Anoctamin 5 regulates the cell cycle and affects prognosis in gastric cancer
2022-09-14TomoyukiFukamiAtsushiShiozakiToshiyukiKosugaMichihiroKudouHirokiShimizuTakumaOhashiTomohiroAritaHirotakaKonishiShuheiKomatsuTakeshiKubotaHitoshiFujiwaraKazumaOkamotoMitsuoKishimotoYukikoMorinagaEiichiKonishiEigoOtsuji
Tomoyuki Fukami, Atsushi Shiozaki, Toshiyuki Kosuga, Michihiro Kudou, Hiroki Shimizu, Takuma Ohashi,Tomohiro Arita, Hirotaka Konishi, Shuhei Komatsu, Takeshi Kubota, Hitoshi Fujiwara, Kazuma Okamoto,Mitsuo Kishimoto, Yukiko Morinaga, Eiichi Konishi, Eigo Otsuji
Abstract
Key Words: Anoctamin 5; Gastric cancer; Cell cycle; G1/S checkpoint; Cell proliferation
lNTRODUCTlON
The anoctamin (ANO)/transmembrane protein 16 (TMEM16) family is present in numerous eukaryotes,and ten ANO paralogs, ANO1-ANO10 (TMEM16A-H, TMEM16J and K), have been identified in vertebrates[1]. Of these, several function as calcium-activated chloride channels. ANOs comprise a family of plasma membrane proteins that mediate ion transport, phospholipid scrambling, and other membrane protein regulation in numerous cell types[2-6]. Their expression has been detected in both epithelial and non-epithelial tissues types[4]. Although the regulation of ANOs has been extensively examined, the mechanisms by which increased intracellular calcium concentration activates chloride or cation conductance have not been elucidated.
Recent molecular and biochemical studies reported a role for ANOs in human carcinogenesis. For instance, the expression of ANO proteins is upregulated in cancer and associated with a poor patient prognosis[7]. A relationship has been demonstrated between ANO1 and patient prognosis in various cancer types, including gastric, esophageal, breast, lung, and head and neck cancer[8-12]. The upregulation of the genes encoding ANO1 and ANO3 has been associated with several cancer types,specifically gastrointestinal stromal tumors, breast cancer, and squamous cell carcinoma[12,13].Furthermore, ANO6 has been strongly implicated in the metastatic potential of breast cancer[14]. The expression levels of other members of the ANO family are also associated with cell proliferation and cancer development[15-17].
In our previous studies, we identified a crucial role for several chloride ion channels and transporters in patients with gastric cancer (GC); intracellular chloride regulates proliferation and cell cycle progression[18,19], whereas furosemide, a potent inhibitor of the Na+/K+/2Cl-cotransporter, induces G0/G1arrest[20]. Furthermore, leucine rich repeat containing 8 VRAC subunit A regulate the proliferation,apoptosis, migration, and invasion of GC cells[21].
ANO5 has recently been implicated in various cancers, such as thyroid[22] and pancreatic cancer[23];however,limited information is presently available on its involvement in tumor progression in patients with GC or the clinical significance of its expression. Therefore, in the present study, we investigated whether ANO5 contributes to the regulation of cancer growth and evaluated its clinicopathological significance in GC.
MATERlALS AND METHODS
Cell lines, antibodies
MKN7, MKN45, MKN74, HGC27, and NUGC4 human GC cell lines were purchased from the Riken Cell Bank (Tsukuba, Japan). Cells were cultured in RPMI-1640 (Nacalai Tesque, Kyoto, Japan)containing 100 μg/mL of streptomycin, 100 U/mL penicillin, and 10% FBS at 37 °C in a 5% CO2incubator. Rabbit polyclonal anti-ANO5 antibody was obtained from Funakoshi (GTX81161) for immunohistochemical (IHC) analysis and western blotting. Mouse monoclonal anti-β-actin antibody was provided by Sigma-Aldrich (St. Louis, MO, United States) and HRP-conjugated anti-rabbit and mouse secondary antibodies by Cell Signaling Technology (Beverly, MA, United States).
Reverse transcription-quantitative polymerase chain reaction
RNA was extracted from cancer cells using an RNeasy kit (Qiagen, Valencia, CA, United States). The Step One plusTMReal-Time polymerase chain reaction (PCR) System (Applied Biosystems, Foster City,CA, United States) and TaqMan Gene Expression Assays (Applied Biosystems) were employed for reverse transcription-quantitative PCR analysis using the following PCR thermocycling conditions:initial denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.The expression levels of the following genes were assessed:ANO5(Hs01381106_m1),CDKN1A(Hs00355782_m1),CDK2(Hs00608082_m1),CDK4(Hs00175935_m1),CDK6(Hs00608037_m1), cyclin E2(CCNE2; Hs00180319_m1), andE2F1(Hs00153451_m1) (all from Applied Biosystems). The expression of each gene was normalized using the housekeeping gene β-actin (Hs01060665_g1; Applied Biosystems).All assays were performed in triplicates.
Western blotting
The cells were washed twice with ice-cold PBS and harvested in M-PER lysis buffer (Pierce, Rockford,IL, United States) supplemented with protease inhibitors (Pierce Biotechnology). Protein concentrations were measured using a modified Bradford assay (Bio-Rad, Hercules, CA, United States). Cell lysates containing equal amounts of total protein (10 mg/lane) were resolved using 10% SDS-PAGE and subsequently transferred to polyvinylidene fluoride membranes (GE Healthcare, Piscataway, NJ, United States). Membranes were incubated with antibodies for 24 h at 4 °C. Band densities were quantified using ImageJ (version 1.52; National Institutes of Health).
Small interfering RNA transfection
All small interfering RNA (siRNA) reverse transfection procedures were performed using Lipofectamine®RNAiMAX reagent (Invitrogen, Carlsbad, CA, United States) with a final siRNA concentration of 20 nmol/L, according to the manufacturer’s instructions. ANO5 siRNA (Stealth RNAi siRNA;HSS137119, Stealth RNAi siRNA; HSS137120) and control siRNA (Stealth RNAiTM siRNA Negative Control) were obtained from Invitrogen.
Overexpression study
Control-HaloTagRplasmid (Promega, G6591) and ANO5-HaloTagRplasmid (pFN21AE5809) were transfected using P3000TM (Invitrogen) and lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. After passaging, ANO5-expressing cells were used for the cell proliferation assay.
Cell proliferation assay
NUGC4 and MKN45 cells were seeded at densities of 1.0 and 2.0 × 105cells/well, respectively, on sixwell plates and incubated at 37 °C in a 5% CO2incubator. The siRNA was transfected 24 h after seeding.The cells were detached from the plates with trypsin-EDTA 48 h and 72 h after siRNA transfection and counted using a hemocytometer.
Cell proliferation activity was measured using the water-soluble tetrazolium salts-8 assay with Cell Count Reagent SF (Nacalai Tesque). NUGC4, MKN45, and MKN7 cells were seeded at a density of 1.0 ×104, 1.0 × 104, and 1.5 × 104cells/well, respectively, in 24-well plates and were incubated at 37 °C in a 5%CO2incubator. The siRNA was transfected 24 h after seeding. Cell proliferation was evaluated every 24 h by measuring the absorbance at 450 nm using a Thermo Scientific Multiskan FC (Thermo Fisher Scientific).
Cell cycle assay
Cell cycle progression was assessed 48 h after siRNA transfection by flow cytometry. Cells were detached from the plates using trypsin-EDTA and subsequently treated with 0.2% Triton X-100 and stained with propidium iodide with RNase staining buffer (BD Biosciences, San Jose, CA, United States).Flow cytometry data were acquired using a BD Accuri C6 plus flow cytometer (BD Biosciences) to assess DNA content in at least 10000 cells.
Apoptosis assay
Cells were evaluated 72 h after transfection and stained using the ANNEXIN V-FITC Kit (Beckman Coulter, Brea, CA, United States). The frequencies of early and late apoptotic cells among at least 10000 cells were assessed using a BD Accuri C6 plus flow cytometer.
Migration and invasion assays
Migration assays were performed using 24-well cell culture inserts with 8-μm pores (BD Biosciences),whereas invasion assays were performed using Biocoat Matrigel®(BD Biosciences). At 48 h posttransfection, NUGC4 and MKN45 cells were seeded at a density of 3.0 × 105cells/well in serum-free RPMI-1640 in the upper chamber, whereas the lower chamber contained RPMI-1640 with 10% FBS.Matrigel and the cells remaining in the upper chamber after a 48-h incubation were removed. Diff-Quick staining reagents (Sysmex) were used to stain migrated or invaded cells, which were counted in four independent fields of view. Both assays were conducted thrice.
Microarray analysis
NUGC4 and MKN45 cells were transfected with either control or ANO5 siRNA. At 48 h after siRNA transfection, total RNA was extracted using RNeasy kit. Cyanine 3 (Cy3)-labeled cRNA was prepared from 0.1 μg total RNA using the Low Input Quick Amp Labeling Kit (Agilent Technologies, CA, United States) and then subjected to RNeasy column purification (Qiagen). Dye incorporation and cRNA yields were assessed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Subsequently, 0.6 μg Cy3-labeled cRNA was fragmented in a 25 μL reaction volume containing 1 × Agilent fragmentation buffer and 2 × Agilent blocking agent at 60 °C for 30 min. The 2 × Agilent hybridization buffer (25 μL)was then added, and hybridization to SurePrint G3 Human GE 8 × 60K Microarray Ver3.0 (Agilent Technologies) was conducted at 65 °C for 17 h in a rotating Agilent hybridization oven. The microarrays were then washed with GE Wash Buffer 1 (Agilent Technologies) at room temperature for 1 min,followed by GE Wash buffer 2 (Agilent Technologies) at 37 °C for 1 min.
Microarray data processing
Slides were scanned using the Agilent SureScan Microarray Scanner (G2600D) with the one color scan setting for 8 × 60k array slides. The scanned images were analyzed with Feature Extraction Software(Agilent Technologies) using the default parameters to obtain background-subtracted and spatially detrended processed signal intensities. Microarray data were analyzed using ingenuity pathway analysis software (Ingenuity Systems, Redwood City, CA).
Patients and primary tissue samples
Histologically proven primary GC tumor samples were obtained from 195 consecutive patients who underwent curative gastrectomy between 2011 and 2013 at Kyoto Prefectural University of Medicine,Japan. For mRNA analysis, frozen tissue samples of normal stomach and tumors were collected from surgical specimens and stored at -80 °C. Written informed consent was obtained from all patients prior to enrollment. Patients with noncurative resection or preoperative chemotherapy were excluded from the study. Tumor staging was conducted according to the International Union Against Cancer/TNM Classification of Malignant Tumors (8thedition)[24]. The present study was approved by the Institutional Review Board of the Kyoto Prefectural University of Medicine (ERB-C-1195).
IHC
The Vectastain avidin-biotinylated peroxidase complex Elite Kit (Vector Laboratories, Burlingame, CA,United States) was employed for IHC staining using the avidin-biotinylated peroxidase complex method. After deparaffinization in xylene, sections were rehydrated in a graded series of ethanol solutions. The sections were then incubated in 0.3% H2O2for 30 min to block endogenous peroxidase activity. Endogenous biotin, biotin receptors, and avidin-binding sites were also blocked using an Avidin/Biotin Blocking Kit (Vector laboratories). Sections were incubated with ANO5 antibody diluted 1:100 at 37 °C for 1 h and then at 4 °C overnight. Cells were visualized using the standard avidinbiotinylated peroxidase complex method, with hematoxylin as the counterstain.
ANO5 expression levels in immunohistochemically stained samples were semi-quantitatively graded based on the staining intensity and proportion of cytoplasm in the stained cancer cells. The staining intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining).The proportion of stained tumor cells as a percentage of the stained area in the cancer area was scored from 0 to 1.0. IHC scores were calculated as the maximum multiplied product of intensity and proportion scores (0-3.0). IHC diagnosis was based on tumor ANO5 expression assessment, and other IHC parameters were performed by at least two physicians, including an experienced pathologist.
Quantification of intracellular chloride concentration and low chloride stimulation
MQAE reagent, a chloride-sensitive fluorescence probe (Dojindo Laboratories, Kumamoto, Japan) was used to assess intracellular chloride concentrations. NUGC4 and MKN45 cells were seeded in 24-well plates at a density of 3.0 × 104cells/well and then incubated in normal medium at 37 °C with 5% CO2.The medium was then replaced with standard and low-chloride medium in which MQAE was dissolved, and cells were incubated at 37 °C in a CO2incubator for a further 12 h. Following washing with PBS five times, the fluorescence intensity of MQAE was evaluated under a fluorescence microscope(BZ-X800; Keyence, Osaka, Japan). Three fields of view were analyzedpersample at × 100 magnification. Quantification was performed using a BZ-X800 analyzer and accompanying software(BZ-H4C, v.1.1.1.8; Keyence).
A low chloride stimulation experiment was conducted to examine the effects of changes in intracellular chloride concentrations on GC cells. A low-chloride medium supplemented with 10% FBS was prepared in chloride-free RPMI-1640 (chloride replaced with NO3-) (Nacalai Tesque).
c-Jun N-terminal kinase signaling pathway inhibitor treatment
To block the c-Jun N-terminal kinase (JNK) signaling pathway, NUGC4 and MKN45 cells were incubated with the JNK inhibitor SP600125 (10µm, ab120065, Abcam) according to manufacturer’s instructions. The cells were divided into 3 groups: control, ANO5 siRNA, and JNK inhibitor (ANO5 siRNA + SP600125). Cell proliferation was detected every 24 h after ANO5 silencing.
Statistical analysis
Statistical analysis was performed using the Mann-WhitneyUtest for two-group comparisons.Categorical data were analyzed using Fisher’s exact test. The Kaplan-Meier method was used to construct survival curves, and differences in survival were examined using the log-rank test for equality. Prognostic factors were identified using the Cox proportional hazard model. These analyses were performed using the JMP statistical software (version 15; SAS Institute, Cary, NC, United States).Data are presented in the graphs as the mean ± standard error of the mean.P< 0.05 was considered a statistically significant difference.
RESULTS
ANO5 expression in GC cells
ANO5gene and protein expression were first examined in five human GC cell lines, MKN7, MKN45,MKN74, HGC27, and NUGC4, by reverse transcription-quantitative PCR and western blotting. ANO5 expression was detected in several cells in the five GC cell lines (Figure 1A and B). Compared to paired adjacent normal tissue,ANO5 expression was significantly upregulated in GC tissue(P=0.004;n=12;Supplementary Figure 1).
ANO5 expression was knocked down using siRNA in NUGC4 and MKN45 cells, and its effects on tumor progression were assessed.ANO5mRNA (Figure 1C) and protein levels (Figure 1D) were downregulated in NUGC4 and MKN45 cells. We also conducted an overexpression study in MKN7 cells. The ANO5 plasmid increasedANO5mRNA levels (Supplementary Figure 2A, left panel).
ANO5 regulates cell growth and survival in GC cells
The effect of ANO5 siRNA transfection on the proliferation and cell cycle progression of NUGC4 and MKN45 cells were subsequently examined. Compared with the control siRNA, the number of NUGC4 and MKN45 cells was significantly reduced at 48 h and 72 h after the transfection with ANO5 siRNA(Figure 2A, left panel). The results of the cell proliferation assay showed that the relative absorbance of GC cells transfected with the control siRNA (NUGC4 and MKN45) was significantly lower than that of GC cells transfected with ANO5 siRNA (HSS137119) (NUGC4 and MKN45) (Figure 2A, right panel).Whereas, ANO5 plasmid increased the relative absorbance of MKN7 cell (Supplementary Figure 2B).Moreover, ANO5 silencing increased the numbers of NUGC4 and MKN45 cells in the G0/G1phase(Figure 2B). These results indicated that ANO5 regulated the proliferation and cell cycle of GC cells.
To further clarify the role of ANO5, apoptosis assays were performed in NUGC4 and MKN45 cells.ANO5 silencing significantly increased the frequency of early and late apoptotic NUGC4 cells and the frequency of early apoptotic MKN45 cells 72 h after siRNA transfection (Figure 3A). These results indicated that the apoptosis in NUGC4 and MKN45 cells was regulated by ANO5 expression. Another ANO5 siRNA (HSS137120) was used to assess its impact on cell growth and survival, with results similar to those of HSS137119 (Supplementary Figure 3).
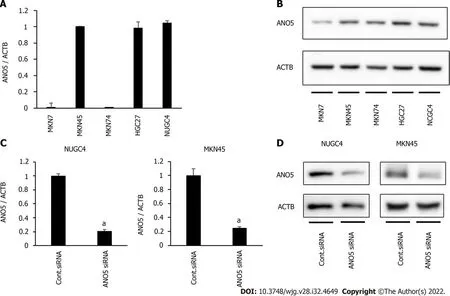
Figure 1 Anoctamin 5 expression in gastric cancer cells. A: Real-time quantitative reverse transcription-quantitative polymerase chain reaction (PCR)showed the expression of anoctamin 5 (ANO5) in various cell lines in gastric cancer; B: Western blotting showed the expression of ANO5 in various cell lines in GC;C: Real-time quantitative PCR revealed that ANO5 small interfering RNA (siRNA) effectively reduced ANO5 mRNA levels in NUGC4 and MKN45 cells; D: Western blotting revealed that ANO5 siRNA effectively reduced ANO5 protein levels in NUGC4 and MKN45 cells. n = 3, mean ± standard error of the mean. aP < 0.05(significantly different from control siRNA). ANO5: Anoctamin 5; ACTB: β-actin; siRNA: Small interfering RNA.
ANO5 promotes the migration and invasion
The effects of the ANO5 silencing on NUGC4 and MKN45 cell migration and invasion were examined using a Boyden chamber assay. The results demonstrated that ANO5 knockdown in NUGC4 and MKN45 cells significantly reduced their migration and invasion (Figure 3B).
Gene expression profiling in ANO5 siRNA-transfected NUGC4 cells
To elucidate the molecular mechanisms underlying the regulation of cellular functions by ANO5, the gene expression profiles of NUGC4 cells transfected with ANO5 siRNA were investigated using microarray. The results revealed that the expression levels of 3491 genes in NUGC4 cells following ANO5 knockdown exhibited fold-changes > 1.8 compared with those in the negative control. Among these, the expression levels of 1802 genes were upregulated, whereas those of 1689 genes were downregulated in NUGC4 cells following ANO5 knockdown. The top 20 genes with significant changes in expression in the ANO5-depleted NUGC4 cells are listed in Tables 1 and 2. Ingenuity pathway analysis showed that ‘Cancer’ was the top-ranked disease and disorder, while ‘DNA Replication,Recombination, and Repair’ and ‘Cell Cycle’ were the two top-ranking molecular and cellular functions(Table 3).
Validation of gene and protein expression
The microarray analysis identified ‘Cell Cycle: G1/S Checkpoint Regulation’ as one of the top-ranking canonical pathways in ANO5-depleted NUGC4 cells (Figure 4A). To confirm these results, seven genes were selected (CDK2,CDK4,CDK6,CDKN1A/p21,CCNE2,E2F1, andRb). These genes were included in‘Cell Cycle: G1/S Checkpoint Regulation,’ andCDK2andCDK6were the two top-ranking downregulated genes in NUGC4 cells following ANO5 knockdown (Table 2). Reverse transcription-quantitative PCR was used to confirm the expression levels of six genes. NUGC4 and MKN45 cells transfected with ANO5 siRNA had significantly lowerCDK2,CDK4,CDK6,CDKN1A,CCNE2, andE2F1expression levels and significantly higherCDKN1Aexpression levels than cells transfected with the control siRNA(Figure 4B). Furthermore, ANO5 plasmid decreasedCDKN1A/p21mRNA levels (Supplementary Figure 2A, right panel). Western blotting revealed that phosphorylated Rb was inhibited following ANO5 knockdown in NUGC4 and MKN45 cells (Figure 4C). Since theCDK2gene is located upstream of Rb and downstream of p21 in the cell cycle transition from G1phase to S phase, the downregulation ofANO5 in GC cells appears to affect the transition from the G1 phase to the S phase by regulating the expression of p21 and its downstream genes in signal pathways.

Table 1 The 20 upregulated genes that displayed the greatest changes in their expression in anoctamin 5-depleted NUGC4 cells

Table 2 The 20 downregulated genes that displayed the greatest changes in their expression in anoctamin 5-depleted NUGC4 cells

SCAMP2 Secretory carrier membrane protein 2 TC1500010047.hg.1-13.05 CMTM7 CKLF like MARVEL transmembrane domain containing 7 TC0300006968.hg.1-13.02 GINS4 GINS complex subunit 4 TC0800007416.hg.1-12.66
In addition, the gene expression profile of MKN45 cells transfected with ANO5 siRNA was investigated by microarray. Changes in gene expression in ANO5-depleted NUGC4 and MKN45 cells are depicted in Supplementary Figure 4. Among the 21440 genes, 7246 genes were upregulated and 6622 genes were downregulated in both cell lines, for a total of 13868 genes (64.7%) with identical expression direction in NUGC4 and MKN45 cells. The direction of gene expression changes of gene related to ‘Cell Cycle: G1/S Checkpoint Regulation’ was consistent in both cell lines (Supplementary Table 1).Furthermore, all 40 genes displayed in Tables 1 and 2 showed the identical expression patterns in ANO5-depleted MKN45 cells (Supplementary Table 2). These results supported that ANO5 affected the cell cycle through similar mechanisms in both NUGC4 and MKN45 cell lines.
ANO5 inhibits the JNK pathway
Effects of low-chloride conditions
To elucidate the molecular mechanisms through which ANO5 affects the cell cycle transition from G1to S phase, changes in the intracellular ion environment were examined. A previous study has reported that intracellular chloride affects cancer growth through the phosphorylation of several key molecules in signal transduction pathways[20]. We previously reported that the culturing in a Cl--replaced medium (replacement of Cl-by NO3-) decreased the intracellular chloride concentration [(Cl-)i] and inhibited cell growth in GC cells[19]. Our previous study also demonstrated that JNK activation under low-chloride conditions inhibited GC cell growth by upregulating p21 expression[18]. Intracellular chloride concentrations in the cells were measured based on the fluorescence intensity of MQAE, a chloride-sensitive fluorescence probe. The results revealed an increases in the fluorescence intensity of MQAE in NUGC4 and MKN45 cells following ANO5 knockdown (Figure 5). Therefore, ANO5 knockdown altered intracellular chloride concentrations in GC cells. Furthermore, low-chloride conditions effectively increased JNK phosphorylation and reduced Rb phosphorylation (Supplementary Figure 6). These results indicated that ANO5 regulated the cell cycleviaJNK signaling by controlling intracellular chloride levels.
IHC analysis of ANO5 expression in human GC tissue
IHC detected the expression of ANO5 in non-cancerous gastric (Figure 6A) and cancerous epithelia(Figure 6B). ANO5 was observed to be expressed in the cell membranes and cytoplasm of GC tissue.The criteria for the staining intensity score were defined as 0 (no staining; Figure 6C), 1 (weak staining;Figure 6D), 2 (moderate staining; Figure 6E), or 3 (strong staining; Figure 6F). The median and mean scores for ANO5 expression were 0.9 (range, 0-2.1) and 0.97 (standard deviation = 0.53), respectively. A cut-off value of 1.3 was used to obtain the smallestPvalue in comparison of 5-year overall survival (OS)rates[26]. The 5-year OS rates for each cut-off value are presented in Table 4.
Patients with GC were divided into low- (ANO5 scores < 1.3,n= 137) and high-ANO5 (ANO5 scores≥ 1.3,n= 58) expression groups based on a cut-off value of 1.3 (Figure 6G). Analysis of the clinicopathological features revealed that ANO5 expression levels were not associated with any of the variables(Table 5). To evaluate the prognostic significance of ANO5 after surgery, the following ten variables were compared: sex, age, tumor location, tumor length, histological type, lymphatic invasion, venous invasion, the pathological T stage, pathological N stage, and ANO5 IHC scores. Univariate analysis showed that patient prognosis was correlated with tumor length, lymphatic invasion, venous invasion,pathological T stage, pathological N stage, and ANO5 IHC scores (P= 0.0020, 0.0002, 0.0126, < 0.0001, <0.0001, and 0.0104, respectively). Multivariate analysis identified high ANO5 expression (≥ 1.3) as an independent prognostic factor (P= 0.0457) (Table 6). Furthermore, the 5-year OS rate was significantly lower in the high expression group (73.9%) than in the low expression group (89.6%). Data obtained from the Kaplan-Meier plotter database also indicated that high ANO5 expression correlates with poorprognosis in GC (Supplementary Figure 7), which was consistent with the present results.

Table 3 lngenuity pathway analysis of anoctamin 5-depleted NUGC4 cells

Table 4 Five-year overall survival rates with cut-off values for anoctamin 5 expression scores
DlSCUSSlON
The ANO family of membrane proteins, also known as TMEM16, play key roles in several physiological functions, including ion transport to phospholipid scrambling[27] and ion channel regulation[28]. While the roles of ANO1 (TMEM16A) and ANO2 (TMEM16B) as calcium-activated chloride channels have been firmly established[29-32], the functions of other family members remain unclear.
Previous studies have evaluated the expression and role of ANO5 during tumor development in various cancer types. Songet al[33] demonstrated that ANO5 (TMEM16E) was widely expressed in the epithelial cells of the human gastrointestinal tract. ANO5 is also expressed in human pancreatic cancer tissues but not in normal pancreatic tissue[23]. Changet al[22] reported that ANO5 expression was downregulated in thyroid cancer, which promoted thyroid cancer cell migration and invasion.However, the expression of ANO5 in human GC tissue and the pathophysiological role of its expression in GC cells have not been demonstrated.
The present study revealed that ANO5 downregulation in GC cells regulates the cell cycle and induces apoptosis, while inhibiting proliferation, migration, and invasion. These results highlight thepotential of ANO5 inhibitors as therapeutic agents for the treatment of GC or other cancer types with high ANO5 expression levels. The present study also indicates that ANO5 plays a key role in the proliferation of GC cells. Cell cycle analysis showed that the number of cells remaining in the G0/G1phase was significantly increased, whereas the number of cells in the S or G2/M phase was decreased in ANO5-depleted NUGC4 and MKN45 cells, suggesting that ANO5 downregulation inhibits GC cell proliferationviacell cycle arrest at the G0/G1phase.

Table 5 Relationships between clinicopathological factors of gastric cancer patients and anoctamin 5 expression
The induction of p21 is dependent on the tumor suppressor protein, p53. However, chloride ions have been shown to play important roles in cell cycle progression by regulating the expression of p21 through a p53-independent pathway in GC cells[19]. It has also been demonstrated that a decrease in chloride induced G0/G1phase arrest by downregulating CDK2 and phosphorylated Rb expression through p21 upregulation. Furthermore, p38 and JNK activation under low-chloride conditions inhibits GC cell viability by upregulating p21 expression[18].
Moreover, ANO5 expression in GC cells affects the transition from the G1to the S phase of the cell cycle by regulating the expression of p21 and its downstream genes through the activation of JNK signaling. To the best of our knowledge, the chloride channel activity of ANO5 has not been confirmed to date. To elucidate the molecular mechanisms underlying the effects of ANO5, we evaluated the changes in the intracellular chloride ion environment in this study. A quantitative analysis of intracellular chloride ion concentrations was conducted based on the fluorescence intensity of MQAE.Immunofluorescent analysis showed that the fluorescence intensity of MQAE increased following ANO5 silencing, indicating a decrease in intracellular chloride concentration. These results suggest that the downregulation of ANO5 induces G0/G1phase arrest by altering the expression of G1/S checkpointrelated genes through the intracellular chloride environment of GC cells. Furthermore, since ANO5 functioned as a chloride channel, it may have the potential to inhibit tumor growth by regulating intracellular chloride concentrations in therapeutic settings.
利用多个教学软件开展实训教学,进一步强化任务驱动教学法的开展,与其他课程互相结合,突出课程重点,呼应其他国际贸易环节。具体可以遵循以下过程,首先使用外贸流程模拟实训平台中的DOC系统对国际结算票据和信用证开证申请书进行填制练习,在此基础上使用国际结算专业实训软件对结算方式的流程进行操作,这样既强化了学生对主要票据和结算方式的掌握,又节省了课内时间,可以在有限的实训学时内,安排更多的练习,让学生完成较多的课程任务。
Although ANO5 was recently implicated in various cancers, its role in tumor progression in patients with GC remains unclear. To demonstrate the clinical significance of ANO5 expression, the survival rate of 195 patients who underwent curative resection for primary GC was investigated. IHC analysisrevealed that high ANO5 expression levels were a poor prognostic factor in patients with GC. Under low-chloride conditions, ANO5 appeared to function as a chloride channel in GC cells. Previous findings showed that various ion transporters function as biomarkers and therapeutic targets[34,35].Targeting ion channels that are activated in cancer cells may be an important strategy for cancer therapy. To the best of our knowledge, the present study is the first to report a relationship between ANO5 expression and the prognosis of patients with GC. Additional functional studies are needed to provide insights into the role of ANO5 in GC progression.
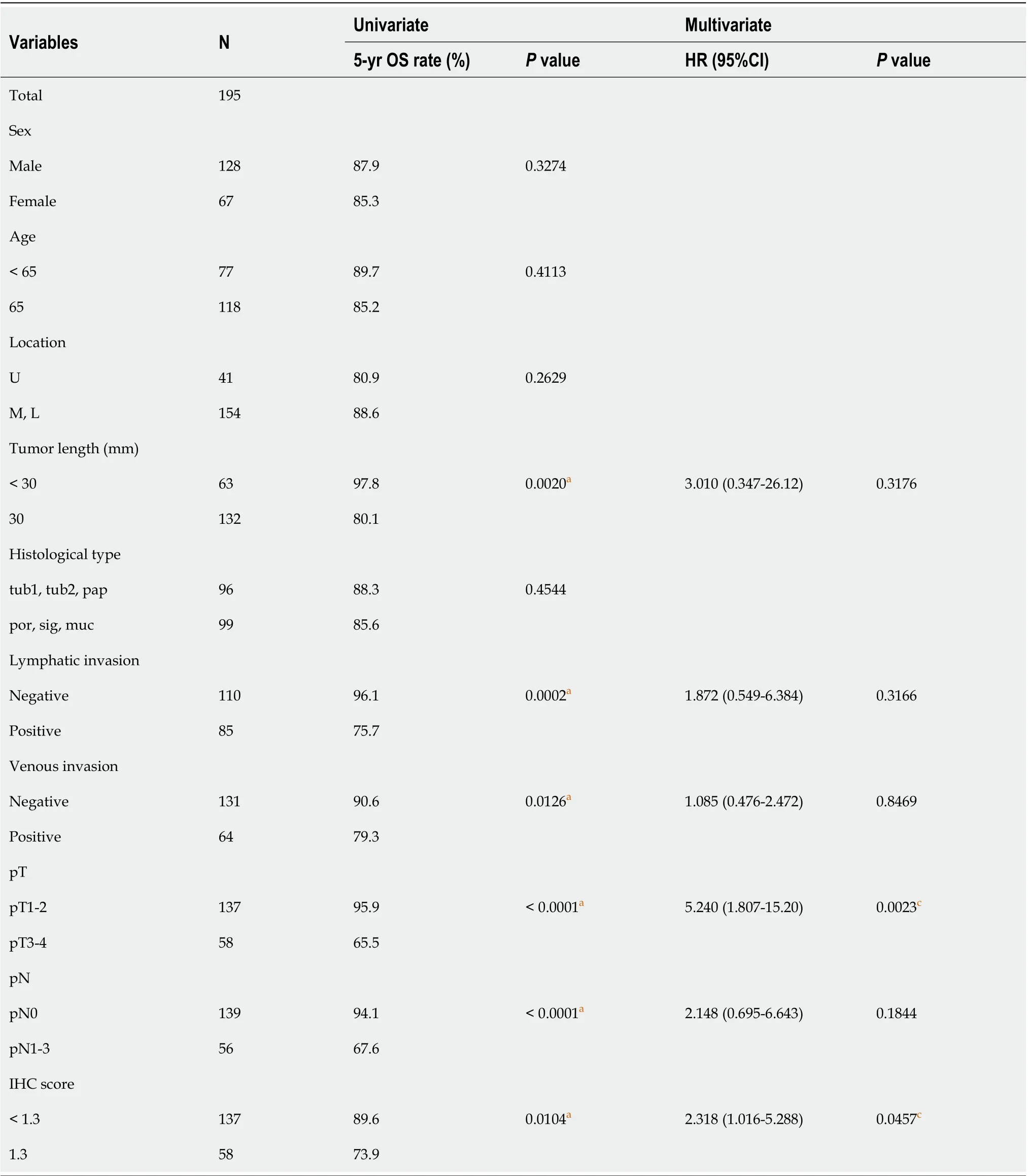
Table 6 Univariate and multivariate analyses of prognostic factors associated with 5-year overall survival

Figure 2 Anoctamin 5 controlled the proliferation and the cell cycle in gastric cancer cells. A: The downregulation of anoctamin 5 (ANO5) inhibited the proliferation of NUGC4 and MKN45 cells. Cell numbers were counted 48 h and 72 h after small interfering RNA transfection (left panel). The proliferative ability of NUGC4 and MKN45 cells was significantly suppressed following ANO5 downregulation (right panel); B: The downregulation of ANO5 increased the number of cells in the G0/G1 phase in NUGC4 and MKN45 cells. Cells transfected with control or ANO5 small interfering RNA were stained with propidium iodide and analyzed by flow cytometry. n = 3, mean ± standard error of the mean. aP < 0.05 (significantly different from control small interfering RNA). ANO5: Anoctamin 5; siRNA: Small interfering RNA; Cont.: Control.

Figure 3 Anoctamin 5 controlled apoptosis, migration, and invasion in gastric cancer cells. A: The downregulation of anoctamin 5 (ANO5)increased the early and late apoptotic cell proportions of NUGC4 and MKN45 cells. Control or ANO5 small interfering RNA-transfected cells were stained with propidium iodide and annexin V and subjected to flow cytometry; B: The downregulation of ANO5 significantly decreased NUGC4 and MKN45 cell migration and invasion, which were evaluated using a Boyden chamber assay. Magnification: × 40. n = 3, mean ± standard error of the mean. aP < 0.05 (significantly different from control small interfering RNA). ANO5: Anoctamin 5; PI: Propidium iodide; siRNA: Small interfering RNA; Cont.: Control.

Figure 4 Network analyses by microarray analysis and ingenuity pathway analysis. A: The signaling map of “Cell Cycle: G1/S Checkpoint Regulation,” one of the top-ranked canonical pathways related to the depletion of anoctamin 5 (ANO5) according to ingenuity pathway analysis. Red and green colors indicate genes with expression levels that were higher or lower, respectively, than reference RNA levels; B: To verify gene expression profiling data, CDK2, CDK4,CDK6, CDKN1A, CCNE2, and E2F1 were examined by real-time reverse transcription-quantitative polymerase chain reaction. The downregulation of ANO5 effectively reduced CDK2, CDK4, CDK6, CCNE2, and E2F1 mRNA levels and increased CDKN1A/p21 mRNA levels in NUGC4 and MKN45 cells; C: The downregulation of ANO5 effectively reduced the phosphorylation levels of Rb protein in NUGC4 and MKN45 cells; D: The downregulation of ANO5 increased the phosphorylation of the c-Jun N-terminal kinase protein in NUGC4 and MKN45 cells. n = 3, mean ± standard error of the mean. aP < 0.05 (significantly different from control small interfering RNA). ANO5: Anoctamin 5; siRNA: Small interfering RNA; Cont.: Control; JNK: c-Jun N-terminal kinase; ACTB: β-actin.
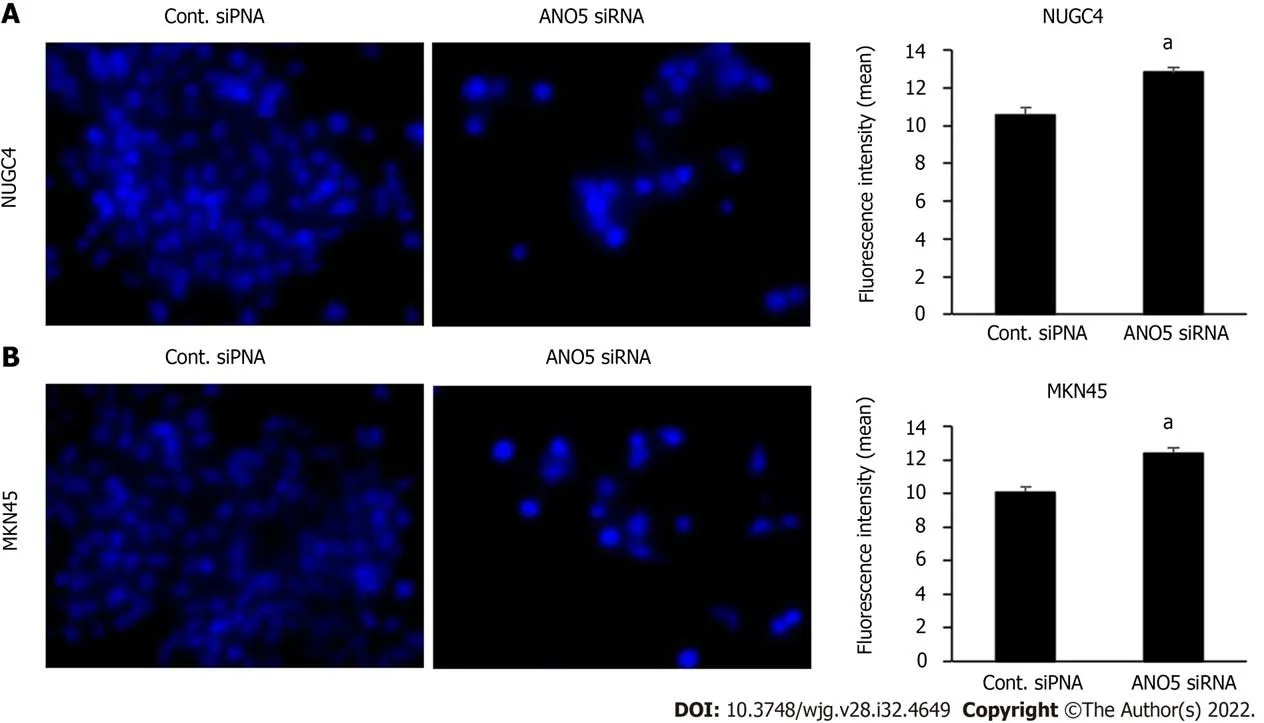
Figure 5 lmmunofluorescent analysis of intracellular chloride concentration using MQAE reagent, a chloride-sensitive fluorescence probe. The downregulation of anoctamin 5 increased the fluorescence intensity of MQAE in NUGC4 and MKN45 cells. A: NUGC4 cell; B: MKN45 cell. n = 3, mean ±standard error of the mean. aP < 0.05 (significantly different from control small interfering RNA). ANO5: Anoctamin 5; siRNA: Small interfering RNA; Cont.: Control.
This study has some limitations that must be addressed. First, it was a retrospective study. Due to the limited sample size, the pathological N stage classification factors were not correlated with the 5-year OS rate. Therefore, further studies are required to confirm our results. Second, in the selection of GC cell lines, we selected five cell lines; however, only three were used in the study.
CONCLUSlON
The present study revealed that ANO5 plays a significant role in cell cycle progression in human GC cells. The results of microarray analysis showed the impact of ANO5 on the expression of G1/S checkpoint-related genes. Furthermore, ANO5 expression significantly affected JNK signaling.Collectively, these results indicate that ANO5 plays an important role in cell cycle progression by regulating the expression of p21 through JNK signaling in human GC cells. The results of the IHC analysis also suggest that high ANO5 expression levels are a poor prognostic factor in patients with GC.The present study may contribute to the identification of ANO5 as a key mediator of tumor progression,with it ultimately being a promising prognostic biomarker or a novel therapeutic target for GC.

Figure 6 Anoctamin 5 protein expression levels in human gastric cancer tissues and a survival analysis based on anoctamin 5 expression. A: Non-cancerous gastric epithelia were immunohistochemically stained using an anti-anoctamin 5 (ANO5) antibody. Magnification: × 100; B: Primary human gastric cancer samples were immunohistochemically stained using an anti-ANO5 antibody. Magnification: × 100; C: The immunohistochemical staining results of ANO5 are shown as intensity 0. Magnification: × 400; D: Intensity 1. Magnification: × 400; E: Intensity 2. Magnification: × 400; F: Intensity 3. Magnification: × 400;G: Gastric cancer patients were classified into two groups based on ANO5 expression: A low ANO5 expression group (< 1.3, n = 137) and high ANO5 expression group (≥ 1.3, n = 58). aP < 0.05 (significant difference). ANO5: Anoctamin 5.
ARTlCLE HlGHLlGHTS
Research background
Anoctamin 5 (ANO5) is a member of a family of calcium-activated chloride channels containing 10 members, also known as transmembrane proteins, and has been reported to be associated with various cancers.
Research motivation
The role of ANO5 in gastric cancer (GC) remains poorly understood. In the present study, we analyzed the relationship between ANO5 expression and tumor progression in GC.
Research objectives
The objectives of the present study were to investigate whether ANO5 contributes to the regulation of cancer growth and to clarify its clinicopathological significance in GC.
Research methods
Knockdown experiments were performed by transfecting human GC cell lines with ANO5 small interfering RNA. Gene expression was then assessed using microarray analysis. Samples from 195 patients with GC were subjected to immunohistochemistry for ANO5, and its relationship with clinicopathological factors and prognosis were examined.
Research results
ANO5 knockdown suppressed the proliferation, migration, and invasion of cells and enhanced apoptosis. Cell cycle analysis showed that ANO5 knockdown suppressed the progression of G1-S phase.The results of microarray analysis showed up- or downregulated expression of genes related to “Cell Cycle: G1/S Checkpoint Regulation” in ANO5 knockdown NUGC4 cells. Survival analysis showed significantly poorer 5-year survival in the ANO5 high expression group (highvslow; 73.9vs89.6%,P=0.0104). Immunohistochemistry multivariate analysis identified the high expression of ANO5 as an independent prognostic factor for 5-year survival in GC patients (P= 0.0457).
Research conclusions
ANO5 plays a significant role in cell cycle progression in human GC cells. The results of the immunohistochemistry analysis suggest that high ANO5 expression levels are a poor prognostic factor in patients with GC.
Research perspectives
The present study may contribute to the identification of ANO5 as a key mediator in tumor progression,with it ultimately being a promising prognostic biomarker or a novel therapeutic target of GC.
FOOTNOTES
Author contributions:Fukami T and Shiozaki A contributed equally to this work; All authors read and approved the final manuscript and agreed to be accountable for all aspects of the report.
Supported byJapan Society for the Promotion of Science, No. 21K08689, No. 21K16456, No. 20K09016, No. 20K09084,No. 19K09202 and No. 19K09182.
lnstitutional review board statement:The study was reviewed and approved by the Kyoto Prefectural University of Medicine Institutional Review Board, No. ERB-C-1195.
lnformed consent statement:Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Conflict-of-interest statement:All the authors report no relevant conflicts of interest for this article.
Data sharing statement:Technical appendix, statistical code, and dataset available from the corresponding author at shiozaki@koto.kpu-m.ac.jp. Participants gave informed consent for data sharing.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:Japan
ORClD number:Tomoyuki Fukami 0000-0002-2585-3611; Atsushi Shiozaki 0000-0003-3739-160X; Toshiyuki Kosuga 0000-0002-1657-7272; Michihiro Kudou 0000-0003-3518-528X; Hiroki Shimizu 0000-0002-6463-8498; Takuma Ohashi 0000-0003-0939-3739; Tomohiro Arita 0000-0001-7127-6504; Hirotaka Konishi 0000-0002-4899-8944; Shuhei Komatsu 0000-0001-6074-7614; Takeshi Kubota 0000-0002-2246-3028; Hitoshi Fujiwara 0000-0002-6507-4313; Kazuma Okamoto 0000-0002-8270-4217; Mitsuo Kishimoto 0000-0002-7407-9044; Yukiko Morinaga 0000-0003-2311-1830; Eiichi Konishi 0000-0002-7122-2271; Eigo Otsuji 0000-0002-3260-8155.
S-Editor:Fan JR
L-Editor:Filipodia
P-Editor:Cai YX
猜你喜欢
杂志排行
World Journal of Gastroenterology的其它文章
- The mechanism of Yinchenhao decoction in treating obstructivejaundice-induced liver injury based on Nrf2 signaling pathway
- Effects of Granule Dendrobii on chronic atrophic gastritis induced by N-methyl-N'-nitro-N-nitrosoguanidine in rats
- Machine learning predicts portal vein thrombosis after splenectomy in patients with portal hypertension: Comparative analysis of three practical models
- Sirolimus increases the anti-cancer effect of Huai Er by regulating hypoxia inducible factor-1α-mediated glycolysis in hepatocellular carcinoma
- lnternational patterns in incidence and mortality trends of pancreatic cancer in the last three decades: A joinpoint regression analysis
- Expanding beyond endoscopy: A review of non-invasive modalities in Barrett’s esophagus screening and surveillance
