SARS-CoV-2 Omicron variant (B.1.1.529): A concern with immune escape
2022-06-16AdekunleSanyaoluAleksandraMarinkovicStephaniePrakashNafeesHaiderMartinaWilliamsChukuOkorieOlanrewajuBadaruStellaSmith
Adekunle Sanyaolu, Aleksandra Marinkovic, Stephanie Prakash, Nafees Haider, Martina Williams, Chuku Okorie, Olanrewaju Badaru, Stella Smith
Adekunle Sanyaolu, Department of Public Health, Federal Ministry of Health, Abuja 0000,Nigeria
Aleksandra Marinkovic, Stephanie Prakash, Martina Williams, Department of Basic Sciences,Saint James School of Medicine, The Valley 0000, Anguilla
Nafees Haider, Department of Basic Sciences, All Saints University School of Medicine, Roseau 0000, Dominica
Chuku Okorie, Department of Allied Sciences, Union County College, Plainfield, NJ 07060,United States
Olanrewaju Badaru, Department of Public Health, Nigeria Centre for Disease Control, Abuja 0000, Nigeria
Stella Smith, Department of Molecular Biology and Biotechnology, Nigerian Institute of Medical Research, Lagos 100001, Nigeria
Abstract Omicron, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant that is now spreading across the world, is the most altered version to emerge so far, with mutations comparable to changes reported in earlier variants of concern linked with increased transmissibility and partial resistance to vaccineinduced immunity. This article provides an overview of the SARS-CoV-2 variant Omicron (B.1.1.529) by reviewing the literature from major scientific databases. Although clear immunological and clinical data are not yet available, we extrapolated from what is known about mutations present in the Omicron variant of SARS-CoV-2 and offer preliminary indications on transmissibility, severity, and immune escape through existing research and databases.
Key Words: SARS-CoV-2; COVID-19; Omicron; B.1.1.529; Variant of concern; Emerging variants
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant, Omicron (B.1.1.529), was first reported to the World Health Organization (WHO) from South Africa on November 24, 2021[1]. The Omicron infection was first confirmed from a sample collected on November 9, 2021[1,2]. The variant was also detected in Botswana in samples collected on November 11, 2021[1,3]. As of January 10, 2021, B.1.1.529 had spread across 105 countries, with most states and territories in the United States testing positive for the variant[3,4]. The Centers for Disease Control and Prevention (CDC) reported that of the 43 Omicron cases initially detected in the United States, 34 had been fully vaccinated, and 25 cases were adults aged 18 years to 39 years[5,6]. By the week of December 25, 2021, the Omicron variant accounts for approximately 95.4% of circulating SARS-CoV-2 strains, while Delta accounts for 4.6%[3].
Many of the cases included mild symptoms such as coughing, congestion, and fatigue; among the less frequently reported symptoms are nausea and vomiting, diarrhea, shortness of breath, difficulty breathing, and loss of smell or taste[6]. As of November 28, 2021, there is no evidence that the symptoms linked with Omicron are distinct from those associated with other variants, according to the WHO[1]. The severity of the condition, as well as its precise signs and symptoms, are still unknown[3].
Omicron has been labeled a variant of concern (VOC) by the WHO and European Center for Disease Prevention and Control (ECDC) on November 26, 2021, because it contains genetic changes that are predicted to increase transmissibility and decrease the effectiveness of social and public health measures along with available vaccines and therapeutics[7,8]. Its genetic profile consists of 26 unique mutations that make it significantly different from other existing variants and indicate that it is a new lineage of SARS-CoV-2[9]. This variant carries 32 mutations in the spike protein alone[7]. Omicron poses an issue because vaccines that have been created to mitigate SARS-CoV-2 infections target spike proteins. Studies in Germany, South Africa, Sweden, and Pfizer have shown a 25 to 40 times decrease in the ability of antibodies created by the Pfizer BioNTech vaccine to neutralize the variant after two doses[10,11]. However, severe coronavirus disease 2019 (COVID-19) can still be managed with the use of corticosteroids to induce T-cell apoptosis and act as an NF-KB inhibitor, and interleukin 6 (IL-6) receptor blockers, which act by targeting the IL-6/IL-6R/JAK pathway to suppress the overreaction of the immune system in COVID-19 patients and blocking the binding of IL-6 to its receptor[1]. Other studies underway to assess treatment efficacy against the Omicron variant include British drugmaker GSK and its United States partner Vir Biotechnology. According to data from their investigation, all spike mutations are effectively treated by their antibody-based COVID-19 therapy[12]. Although science and knowledge about this variant keep changing as they emerge, this report evaluates the literature from key scientific databases to provide an overview of the SARS-CoV-2 variant Omicron (B.1.1.529).
GLOBAL EPIDEMIOLOGY OF THE OMICRON VARIANT
Despite efforts to better understand viral neutralization and how antibodies and T-cells respond to the SARS-CoV-2 variant, Omicron remains a mystery[13]. On November 11, 2021, the variation was discovered in samples collected in Botswana and then in South Africa by November 14, 2021[3,8,13]. Depicted in Figure 1, most countries and territories have been affected by the Omicron variant, with the United Kingdom, United States, Denmark, France, and Germany most severely impacted, as this variant is presumed to spread more easily, even among the vaccinated population and those who do not show symptoms; therefore, increasing the overall proportion of COVID-19 cases[3,4].
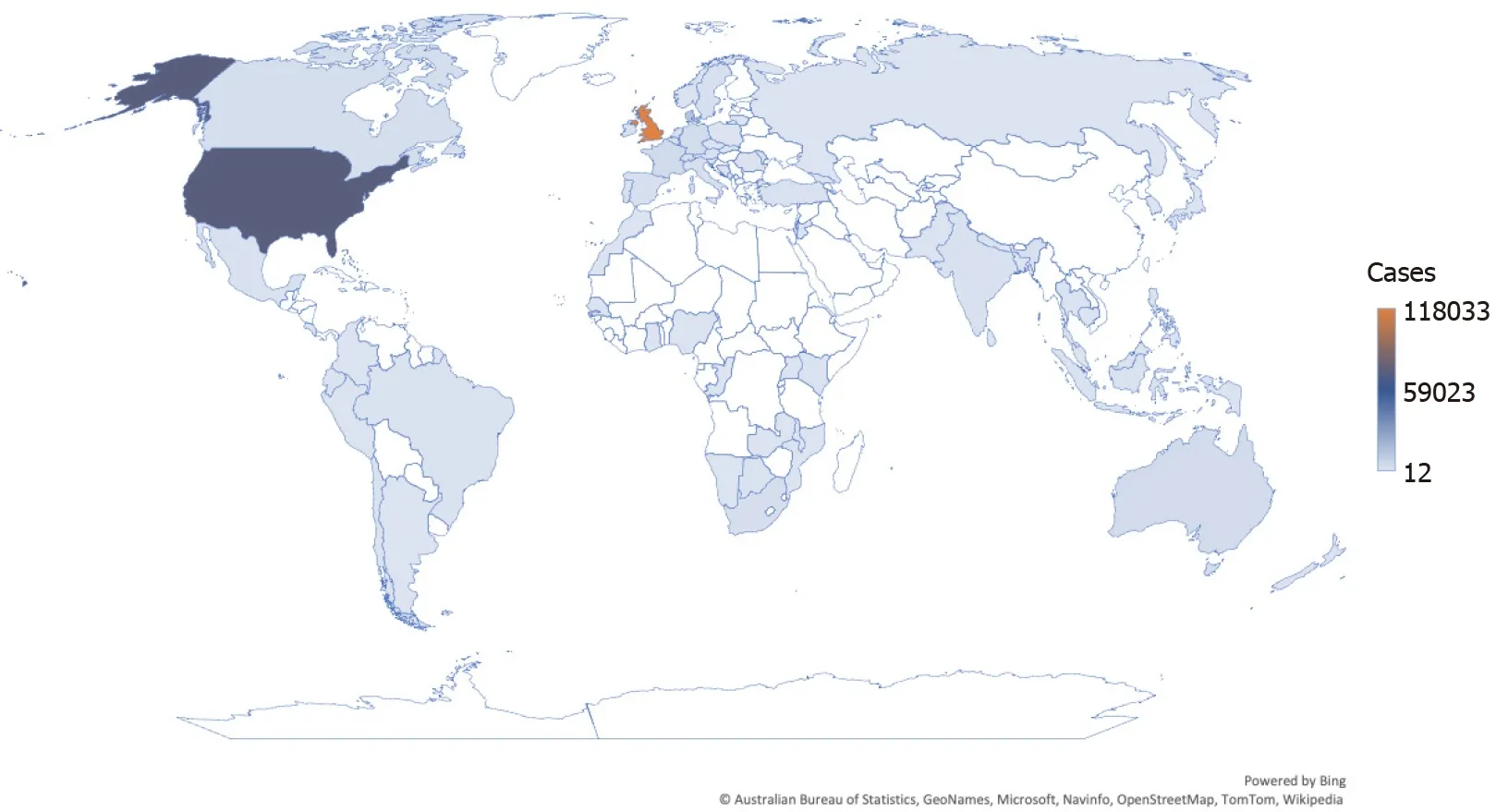
Figure 1 Confirmed Omicron cases worldwide. Data recreated and reported by GISAID as of January 10, 2022, with 242159 Omicron genome sequences reported across 105 countries[4].
Genomic sequence
As a result of genomic surveillance, thousands of mutations have been found in the SARS-CoV-2 genome[14-16]. Numerous viral variants with mutations in the spike protein, including Alpha, Beta, and Delta, have been found[17]. These variants exhibited alterations in the receptor-binding domain (RBD), and the 25 amino acids connected to the spike protein showed an increased affinity for the angiotensinconverting enzyme 2 (ACE2) receptor, boosting transmissibility[14,18].
A recent report presented by Dejnirattisaiet al[14] compared neutralization titers of the SARS-CoV-2 Omicron variant with the titers of the Victoria, Beta, and Delta variants[14,19]. Sera were acquired from individuals who received the AstraZeneca or Pfizer vaccine, both of which were administered in two doses[14,20]. According to the findings, there was a considerable decrease in neutralization titers, with evidence that some individuals were unable to neutralize at all; this can lead to breakthrough vaccine infections in previously infected patients or those who completed double doses of vaccination[21-23].
Although the amino acid sequence of the Omicron spike protein can be altered by nine different mutations (S: N440K, S: G446S, S: S447N, S: T4+78K, S: E484A, S: Q493R, S: G496S, S: Q298R, and S: N501Y), the research found that antibodies can still adhere to the mutated spike protein[24]. The Omicron variant mutations do not show any structural changes that would suggest antibody evasion; nevertheless, alterations in amino acid attachments to various locations of the binding site can cause interference when engaging with antibodies[24].
Mutations
Approximately 30 mutations in the viral spike protein have been discovered, including three small deletions and one small insertion[8]. Roughly half of the mutations affect the RBD, which serves as the virus's principal site of interaction of the virus with human cells and the target protein for several current COVID-19 vaccines[8,13]. Previously, many SARS-CoV-2 variant strains revealed distinct mutations; however, the Omicron variant shows numerous types of mutations, as well as novel mutations[13]. Although the actual origin of Omicron is unknown, numerous possibilities are now being pursued, including evolution in animal reservoirs and human reinfection, or co-infection with seasonal human coronaviruses (HCoVs), such as HCoV-229E[25-27]. Chronically infected individuals are suggested to be the source of origin, as evidenced by viral sequencing[25]. Additional research revealed that when faced with a strong immune response, SARS-CoV-2 may acquire the ability to avoid antibodies through two deletions in the N-terminal domain and a mutation in the spike protein[28]. Finally, it has been proposed that natural selection can arise as a result of mutations that increase viral infectivity, antibody resistance, and vaccine breakthrough[25,29-32]. Evolutionary descent of the Omicron lineages showed that mutations arose under selection pressure due to antibodies elicited by infection, vaccination, or both, in the human population on a large scale. As of February 2022, the Omicron variant has mutated into three lineages: BA.1, BA.2, and BA.3. A sub-lineage of BA.1 with an R346K substitution in the spike protein is classified as BA.1.1. BA.1 emerged first, which was followed by BA.2 and BA.3. Like BA.1, the earlier strains of BA.2, BA.3, and BA1.1 were detected in the Gauteng Province in South Africa. It thus suggests that the diversification of Omicron occurred in South Africa. Although BA.1 is spreading quicker than BA.2, the BA.2 lineage has become more prominent in several nations after January 2022. The genetic sequence in the spike protein of the BA.2 lineage differs from the BA.1 lineage suggesting it may confer greater immune resistance against antibodies[33-35].
Containment strategy
The U.S. Food and Drug Administration (FDA) Emergency Use Authorization (EUA) diagnostic developer, DTPM, identifies and develops assays capable of diagnosing COVID-19[36]. However, due to a nine-nucleotide deletion in the N gene, exclusive to the Omicron variant, this single target test known as the reverse transcription-polymerase chain reaction (RT-PCR) of DTPM is predicted to fail, resulting in false-negative findings in patients[36]. The specific deletion of nine nucleotides is unique to the Omicron variant and poses a potential diagnostic problem, although previously detected variants should not be affected[36].
Mutations have the potential to change the accuracy of these tests, resulting in unpredictable analytical performance characteristics and false-negative results. Using a widely available commercial assay, a G-to-U transversion (nucleotide 26372) was found in the SARS-CoV-2 E gene in three cases with low viral detection efficiency[37]. Current SARS-CoV-2 PCR tests still detect the Omicron variant[7,36]. According to reports, one of the three target genes is not detected in a commonly used PCR test[7]. This targeted gene is referred to as an S gene dropout or S gene target failure[7]. As a result, pending sequencing confirmation, this test can be utilized as a marker for the Omicron variant[7]. Furthermore, the FDA is continuing to assess the impact of Omicron on SARS-CoV-2 diagnostic tests in partnership with government authorities and test producers[36]. The FDA’s current investigation shows that the performance of some EUA-authorized molecular tests (i.e., PCR) may be affected by the mutations in the SARS-CoV-2 Omicron variant[36]. As a response, the FDA has classified the different tests into two categories: those that are predicted to fail to identify the Omicron variant and those that are expected to detect the variant using a unique gene dropout detection pattern[36]. In addition to molecular diagnostics (i.e., PCR), early evidence suggests that antigen tests can detect the SARS-CoV-2 Omicron form, although that sensitivity may be low[36].
There is much to learn about the reinfected population and effective treatment and management procedures with the Omicron variant, which has led many healthcare providers to doubt existing treatment modalities[38]. Mayeret al[38] conducted a recent case series investigation after a rise in people with mild respiratory symptoms of SARS-CoV-2 infections in the Western Cape province. After the patients received confirmation of their COVID-19 using molecular assays, they were placed in isolation and required a daily diary to record their symptoms[38]. A total of 7 patients were studied; of which, 6 of the 7 were fully vaccinated with a respective booster shot, and 5 of the 7 presented with the Omicron genome sequence[38]. Although the study reported breakthrough infections experienced by completely vaccinated patients and some who had also received a booster vaccine, all cases had increased levels of antibodies against the spike protein, a common finding in patients vaccinated with a booster dose[38,39]. Despite the inability to get accurate RNA viral loads, it is hypothesized that these individuals will have an increase in viral loads, suggesting that the Omicron variant could evade vaccine-induced immunity[38]. In another study on naive individuals following a booster shot (third dose), a 14-fold reduction in neutralizing activity against Omicron was observed; thus, the findings suggest the need for a third dose vaccination to provide robust neutralizing antibody responses against the Omicron variant[40].
Most COVID-19 vaccines have remained successful in preventing severe COVID-19, hospitalization, and death for all preceding variants, due to T-cell immune responses being more significant than antibodies[2]. In a matched study of more than 9000 Omicron cases in Ontario, the risk of hospitalization or death was lower for Omicron cases when compared with Delta cases[41]. Importantly, the implications of the remaining Omicron mutations are unknown, leaving a great deal of ambiguity about how the complete mix of deletions and mutations may affect viral behavior and vulnerability to natural and vaccine-mediated immunity[2]. Furthermore, a brief clinical course indicated that fully vaccinated patients who had received a booster dose retained sufficient protection against severe COVID-19 infections; thus, this supported the continued use of booster doses to help combat the spread of the Omicron variant[38,42].
COVID-19 has presented different lessons and challenges to various regions and countries of the world, and long-term data will be needed to assess vaccine efficacy in the face of the potential appearance of novel variants like Omicron[43]. Despite some evidence that vaccination alone may not be enough to prevent symptomatic infection, non-pharmaceutical practices such as continued use of face masks in the public despite vaccination and booster status of the vaccine, proper hygiene precautions, and social distancing, as well as genomic surveillance, are required to successfully combat this variant[38,44].
CONCLUSION
The emergence and global spread of Omicron, which may be antibody-resistant and appears to be highly transmissible, emphasize the importance of genomic surveillance in conjunction with immune profiling. Reduced antibody titers may impair the ability of vaccines to prevent infection, but protection against severe disease is likely to be maintained. To avoid or minimize further spread and mutations, preventive measures such as adequate patient care management, early detection of suspicious cases, outbreak tracing, isolation protocols for the infected, continued adherence to social distancing, wearing a face mask, and vaccination must be accepted by the public and encouraged by public health professionals, government officials, and community leaders.
FOOTNOTES
Author contributions:Sanyaolu A, Marinkovic A, Prakash S, Haider N, Williams M, Okorie C, Badaru O and Smith S contributed to the design, writing and final approval of the manuscript.
Conflict-of-interest statement:The authors have no conflict of interest to this manuscript.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:Nigeria
ORCID number:Adekunle Sanyaolu 0000-0002-6265-665X; Aleksandra Marinkovic 0000-0002-3672-0777; Stephanie Prakash 0000-0003-2664-9775; Nafees Haider 0000-0003-4449-0905; Martina Williams 0000-0001-5136-4179; Chuku Okorie 0000-0001-5483-0032; Olanrewaju Badaru 0000-0003-3035-2640; Stella Smith 0000-0003-2163-1189.
S-Editor:Gao CC
L-Editor:A
P-Editor:Gao CC
杂志排行
World Journal of Virology的其它文章
- Severe acute respiratory syndrome coronavirus 2 infection: Role of interleukin-6 and the inflammatory cascade
- Impact of COVID-19 on mental health and emotional well-being of older adults
- Omicron variant and change of electrostatic interactions between receptor binding domain of severe acute respiratory syndrome coronavirus 2 with the angiotensin-converting enzyme 2 receptor
- Educational, psychosocial, and clinical impact of SARS-CoV-2(COVID-19) pandemic on medical students in the United States
