Comparison of 16S rRNA Gene Primers on Studying Microbial Community Composition in Bottom Water and Sediment of Artificial Reefs in Laoshan Bay, China
2022-10-24FANGGuangjieYUHaolinSHENGHuaxiangTANGYanliandLIANGZhenlin
FANG Guangjie, YU Haolin, SHENG Huaxiang, TANG Yanli, *, and LIANG Zhenlin
Comparison of 16S rRNA Gene Primers on Studying Microbial Community Composition in Bottom Water and Sediment of Artificial Reefs in Laoshan Bay, China
FANG Guangjie1), YU Haolin1), SHENG Huaxiang1), TANG Yanli1), *, and LIANG Zhenlin2)
1),,266003,2),,264200,
Marine microorganisms are indispensable regulators of nutrient cycling and energy flow, which are crucial for artificial reefs (ARs) ecosystems. However, little is known about the microbial communities in the bottom water and sediment of ARs. Studies of microbial diversities have greatly advanced due to the development of high-throughput sequencing technologies, whereas the re- sults may vary significantly due to the primers’ choice. This study investigated the influences of two 16S ribosomal RNA gene primer choices (V4 and V3-V4) on microbial community compositions and structures. The results showed that the taxonomic assignment de- tected by primer V3-V4 was higher compared with that obtained by primer V4, whereas microbial community compositions had strong correlations between the two primers. Microbial beta diversities of ARs can be uncovered by both primers, but the relation- ships between communities and environmental parameters were inconsistent. The performances of the two primers for water samples were highly consistent, but the inconformity was evident for sediment samples. Given the relatively lower taxonomic classification of primer V4 for sediment samples, primer V3-V4 was recommended for later studies. With the development and advancement of ARs in China, our findings provide a meaningful reference for ecologists focusing on the microbial diversities and ecological functions of these artificial habitats in the future.
16S rRNA; hypervariable region; ARs; bacteria; archaea
1 Introduction
Marine microorganisms are dominant drivers of global biogeochemical processes, including energy, carbon, andnutrient cycles (Sunagawa, 2015). Despite the impres- sive advances that have been attained in assessing the di- versity of marine microbes, enormous challenges remain due to the scale and variability of marine environments (Ha- mady and Knight, 2009). High-throughput sequencing has facilitated significant improvements in understanding the mechanisms of marine microbial ecology (Langille, 2013). As a predominant and reliable method to assess the diversity of marine microorganisms, 16S ribosomal RNA (rRNA) gene sequencing technology with specific primers has been widely used (Thompson, 2017). However, the results generated by the 16S rRNA gene sequence may be biased owing to the unexpected variable diversity of mi- crobiomes, large scale and complexity of microbial habi- tats, sample and nucleic acid extraction technologies, and diverse sequencing approaches (Hazen., 2013; Per- rine, 2014). With the standardization of sampling te- chnologies and 16S rRNA gene sequencing procedures, the choice of primers is one of the most critical reasons of se- quencing biases (Parada, 2016).
An abundant number of papers compared the perfor- mance of various primers, whereas the choice of an opti- mal primer still is under debate (Nitin., 2017; Fadeev., 2021). The main confusing origins are the incongru- ent taxonomic resolutions and relative abundances obtain- ed by different primers (Claesson, 2010; Birtel, 2015). At present, no standard procedure is available to guide researchers in selecting the optimal primer for their studies. The primer choice mainly depends on scientific questions (Choi, 2017), targeted research species (Pa- rada, 2016), and sequencing technology (Hazen, 2013). Studies of comparisons among primers have been divided into two stages. In the first stage, researchers main- ly focused on the comparisons of microbial identifications of several primers. Youssef(2009) compared the per- formance of eight primers with a nearly full-length primer and suggested that three primers (V4, V5-V6, and V6-V7) were preferred choices in richness estimates. Cai(2013) studied the effectiveness of seven primers for microbial di- versity metrics in municipal wastewater treatment plants, and the results revealed that primer V3-V4 was highly re- commended. Parulekar(2017) analyzed the bacte-rial compositions related to phytoplankton blooms by two primers and advocated that multiple primers should be ap- plied. In the second stage, seeking a better primer to ex-plain specific ecological phenomena was the preferred orien- tation. Primer V5-V7 was advocated for studying the bac- terial communities and functions of aging flue-cured toba- cco (Wang, 2018). Two primers showed significant- ly different microbial communities in the North Atlantic, and primer V4-V5 was recommended because of its recog-nition of numerous archaea (Willis., 2019). Highly con- sistent results profiled by primers V3-V4 and V4 were re- ported when characterizing the taxonomic and functions of cervical microbiota (Onywera and Meiring, 2020). Ac- cordingly, more attention should be paid to select 16S gene primers, given that different primers provide biased diver- sity results (Soergel, 2012), especially in answering particular ecological questions.
Artificial reefs (ARs) are human-made structures deploy- ed in marine waters to provide artificial habitats and shel- ters for aquatic organisms (Lima, 2019). ARs have been universally regarded as an effective tool for fish at- tractions and fishery management (Bohnsack and Suther- land, 1985). Given the rapid decline of fishery resources in coastal water, ARs have been formally deployed to en- hance fishery stock production along the coast of China since 2001 (Sun, 2017). Studies about sediment en- vironment and benthos associated with ARs have become popular in recent years (Chen, 2019). However, stud-ies on bacterial communities in ARs have been ignored. To our knowledge, there is only one paper evaluating the ef- fects of ARson the bacterial communities in the sediment of Bohai Bay (Wang., 2019). Laoshan Bay is located in the southern Shandong Peninsula, Yellow Sea, China, with high-quality aquatic environment and fishery resources. Rock reefs (RR) were deployed near the shore for aqua- culture of sea cucumber () and aba- lone (); concrete reefs (CR) were deployed outside the RR for recreational fishing of black rockfish () and perch (). The types of ARs that we studied are represen- tatives in the Yellow Sea, China, and they can provide mean- ingful suggestions for the choice of primer.
Selecting a suitable primer is crucial to comprehending the microbes in the bottom water and sediment of ARs in Laoshan Bay completely. Hypervariable region V4 is re-garded as the best region for molecular biology studies, es- pecially at the phylum level (Yang, 2016). Based on previous studies on marine microorganisms, two univer-sally used primers, primers V4 and V3-V4, were selected toamplify the 16S rRNA gene. Hypervariable V4 primer isrecommended by ‘The Earth Microbiome Project’ because of its broad coverage of sequences for most bacterial taxa and numerous archaea (Thompson, 2017; Liu, 2019). Hypervariable V3-V4 primer is applied universally because it can lead to rich information and high accu- racyof taxonomic classification (Cai, 2013; Li, 2020). In this study, the main objectives were 1) to com- pare the microbial diversities and community structures obtained by the two primers; 2) to exhibit the properties and suitability of primers applied for the microbial ana- lysis in ARs and to provide suggestions for the future pri- mer choice.
2 Materials and Methods
2.1 Study Sites and Sample Collection
All water and sediment samples were collected from ARs in Laoshan Bay, China, in May 2020 (Fig.1). At each site, triplicate bottom water and sediment subsamples were ran-domly obtained. A total of 16L bottom water and sufficient sediment samples were obtained from four sites, namely, RR, transition area (TA), CR, and adjacent area (AA), by a water sampler and grab sediment sampler. These samples were ample to process DNA extraction and physicoche- mical parameter assay. For molecular sequencing analysis, 12L bottom water was filtered through 0.2μm pore size, 47mm polycarbonate filters (Millipore) and then stored at −80℃ for later DNA extraction. Other water samples were applied to measure the physical and chemical properties. Sediment samples were divided into two parts: one was stored at −80℃ for 16S rRNA gene sequencing, and the other was stored at −20℃ for the analysis of environmen- tal parameters.
Fig.1 Sampling area of ARs in Laoshan Bay, Yellow Sea, China (In the minimap is the rectangular box in red).
2.2 Measurements of Environmental Parameters
Water environmental parameters, such as sampling depth, temperature, salinity (Sal), dissolved oxygen, pH, and chlo- rophyll-, were measured by YSI PRODSS (USA). Water transparency (Trans) was measured with a Secchi Disc. Water turbidity (Turb) was measured by a Turb 430 IR (Xylem Analytics, Germany). Chemical oxygen demand (COD), and five nutrients in seawater, including nitrate (NO3-N), nitrite (NO2-N), ammonium (NH4-N), active pho- sphate, and active silicate (SiO3), were determined based on the GB/T 12763-2007 (State Bureau of Quality and Te- chnical Supervision of China, 2007). For the sediment sam- ples, the mean particle size (Par) was measured by a Mas- tersizer 3000 (Malvern, UK). The sediment was oven driedat 105℃ for 72h, and bulk density (BD) was measured by dividing the dried mass by the volume of the sediment (Lee, 2021). Water content (WC) was obtained by mea- suring the weight loss after the sediment was dried at 70℃ for at least 72h until a constant weight was observed. Or- ganic matter content (OM) was measured by burning the sediment to ash at 550℃ for 4h (Heiri, 2001). Mud content (MC) was determined following the method forMC-dry sediment (Eleftheriou, 2013). The sediment pH, Sal, and electrical conductivity (EC) were tested by examining the mixture of deionized water and sediment with a pro- portion of 1:2.5 (W/V) (Liao, 2014).
2.3 DNA Extraction, Amplification, and Sequencing
The genomic DNA of water and sediment samples in the ARs was extracted with the FastDNA SPIN Kit for Soil (MP Biomedicals, USA) following the manufacturer’s guidelines. After checking the DNA extraction by 1% aga- rose gel electrophoresis, the concentration of samples was measured by a NanoDrop 2000 (Thermo Scientific, USA). Two universal primers for polymerase chain reaction (PCR) were selected to amplify the DNA samples. One primerpair was 515F (5’-GTGYCAGCMGCCGCGGTAA-3’) and806R (5’-GGACTACNVGGGTWTCTAAT-3’) with 291 bp for the V4 hypervariable region, and the second primer pair was 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) with 468bp for the V3-V4 hypervariable region. The PCR amp- lification procedures were conducted, complying with the standard guidelines of the sequencing protocol. Sequenc- ing data are publicly available in the National Center for Biotechnology Information Sequence Read Archive data- base under accession numbers PRJNA 687879 and PRJNA 687880.
2.4 16S rRNA Gene Sequence Classification
Low-quality reads with Q<20 or shorter than 200bp and showing significantly mismatched bases in barcodes or pri-mers were discarded using QIIME 1.9 (Caporaso, 2010). In accordance with the 97% similarity principle, raw reads were clustered to the operational taxonomic units (OTUs) using UPARSE 7.0; UCHIME was used to clas-sify and delete chimeric sequences (Edgar, 2013). The comparison of obtained OTUs with Silva v138 16S rRNA gene database was executed to ensure the accuracy of taxo-nomy sorting; annotation was performed by using the RDPClassifier 11.5 with a 0.7 threshold (McDonald, 2012).Aiming to compare the differences among total samplesequivalently, random subsets with the lowest sequencingnumber (27801 sequences) in all samples were selected for the downstream analyses.
2.5 Statistical Analysis
The richness of OTUs and alpha diversities (Shannon, Simpson, Chao 1, and Good’s coverage) were calculated and compared between two primers. Principal coordinate ana- lysis (PCoA) was performed to compare the differences in microbial communities based on a Bray-Curtis distance ma- trix using the ‘’ package. The significances of the differences among communities were evaluated by the ana-lysis of similarity (ANOSIM). Permutation multivariate ana- lysis of variance (PERMANOVA) was used to assess whe-ther the prior groups of primers, habitats, sites, and their in-teractions resulted in different community compositions (Anderson, 2010). Differentially abundant taxa at all taxo-nomic levels were distinguished and visualized using linear discriminant analysis (LDA) values (Segata, 2011). The relationships between environmental parameters and com- munity compositions were explored by distance-based redun- dancy analysis (db-RDA) using the ‘’ package. Dataanalysis was conducted in R programming (version 4.0.2).
3 Results
3.1 Comparison of Abundance and Diversity of Microbial Taxa
A total of 13477 and 8633 OTUs were recognized by the universal primers V4 and V3-V4, respectively. The propor- tions of reads identified from phylum to genus were obtain-ed to compare the taxonomic resolutions of the two primers. The taxonomic resolutions of primer V3-V4 were higher than that of primer V4 in the ARs (<0.05). In the bottom water samples, the identifications at the phylum and genus levels for primer V4 were 99.35% and 96.45%, respective- ly. Meanwhile, those for primer V3-V4 were 99.86%(phy- lum) and 97.92% (genus) (Fig.2A). Rickettsiales (31.80%) and norank_clade (30.99%) were the most dominant among the unclassified families for primersV3-V4 and V4. The classifications were 98.06% and 90.48% for primer V4 at the phylum and genus levels in the sediment samples, re- spectively, in contrast to the higher identifications of 98.87%and 95.02% for primer V3-V4 (Fig.2B). Bacteroidales (22.40%) and Gammaproteobacteria (25.85%) were the most dominant among the unclassified families for primers V3-V4 and V4, respectively. Given the taxonomic resolu- tions at the genus level, the proportions of OTUs affiliated to specific genera for primers V4 and V3-V4 were 48.6% and 52.5%, respectively. For the bottom water samples, theaforementioned proportions (genera) for primers V4 andV3-V4 were 49.8% and 51.6%. Meanwhile, in the sediment samples, the identified proportions for primers V4 and V3-V4 were 45.8% and 48.2%, respectively.
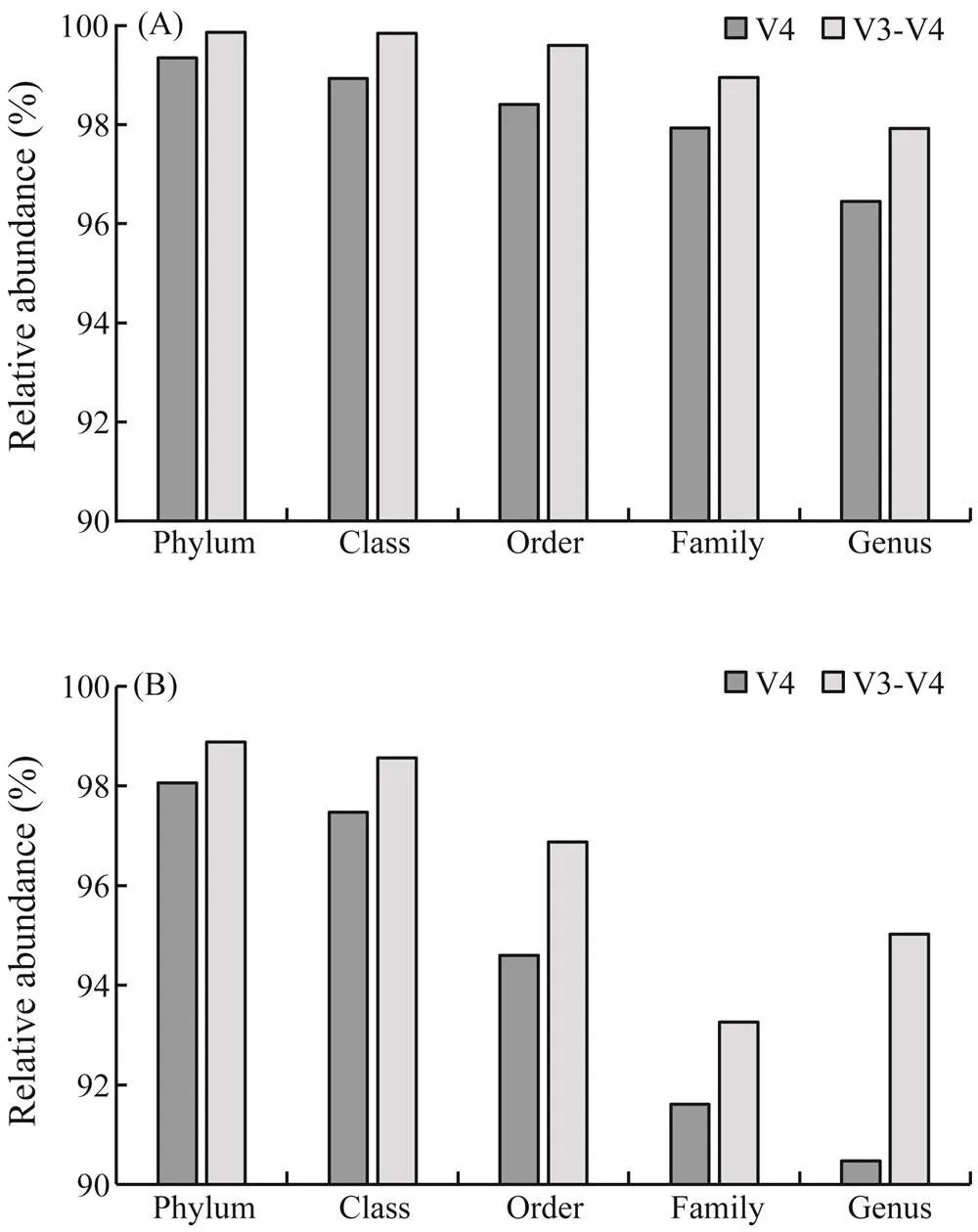
Fig.2 Proportions of classified reads for primers V4 and V3- V4 at different taxonomic levels in the (A) bottom water and (B) sediment. The classifications of relative abundances in the y-axis starts from 90%.
A total of 22 shared phyla were identified between the two primers, and their linear relationships of relative abun- dances were compared (Fig.3). Pearson’s correlations () of microbial abundances between the two primers in the bot- tom water and sediment were 0.97 (<0.001) and 0.94 (<0.001), respectively. Firmicutes and Planctomycetota were the prominent outliers for the bottom water and sediment samples.
Overall, 20 dominant phyla were detected with relative abundances of at least 1% in the ARs, with 8 and 18 phyla in the bottom water and sediment, respectively (Fig.4). How- ever, the relative abundances of phyla in the ARs assessed by the two primers were different. In the bottom water, se- ven and five dominant phyla were detected by primers V4 and V3-V4, respectively. Similarly, the relative abundances of Proteobacteria assessed by the two primers were both over 50%. The proportions of Bacteroidota, Thermoplasma-tota, Verrucomicrobiota, and Planctomycetota were higher for primer V4. Meanwhile, the proportions of Cyanobac- teria, Actinobacteriota, and Firmicutes were higher for pri- mer V3-V4. In the sediment, 15 and 16 dominant phyla were detected by the two primers. The relative abundances of Proteobacteria were both over 28%, as detected with the two primers. The proportions of Planctomycetota, NB1-j, Verrucomicrobiota, and Crenarchaeota were higher for pri- mer V4, but the proportions of Desulfobacterota, Acido- bacteriota, Actinobacteriota, Chloroflexi, Gemmatimona- dota, and Firmicutes were higher for primer V3-V4.

Fig.3 Comparison of the relative abundances of the phyla measured by primers V4 and V3-V4 in the (A) bottom water and (B) sediment.
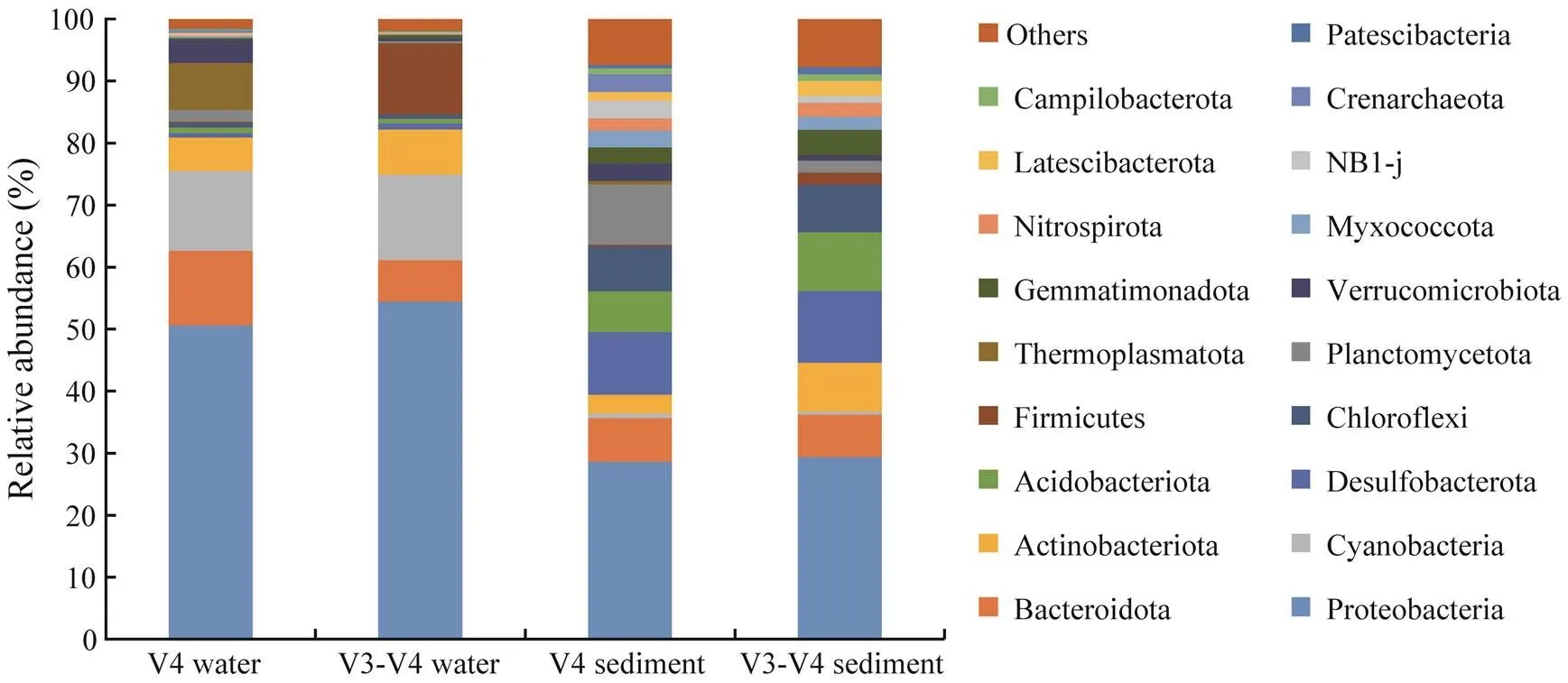
Fig.4 Relative abundances of microbial phyla sequenced by primers V4 and V3-V4 in the bottom water and sediment of ARs. The dominant phyla with more than 1% relative abundance of the entire community are shown.
The richness of OTUs and alpha diversity indices of the two primers were obtained (Table 1). The number of OTUssequenced by primer V4 was significantly higher than that of primer V3-V4 in the bottom water and sediment of ARs.In the bottom water, the Shannon and Simpson indiceswere similar between the two primers. The Chao 1 index of primer V4 was significantly higher than that of primer V3- V4 (<0.001), whereas the coverage index of primer V4 was notably lower (<0.001). In the sediment, the Shan-non and Chao 1 indexes of primer V4 were significantly higher (<0.001) than those of primer V3-V4, but the Sim-pson index and coverage index (<0.001) of primer V4 were remarkably lower.
3.2 Microbial Community Compositions and Structures
The whole sequences obtained by the two primers were affiliated to genus rank and then combined as one dataset to compare the dissimilarity of microbial groups. PCoA wasconducted to compare the microbial community composi- tions in the bottom water and sediment of ARs. Evident dis- tinctions between the two habitats and two primers were observed. The first and second principal coordinates ex- plained 61.22% and 16.78% of the variations in the mi- crobial community compositions, respectively (ANOSIM,=0.001, Figs.5A and 5B). The PCoA of the bottom wa- ter samples showed that the two primers formed two dif- ferent community groups, and the differences among four sites for primer V4 were lower (Fig.5C). In the sediment, the distinctions of communities among the four sites were more evident for primer V3-V4 (Fig.5D). The results of PERMANOVA showed that the microbial communities eva- luated by the two primers were significantly different (<0.001, Table 2). The differences among the four sites in the sediment were higher than those in the bottom water, al- though both showed no statistically significant differences. The relationship between the primer and site affected the microbial community composition analysis results signifi- cantly (<0.001).
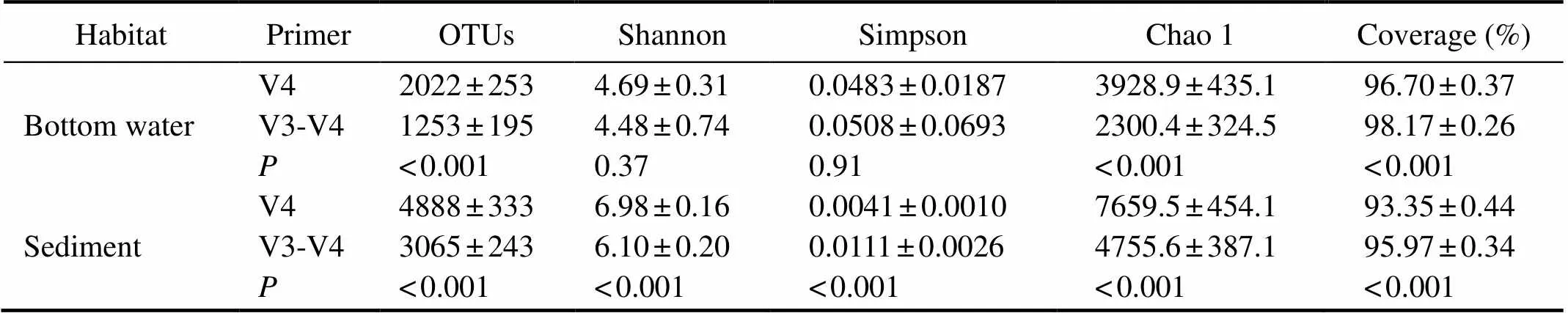
Table 1 OTUs and alpha diversity (mean±SD) of microbial communities based on the two 16S rRNA gene primers in the bottom water and sediment of ARs
Notes: OTUs, operational taxonomic units; SD, standard deviation;, significant difference between the two primers tested.
The differentially abundant microbial taxonomic levels in the ARs for the two primers were identified with LDA values of more than 4.0 (Fig.6). More differential species were observed in the bottom water than in the sediment. In the bottom water, the phyla Thermoplasmatota, Bacteroido, ta and Verrucomicrobiota were detected with higher abun- dances when using primer V4. However, phyla Firmicutes and Actinobacteriota were more enriched when detected with primer V3-V4. In the sediment, phyla Planctomycetota and Crenarchaeota were observed with greater abundance using primer V4, whereas Actinobacteriota and Acidobac- teriota were more abundant when using primer V3-V4.
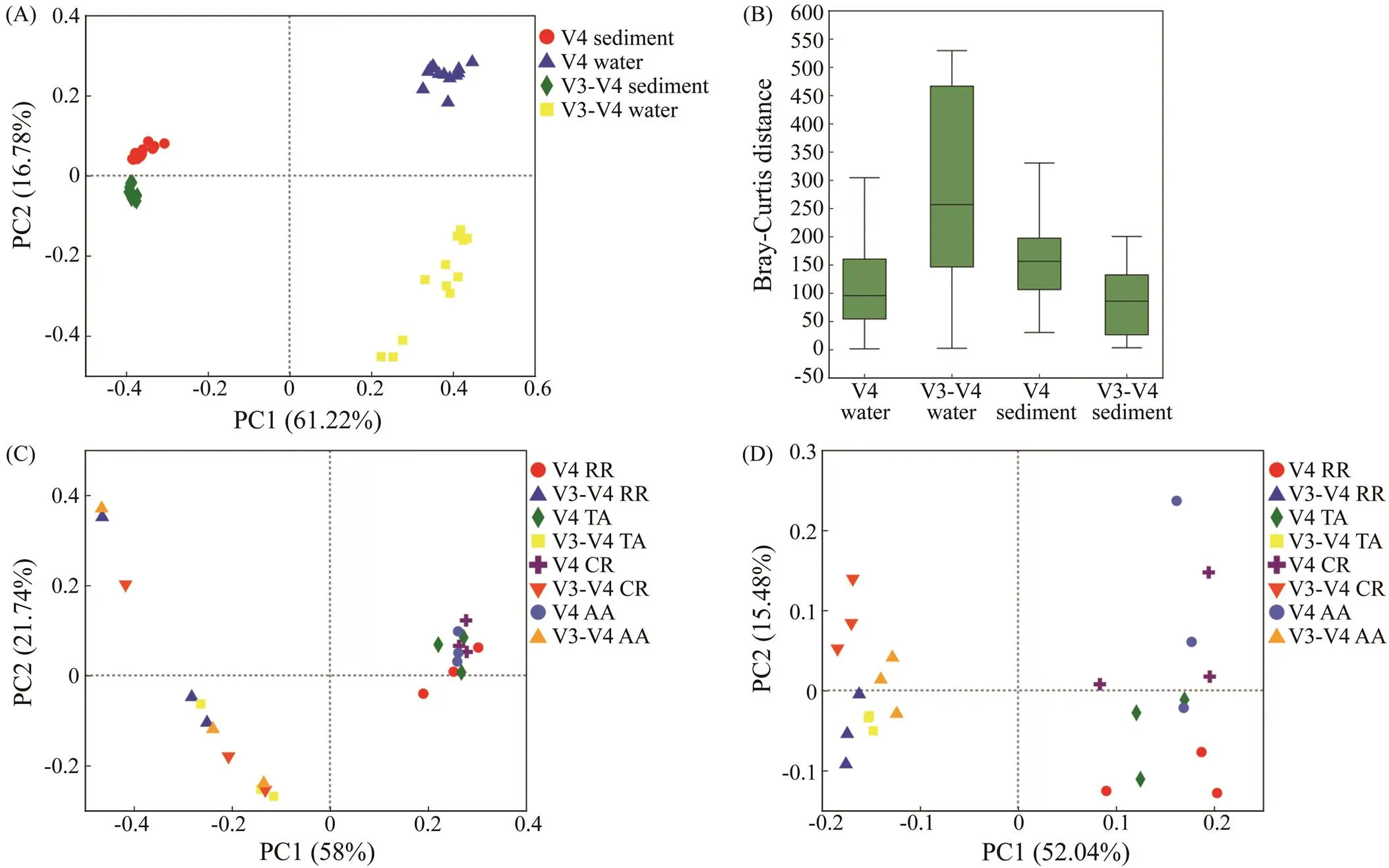
Fig.5 Comparison of microbial communities in the ARs sequenced by the 16S rRNA gene primers V4 and V3-V4. (A) PCoA plot of microbial communities sequenced by the primers V4 and V3-V4 in the bottom water and sediment. (B) Bray-Curtis distance box plot evaluated by the ANOSIM. (C) PCoA plots of microbial communities sequenced by the two primers in the bottom water. (D) PCoA plots of microbial communities sequenced by the two primers in the sediment. Sites: RR, TA, CR, and AA.
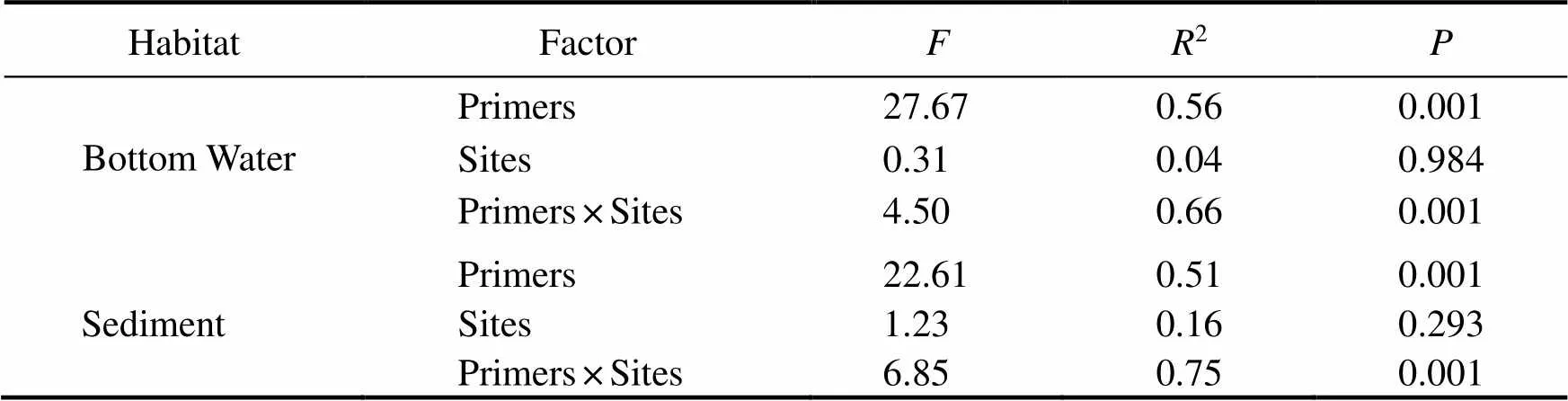
Table 2 Effects of primers, sampling sites, and their interactions on the differentiations of microbial communities based on the PERMANOVA
Notes: Primers included primer V4 and V3-V4; sampling sites included RR, TA, CR, and AA.
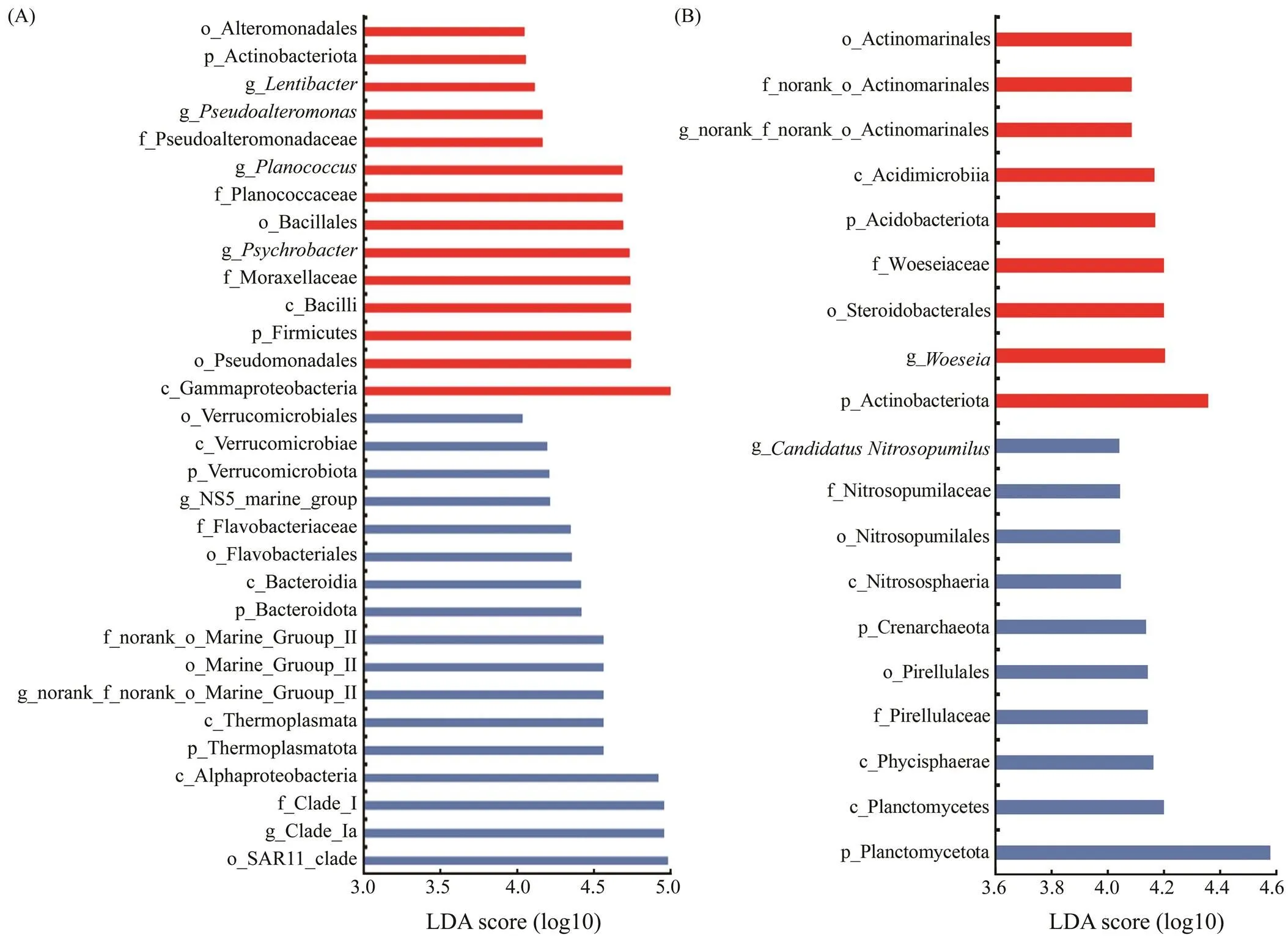
Fig.6 Differentially abundant microbial taxonomic levels of primers V4 and V3-V4 with LDA values of more than 4.0 in the (A) bottom water and (B) sediment. Red and blue bars represent primers V3-V4 and V4, respectively.
3.3 Relationships Between Microbial Communities and Environmental Factors
The results of db-RDA showed the inconsistent influences of environmental parameters on the microbial com- munity compositions, as analyzed by the two primers (Fig.7). The first and second axes mutually explained 33.64% and 46.24%, respectively, of the variations in microbial com- munities in the bottom water for primers V4 and V3-V4. In the sediment, the variations in microbial community com- positions explained by both the two axes were 28.33% and 28.76% for the two primers. In the bottom water, the main influencing parameters were Turb (<0.05) and Trans (=0.09) for primer V4, whereas NO2-N (<0.05) was the main influencing parameter for primer V3-V4. In the sedi- ment, the main influencing parameters were EC (<0.05) and OM (<0.05) for primer V4 and Par (<0.05) for pri- mer V3-V4.
4 Discussion
ARs have been proven to be excellent in maintaining coastal habitats and conserving marine biodiversity (Lima., 2019), whereas our understandings of microbiota in these artificial habitats are limited. Despite the rapid de- velopment and advancement of sequencing technology, we should thoroughly evaluate and select the 16S rRNA gene sequencing primers. A best resolution for the microbial ta- xonomic identification could be obtained by full length 16S rRNA gene sequences, while the limitations on the length of sequence reads and high cost at present hinder the ap- plication of this method (Yang., 2016). Thus, univer- sal primers link with the targeted conserved regions in 16S rRNA were applied to provide phylogenetic information ofmicrobiome. However, deviations caused by the choices of primers (different conserved regions) got more and more attention (Fadeev., 2021). In the present study, we com- pared the effects of two primers (V4 and V3-V4) on the microbial diversity and community structures in ARs. Our analysis showed highly consistent results, as determinedby the two primers, whereas several significant differences were also observed.
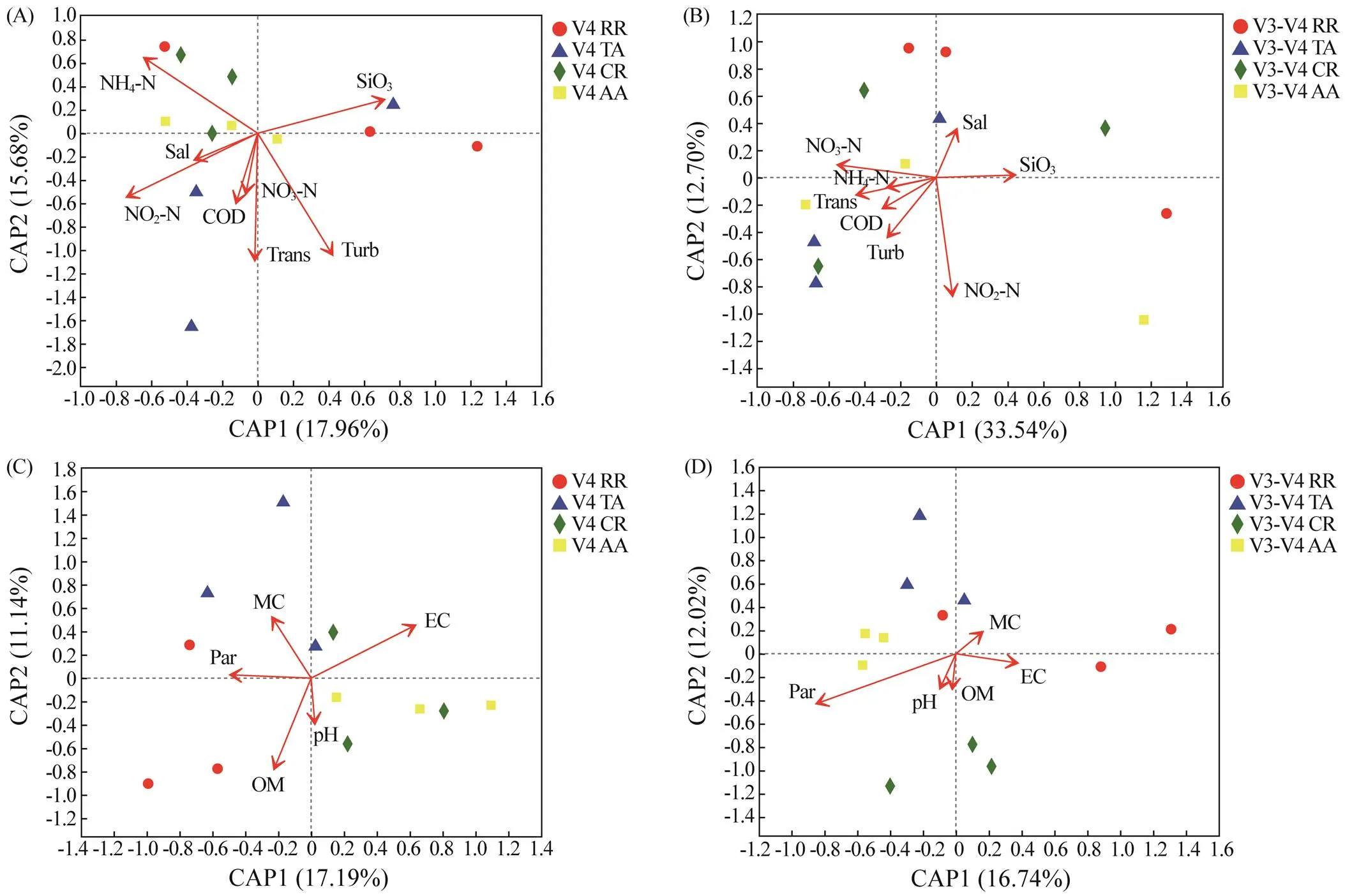
Fig.7 db-RDA of the microbial communities with environmental parameters in the ARs. Microbial communities in the bottom water sequenced by primers (A) V4 and (B) V3-V4. Microbial communities in the sediment sequenced by primers (C) V4 and (D) V3-V4.
The microbial composition analyses in the bottom wa- ter and sediment of ARs were affected by the choice of pri-mers. The richness of OTUs and phylum of the microbiomes detected by primer V4 were higher than those detected by primer V3-V4. On the contrary, the identification resolu- tions from the phylum to genus levels of primer V4 were lower than those of primer V3-V4 (Fig.2). Onywera and Meiring (2020) reported similar results. Primer V4 has short- er reads and less sequence information compared with pri- mer V3-V4, and the taxonomic resolutions of primer V4 are relatively lower (Michael., 2014). Thus, the high- er numbers of OTUs and phyla of the microbiomes ob- served by primer V4 might have resulted from mistaken sequences (Loman, 2012). Possibly, OTUs from close- ly related bacteria are difficult to detect by primers with short sequences (Michael., 2014). However, an oppo- site view was put forward. Willis(2019) revealed the contrasting diversities of dominant and rare taxa detected by different primers. Therefore, a higher diversity observedby the primer V4 is probably meaningful in the study of rare microbiomes. As for the classification resolutions, the performances of the two primers in the water and sediment samples were discordant. The proportion of genera identi- fied for primer V3-V4 was 1.81% higher than that for pri- mer V4 in the bottom water samples, whereas in sediment samples, this proportion was 2.23%. In addition, the dif- ference in microbial alpha diversities observed by the two primers in the sediment samples was notably higher than that in the bottom water samples. These results indicated that the accuracy of primer V4 for the sediment samples was lower than that of primer V3-V4.
Although a high correlation was observed between the relative abundances measured by the two primers, several divergences were observed (Fig.4). In the bottom water, the relative abundances of three phyla, Firmicutes (primer V3-V4, 11.5%), Thermoplasmatota (primer V4, 7.6%), and Ver- rucomicrobiota (primer V4, 3.7%) varied significantly be- tween two primers. Thermoplasmatota is an archaea clade belonging to Crenarchaeota, which is often detected in ma- rine hot springs or deep-sea hydrothermal vents (DeLong, 1992). The study area is adjacent to the Jimo ocean hot spring, which may be the source of Thermoplasmatota. In the sediment, the abundances of more than half of the phy- la sequenced by the two primers were different (Fig.4). Phy- la Planctomycetota and Crenarchaeota, as assessed by the primer V4, were significantly higher, and both play a vital role in the procedure of anaerobic ammonium oxidation. Actinobacteriota, which decomposes various kinds of OM, is widely observed in marine sediment. The differences in dominant phylum abundances in the sediment between the two primers were significantly higher than those in the bot- tom water. The spatial heterogeneity of microbial commu- nities in the sediment was higher than that in water. In ad- dition, the mixture of microbial communities in the sedi-ment was lower, which probably resulted in the above phe- nomenon (Perkins, 2014). The characteristic of co- vering several important archaeal clades is an important ad- vantage for primer V4 compared with primer V3-V4. Al- though the relative abundance of archaea was high in the marine water (Beman, 2008), the understanding oftheir ecological roles is limited given the lack of focus (Needham and Fuhrman, 2016). With the increased under- standing of archaea, ecologists can pay more attention to archaea’s abundance changes and ecological roles. The‘Earth Microbiome Project’ advocated primer V4, and one of the most important reasons is its improved efficiency in detecting archaea (Thompson., 2017). Fadeev(2021) also suggested the use of primer V4-V5 for both bacterial and archaeal community dynamics in the Arctic because of its better coverage of the archaeal domain. Al- together, primer V4 has a nonnegligible superiority of de- tecting archaea taxa from water samples, and this property might be one of the reasons for the differences between the two primers.
In spite of the observed differences in taxonomic ranks, the influences of primer choices on the analysis results of microbial community compositions and beta diversities are limited. The PCoA plots obtained by the two primers re- vealed that two distinct microbial communities had form- ed between the bottom water and sediment (Fig.5). Pre- vious studies also reported similar results (Barb., 2016;Onywera and Meiring, 2020). These consistent beta diver- sities suggested that the influence of primer choices on com- munity similarities between different environments is low but has a significant influence on the studies of specific taxa (Willis, 2019). However, several detailed beta diversity differences between the two primers were found. The variations in microbial communities between the bot- tom water and sediment for primer V3-V4 were higher than those for primer V4 (Figs.5C and 5D). The results of PCoA were based on the Bray-Curtis distance, and to a certain extent, a relatively low resolution of primer V4 in the ge- nus gave rise to this result. In the water samples, differ- ences among four sites of the primer V3-V4 dataset were higher than that of the primer V4 dataset (Fig.5C). In the sediment samples, the total dissimilarity among four sites observed by primer V4 was higher than that obtained with primer V3-V4, whereas the dissimilarities between groups exhibited by the primer V3-V4 dataset were more signi- ficant (Fig.5D). These different performances indicated that primer V3-V4 is more helpful in distinguishing microor- ganisms in different habitats. Although matching beta di- versity results were derived from the two primers, db- RDA findings showed that microbial communities were affected by different environmental parameters. However, a limited number of papers illustrate these mismatches. A similar study illustrated that the PCR-denaturing gradient gel electrophoresis (DGGE) and terminal restriction frag- ment length polymorphism may have disparate ecological explanations for the same samples (Zhang., 2018), whereas Zhang’s results were inconsistent with those of Yu. (2015), despite using the same DGGE method. These observed inconsistencies can be ascribed to the high en- vironmental variability rather than the choices of primers and sampling technologies. Some important but uncollect- ed environmental variables such as water flow-induced dis- criminations could also induce these conflicting results. In particular, environment significantly shapes microbial com- munities, but cannot impact the results of microbial com- positions obtained by primers. Of course, complex and di- verse environmental characteristics greatly bring about dif- ficulties and differences in analyzing the relationships be- tween microbial communities and environmental factors based on different primers.
Rapid advances in various sequencing techniques have been achieved, and primer choices may not be noteworthy in the future. However, assessing their performance in ex- plaining micro-ecology questions and predicting microbial community dynamics are indispensable at present. The com- parisons and evaluations of different primers were carried out through many aspects. For example, some applied mockcommunities with known full length sequences (Barb., 2016); some analyzed the performances of primers in(Klindworth., 2013); some assessed the unifor- mity of conclusions using different primers (Wang., 2018). In this study, the third method was selected to eva- luate the performances of two primers for profiling micro- bial diversities and community compositions. The results suggested that primer V4 generated a comprehensive result of microbial community structure in the bottom water of ARs;primer V3-V4 provided an accurate profile of bacterial com- munity compositions in the sediment. For the water sam- ples of ARs, given that the communities are highly con- sistent, as analyzed by the two primers, whereas the rela- tive abundance of archaea was nonnegligible (8%), primer V4 is preferred. For the sediment samples of ARs, with higher taxonomic classifications and more distinct parti- tions exhibited, primer V3-V4 is more preferred. When stu- dying microbial communities in the bottom water and se- diment simultaneously, primer V3-V4 was the better op- tion. Notably, several limitations existed in this study, such as the mock community which was not analyzed with en- vironmental samples simultaneously. Mock community is recommended to assess the accurate assignment of se- quences from primers with known full-length sequences (Yeh., 2018). However, our understanding of micro- bial community in the ARs is limited, which is a tough pro- blem for the construction of a representative mock com- munity. Additionally, evaluating the performances of pri- mers in the field samples is another path to select the op- timum primer (Parada., 2016). The congruence of re- sults achieved from varied primers should be assessed for unknown samples. With careful analysis of the two pri- mers applied for microbial communities in the ARs, this study can provide meaningful references for later studies with the mock community.
5 Conclusions
ARs are important means for the recovery of the ma- rine environment in China, whereas little is known about microbial communities living in these particular habitats. A robust 16S rRNA gene primer is necessary to study the microbial diversity and community structure in the ARs. In the present study, we compared the performance of two universal primers (V4 and V3-V4) on studying the micro- bial communities in the water and sediment of ARs. The results showed that microbial diversities, community com- positions, and beta diversities detected by the two primers were generally comparable, which suggested that both pri-mers can provide reliable profiling. However, unconfor- mity in identifying specific taxonomic compositions and explaining the relationships between the microbial com- munities and environmental factors were observed. In addi- tion, several archaeal clades were available for primer V4, which amplified the difference between the two primers. Given the general similarities and partial discrepancies, we proposed to profile the microbial community in the ARs using primer V3-V4 because of its relatively higher classi- fication efficiency. To further understand the suitability of primer, evaluating the quantitative performance using me- tagenomics and mock community is recommended.
Acknowledgements
This study was funded by the Project of National Key R&D Program of China (No. 2019YFD0901302). In ad-dition, we thank all scientific staff and crew members of Qingdao Longpan Company for their assistance in surveys.
Anderson, M. J., 2010. A new method for non-parametric mul- tivariate analysis of variance., 26 (1): 32-46, DOI: 10.1111/j.1442-9993.2001.01070.pp.x.
Barb, J. J., Oler, A. J., Kim, H. S., Chalmers, N., Wallen, G. R., and Cashion, A., 2016. Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA us-ing mock samples., 11 (2): e0148047, DOI: 10.1371/ journal.pone.0148047.
Beman, J. M., Popp, B. N., and Francis, C. A., 2008. Molecular and biogeochemical evidence for ammonia oxidation by ma- rine Crenarchaeota in the Gulf of California., 2 (4): 429-441, DOI: 10.1038/ismej.2007.118.
Birtel, J., Walser, J. C., Pichon, S., Burgmann, H., and Matthews, B., 2015. Estimating bacterial diversity for ecological studies: Methods, metrics, and assumptions., 10 (4): e0125356, DOI: 10.1371/journal.pone.0125356.
Bohnsack, J. A., and Sutherland, D. L., 1985. Artificial reef re- search: A review with recommendations for future priorities., 37 (1): 11-39, DOI: 10.1515/botm. 1985.28.11.501.
Cai, L., Ye, L., Tong, A. H., Lok, S., and Zhang, T., 2013. Bi- ased diversity metrics revealed by bacterial 16S pyrotags de- rived from different primer sets., 8 (1): e53649,DOI: 10.1371/journal.pone.0053649.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bu- shman, F. D., Costello, E. K.,., 2010. QIIME allows ana- lysis of high-throughput community sequencing data., 7 (5): 335-336, DOI: 10.1038/nmeth.f.303.
Chen, Q., Yuan, H., and Chen, P., 2019. Short-term effects of ar- tificial reef construction on the taxonomic diversity and eco- exergy of the macrobenthic faunal community in the Pearl Ri- ver Estuary, China., 98: 772-782, DOI: 10.1016/j.ecolind.2018.12.001.
Choi, C. J., Bachy, C., Jaeger, G. S., Poirier, C., Sudek, L., Sarma,V.,., 2017. Newly discovered deep-branching marine plas- tid lineages are numerically rare but globally distributed., 27 (1): R15-R16, DOI: 10.1016/j.cub.2016.11. 032.
Claesson, M. J., Wang, Q., O’Sullivan, O., Greene-Diniz, R., Cole, J. R., Ross, R. P.,., 2010. Comparison of two next-gene- ration sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions., 38 (22): e200, DOI: 10.1093/ nar/gkq873.
DeLong, E. F., 1992. Archaea in coastal marine environments., 89: 5685- 5689, DOI: 10.1073/pnas.89.12.5685.
Edgar, R. C., 2013. UPARSE: Highly accurate OTU sequences from microbial amplicon reads., 10: 996-998, DOI: 10.1038/NMETH.2604.
Eleftheriou, A., 2013.. John Wiley & Sons, Ltd., Chichester, 477pp.
Fadeev, E., Cardozo-Mino, M. G., Rapp, J. Z., Bienhold, C., and Boetius, A., 2021. Comparison of two 16S rRNA primers (V3- V4 and V4-V5) for studies of arctic microbial communities., 12: 637526, DOI: 10.3389/fmicb. 2021.637526.
Hamady, M., and Knight, R., 2009. Microbial community profi- ling for human microbiome projects: Tools, techniques, and challenges., 19 (7): 1141-1152, DOI: 10. 1101/gr.085464.108.
Hazen, T. C., Rocha, A. M., and Techtmann, S. M., 2013. Ad- vances in monitoring environmental microbes., 24 (3): 526-533, DOI: 10.1016/j.copbio. 2012.10.020.
Heiri, O., Lotter, A. F., and Lemcke, G., 2001. Loss on ignition as a method for estimating organic and carbonate content in sedi- ments: Reproducibility and comparability of results., 25: 101-110, DOI: 10.1023/A:1008119611481.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M.,., 2013. Evaluation of general 16S ribosomal RNA genePCR primers for classical and next-generation se- quencing-based diversity studies., 41 (1): e1, DOI: 10.1093/nar/gks808.
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A.,., 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences., 31 (9): 814-821, DOI: 10.1038/nbt.2676.
Lee, H., Heo, Y. M., Kwon, S. L., Yoo, Y., Kim, D., Lee, J.,.,2021. Environmental drivers affecting the bacterial community of intertidal sediments in the Yellow Sea., 755: 142726, DOI: 10.1016/j.scitotenv.2020.142726.
Li, N., Zhao, H., Jiang, G., Xu, Q., Tang, J., Li, X.,., 2020. Phylogenetic responses of marine free-living bacterial commu- nity to phaeocystis globosa bloom in Beibu Gulf, China., 11: 1624, DOI: 10.3389/fmicb.2020.01624.
Liao, X., Zhang, C., Yao, L., Li, J., Liu, M., Xu, L.,., 2014. Sorption behavior of nonylphenol (NP) on sewage-irrigated soil: Kinetic and thermodynamic studies.,473-474 (6): 530-536, DOI: 10.1016/j.scitotenv.2013.12. 055.
Lima, J. S., Zalmon, I. R., and Love, M., 2019. Overview and trends of ecological and socioeconomic research on artificial reefs., 145: 81-96, DOI: 10. 1016/j.marenvres.2019.01.010.
Liu, J., Zheng, Y., Lin, H., Wang, X., Li, M., Liu, Y.,., 2019. Proliferation of hydrocarbon-degrading microbes at the bot- tom of the Mariana Trench., 7 (1): 47, DOI: 10. 1186/s40168-019-0652-3.
Loman, N. J., Misra, R. V., Dallman, T. J., Constantinidou, C., Gharbia, S. E., Wain, J.,., 2012. Performance comparison of benchtop high-throughput sequencing platforms., 30 (5): 434-439, DOI: 10.1038/nbt.2198.
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., De- Santis, T. Z., Probst, A.,., 2012. An improved Greenge- nes taxonomy with explicit ranks for ecological and evolu- tionary analyses of bacteria and archaea., 6 (3): 610-618, DOI: 10.1038/ismej.2011.139.
Michael, C. N., Hilary, G. M., Jacquelynn B., Sharon, L. G., and Joerg G., 2014. Analysis, optimization and verification of il- lumina-generated 16S rRNA gene amplicon surveys., 9 (4): e94249, DOI: 10.1371/journal.pone.0094249.
Needham, D. M., and Fuhrman, J. A., 2016. Pronounced daily succession of phytoplankton, archaea and bacteria followinga spring bloom., 1: 16005, DOI: 10.1038/ nmicrobiol.2016.5.
Nitin, P. N., Pandurang, K., Andrew, J., Synne, K., Hans, U., and Anette, J., 2017. Characterization of bacterial community as- sociated with phytoplankton bloom in a eutrophic lake in SouthNorway using 16S rRNA gene amplicon sequence analysis., 12 (3): e0173408, DOI: 10.1371/journal.pone.017 3408.
Onywera, H., and Meiring, T. L., 2020. Comparative analyses of Ion Torrent V4 and Illumina V3-V4 16S rRNA gene metabar- coding methods for characterization of cervical microbiota: Taxonomic and functional profiling., 7: e00278, DOI: 10.1016/j.sciaf.2020.e00278.
Parada, A. E., Needham, D. M., and Fuhrman, J. A., 2016. Every base matters: Assessing small subunit rRNA primers for ma- rine microbiomes with mock communities, time series andglobal field samples., 18 (5): 1403- 1414, DOI: 10.1111/1462-2920.13023.
Parulekar, N. N., Kolekar, P., Jenkins, A., Kleiven, S., Utkilen, H., Johansen, A.,., 2017. Characterization of bacterial commu- nity associated with phytoplankton bloom in a eutrophic lake in South Norway using 16S rRNA gene amplicon sequence ana- lysis., 12 (3): e0173408, DOI: 10.1371/journal.pone. 0173408.
Perkins, T. L., Clements, K., Baas, J. H., Jago, C. F., Jones, D. L., Malham, S. K.,., 2014. Sediment composition influ- ences spatial variation in the abundance of human pathogen indicator bacteria within an estuarine environment., 9 (11): e112951, DOI: 10.1371/journal.pone.0112951.
Perrine, C., Adrien, V., Céline, L. M., Pierre, E. C., Anne, G., and Marie-Anne, C. B., 2014. Influence of DNA extraction me- thod, 16S rRNA targeted hypervariable regions, and sample origin on microbial diversity detected by 454 pyrosequencing in marine chemosynthetic ecosystems., 80 (15): 4626-4639, DOI: 10.1128/AEM. 00592-14.
Segata, N., Izard, J., Waldron, L. D., Gevers, D., Miropolsky, L., Garrett, W. S.,., 2011. Metagenomic biomarker discovery and explanation., 12 (6): R60, DOI: 10.1186/ gb-2011-12-6-r60.
Soergel, D. A., Dey, N., Knight, R., and Brenner, S. E., 2012. Se- lection of primers for optimal taxonomic classification of en- vironmental 16S rRNA gene sequences., 6 (7): 1440-1444, DOI: 10.1038/ismej.2011.208.
State Bureau of Quality and Technical Supervision of China, 2007..China Standards Press, Beijing (in Chinese).
Sun, P., Liu, X., Tang, Y., Cheng, W., Sun, R., Wang, X.,., 2017. The bio-economic effects of artificial reefs: Mixed evi- dence from Shandong, China., 74 (8): 2239-2248, DOI: 10.1093/icesjms/fsx058.
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., La- badie, K., Salazar, G.,., 2015. Ocean plankton. Structure and function of the global ocean microbiome., 348 (6237): 1261359, DOI: 10.1126/science.1261359.
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J.,., 2017. A communal catalogue reveals Earth’s multiscale microbial diversity., 551 (7681): 457-463, DOI: 10.1038/nature24621.
Wang, F., Men, X., Zhang, G., Liang, K., Xin, Y., Wang, J.,., 2018. Assessment of 16S rRNA gene primers for studying bac-terial community structure and function of aging flue-cured tobaccos., 8 (1): 182, DOI: 10.1186/s13568-018- 0713-1.
Wang, Y., Sun, J., Fang, E., Guo, B., Dai, Y., Gao, Y.,., 2019. Impact of artificial reefs on sediment bacterial structure and function in Bohai Bay., 65(3): 191-200, DOI: 10.1139/cjm-2018-0157.
Willis, C., Desai, D., and LaRoche, J., 2019. Influence of 16S rRNA variable region on perceived diversity of marine micro- bial communities of the Northern North Atlantic., 366 (13): fnz152, DOI: 10.1093/femsle/ fnz152.
Yang, B., Wang, Y., and Qian, P. Y., 2016. Sensitivity and corre- lation of hypervariable regions in 16S rRNA genes in phylo- genetic analysis., 17 (2016): 135, DOI: 10. 1186/s12859-016-0992-y.
Yeh, Y. C., Needham, D. M., Sieradzki, E. T., Fuhrman, J. A., and Gregory, C. J., 2018. Taxon disappearance from micro- biome analysis reinforces the value of mock communities as a standard in every sequencing run., 3 (3): e00023-18, DOI: 10.1128/mSystems.00023-18.
Youssef, N., Sheik, C. S., Krumholz, L. R., Najar, F. Z., Roe, B. A., and Elshahed, M. S., 2009. Comparison of species rich- ness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys., 75 (16): 5227-5236, DOI: 10.1128/AEM. 00592-09.
Yu, L. Y., Zhang, W. J., Liu, L. M., and Yang, J., 2015. Deter- mining microeukaryotic plankton community around Xiamen Island, southeast China, using Illumina MiSeq and PCR-DGGETechniques., 10 (5): e0127721, DOI: 10.1371/jour nal.pone.0127721.
Zhang, W., Mo, Y., Yang, J., Zhou, J., Lin, Y., Isabwe, A.,., 2018. Genetic diversity pattern of microeukaryotic communi- ties and its relationship with the environment based on PCR- DGGE and T-RFLP techniques in Dongshan Bay, southeast China., 164: 1-9, DOI: 10.1016/j. csr.2018.05.006.
March 2, 2021;
May 19, 2021;
October 5, 2021
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
. E-mail: tangyanli@ouc.edu.cn
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Variations in Dissolved Oxygen Induced by a Tropical Storm Within an Anticyclone in the Northern South China Sea
- Intensity of Level Ice Simulated with the CICE Model for Oil-Gas Exploitation in the Southern Kara Sea, Arctic
- Learning the Spatiotemporal Evolution Law of Wave Field Based on Convolutional Neural Network
- Development and Control Strategy of Subsea All-Electric Actuators
- Acoustic Prediction and Risk Evaluation of Shallow Gas in Deep-Water Areas
- In Situ Observation of Silt Seabed Pore Pressure Response to Waves in the Subaqueous Yellow River Delta
