantly higher than those of the wild type(without the T insertion)at Arahy.04:100184540(Fig.3A-D;Table 2).The same trend was observed in the indoor germination test of the verification group(Fig.3E;Table 2).The corresponding nucleotide in the low-FSG parent wt09-0023 at Arahy.04:101089930 changed from T to G,resulting in the alteration of the putatively encoded amino acid residue from asparagine(N)to lysine(K)(Table 2).The T insertion in the YZ9102 type at Arahy.04:100184540 resulted in a frame shift without premature termination of translation.

Fig.3.KASP marker validation of SNP(Arahy.04:101089930)and InDel(Arahy.04:100184540).(A-C)Phenotypic difference between two base types at the SNP and InDel loci in the 200 RILs in Zhengzhou 2020,Ledong 2020,and Xinxiang 2021 respectively.(D)Phenotypic difference between two base types at the SNP and InDel loci in the 200 RILs in the field germination experiment.(E)Phenotypic difference between two base types at the SNP and InDel loci in the 96 cultivars of the verification group.
4.Discussion
This study was performed by leveraging an existing highdensity linkage genetic map recently constructed in our laboratory from the peanut RIL population that has 5120 markers spanning 3184 cM and a mean marker interval size of 0.6 cM[29].The population of 521 RILs is larger than those used in previous studies[5,6,19-24].Given that germination of fresh peanut seeds depends on a moist and warm environment[5,6],the germination tests conducted in this study under both laboratory and field conditions enabled a comprehensive analysis and quantification of FSG.
In the present study,24 QTL associated with peanut FSG have been detected at different germination stages in multiple environments.Except for qFSGA05 on A05,the other 22 QTL were all located on both A and B sub-genomes(Table S2),for example,A01 and A11,A02 and A12,A04 and A14,A06 andA16,A07 and A17,A10 and A20.This represents a new advance following two previous studies on peanuts that revealed four putative QTL intervals for fresh-seed dormancy using a F2and its derived RILs populations[5,6],and in cereals such as wheat[8],barley[12],and rice[13]where multiple QTL have also been found to be associated with PHS and seed dormancy traits.Consistent with these previous studies,our present study shows that the fresh-seed dormancy,in situ germination,and FSG traits are quantitative traits that are regulated by multiple genes.
In our study,a stable major QTL interval was located on Arahy.04.This is despite the previous findings in Spanish-type peanut that two QTL were mapped onto A05 and B06 in an F2population[5]while two other QTL were detected on A09 and B05 in its derived RIL population[6].Recently[33],a QTL mapping study of peanut seed dormancy revealed two major QTL on A04 and A05 in a RIL population of 164 lines.The location of this major QTL on A04 is similar to our findings in the present study.Herein we have further designed two KASP markers based on one SNP and one InDel in candidate genes associated with FSG,which were subsequently validated as highly correlated with FSG in both the RIL population and verification group,indicating their potential for use in marker-assisted breeding for FSG amelioration in peanuts.
The regulation of FSG is a complex process with many influencing factors.Numerous studies[34]have shown that the germination of fresh seeds during harvest is affected by intrinsic factors such as seed phytohormones,carbohydrates,reactive oxygen species(ROS),and nitric oxide(NO),as well as exogenous factors such as environmental humidity,temperature,and oxygen concentration.In the present study,KASP markers designed for two candidate genes,Arahy.N8MLZ0 and Arahy.ISSF38,were validated.The gene Arahy.ISSF38 has not been annotated with a function and the other gene,Arahy.N8MLZ0,has been annotated as encoding a ribosomal RNA small subunit methyltransferase NEP1 that is involved in ribosome biogenesis by directly or indirectly interfering with methylation reactions in the early steps of pre-rRNA processing necessary for the generation of 40S ribosomal subunits[35].The connection of NEP1 with FSG invites further study.
5.Conclusions
A major stable QTL qFSG04 for peanut FSG was identified.Within its confidence interval,one SNP and one InDel were identified in candidate genes,and their association with FSG was validated using KASP markers.These markers could be applied to genetic improvement of peanut FSG resistance.The candidate genes could be used for genetic engineering to improve seed germination traits.
CRediT authorship contribution statement
Maoning Zhang:Investigation,Formal analysis,Validation,Visualization,Writing-Original Draft.Qing Zeng:Investigation,Writing-Review & Editing.Hua Liu:Resources,Software,Data Curation.Feiyan Qi:Resources,Software,Data Curation.Ziqi Sun:Resources,Software,Data Curation.Debing Liu:Investigation.Lijuan Miao:Investigation.Xiaona Li:Investigation.Chenyu Li:Investigation.Junjia Guo:Investigation.Mengyuan Zhang:Investigation.Lei Shi:Investigation.Mengdi Tian:Investigation.Jing Xu:Resources.Wenzhao Dong:Resources.Bingyan Huang:Conceptualization,Methodology,Writing-Review & Editing,Supervision,Project administration,Funding acquisition.Xinyou Zhang:Conceptualization,Methodology,Writing-Review &Editing,Supervision,Project administration,Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by China Agriculture Research System(CARS-13),Henan Provincial Agriculture Research System,China(S2012-5),Major Science and Technology Projects of Henan Province(201300111000),and the Henan Provincial R&D Projects of Interregional Cooperation for Local Scientific and Technological Development Guided by Central Government(YDZX20214100004191).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2022.03.012.
杂志排行
The Crop Journal的其它文章
- Profiling seed soluble sugar compositions in 1164 Chinese soybean accessions from major growing ecoregions
- The mitochondria-localized protein OsNDB2 negatively regulates grain size and weight in rice
- Integrated linkage mapping and genome-wide association study to dissect the genetic basis of zinc deficiency tolerance in maize at seedling stage
- Delaying application time of slow-release fertilizer increases soil rhizosphere nitrogen content,root activity,and grain yield of spring maize
- Effects of increasing panicle-stage N on yield and N use efficiency of indica rice and its relationship with soil fertility
- Yield sustainability of winter wheat under three limited-irrigation schemes based on a 28-year field experiment
