斑点叉尾全基因组微卫星分布特征分析*
2022-04-11唐荣叶苏孟园杨汶珊徐杰杰尹绍武
唐荣叶 苏孟园 杨汶珊 徐杰杰 王 涛 尹绍武
唐荣叶 苏孟园 杨汶珊 徐杰杰 王 涛①尹绍武①
(南京师范大学海洋科学与工程学院 江苏省特色水产育种与绿色高效养殖技术工程研究中心 江苏 南京 210023)


微卫星又称简单序列重复(simple sequence repeats, SSRs),广泛分布于真核、原核生物以及病毒中,是由1~6 bp碱基为单元重复串联而成的DNA序列。根据核心序列的排列差异,又分为完整型和不完整型(Morgante, 2001)。其中,完整型指重复序列中不存在其他碱基或重复序列的情况,不完整型指重复序列中存在错配情况。目前,对微卫星的研究主要集中于完整型(郑燕等, 2012),因其具有多态性高、共显性遗传等特点,目前被广泛应用于遗传图谱构建(Shen, 2007; Xia, 2010)、遗传多样性分析和种质资源保护(Narasimhamoorthy, 2008)等研究中。

1 材料与方法
1.1 全基因组来源
1.2 数据处理方法

2 结果与分析
2.1 斑点叉尾全基因组微卫星总体分布特征
2.2 斑点叉尾29条染色体不同重复类型微卫星分布特征
2.3 斑点叉尾全基因组微卫星各重复类别特征

图1 斑点叉尾各染色体微卫星数量分布
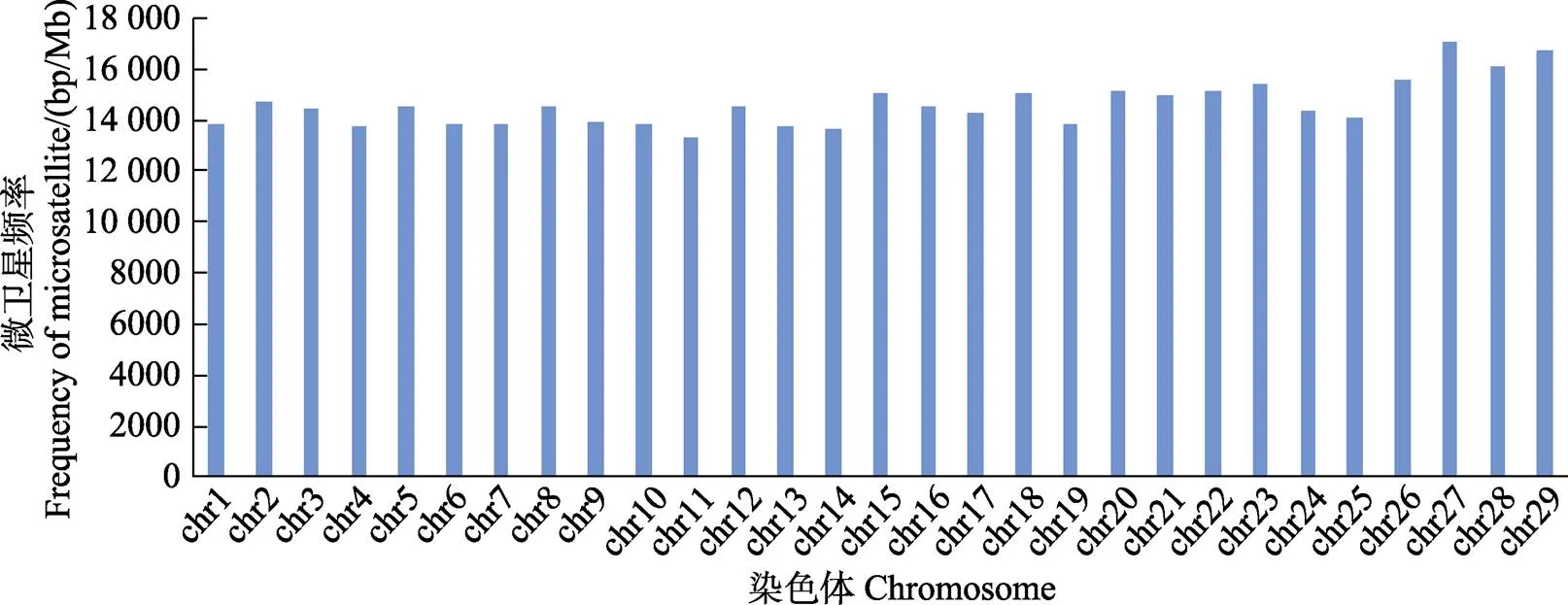
图2 斑点叉尾各染色体微卫星频率分布


Tab.1 Distribution of different types of SSRs in each chromosome of I. punctatus
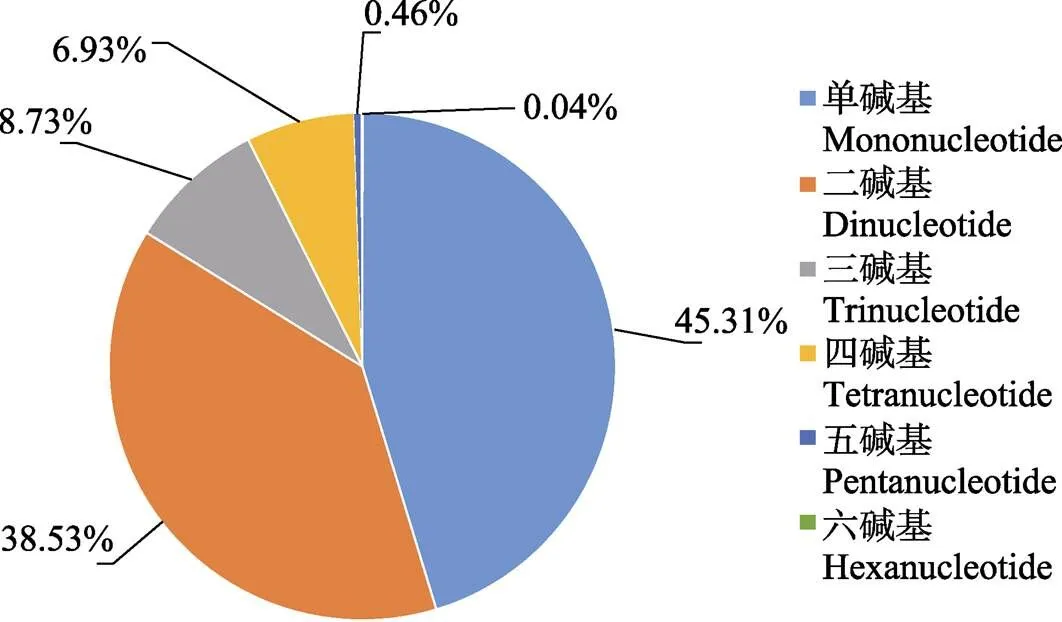
图3 斑点叉尾全基因组微卫星各重复类型占比
2.4 斑点叉尾微卫星各重复类型拷贝数分布特征
3 讨论
3.1 斑点叉尾全基因组微卫星序列分布特征分析
在本研究中,染色体DNA序列的长度与染色体上所含有的微卫星数量具有高度相关性(SPSS,=0.98,<0.01),这与黄杰等(2012)和戚文华等(2013)的研究结果一致。同时也支持了Hancock (1996)的假说:染色体序列越长,微卫星含量越高的趋势。统计各染色体上的各种类型微卫星数量,发现各类型的比例除27号和29号染色体二碱基大于单碱基外,均符合29条染色体整体微卫星分布特征,即单碱基最多,其次是二碱基、三碱基、四碱基、五碱基和六碱基。这个规律与张琳琳等(2008)对赤拟谷盗()的研究结果相似,而与黄杰等(2012)对红原鸡()各染色体微卫星分布情况的研究结果相差甚远。高焕等(2005)认为,同一物种的不同染色体上,各种类型的重复序列分布有很大差异。而本研究的结果表明,各种类型的微卫星在各染色体上的分布符合整体规律,这可能和不同物种的不同类型微卫星的特定功能有关系。


Tab.2 Top 10 repeated copy categories of microsatellites in genomes of I. punctatus
注:占比表示该类别在各自重复类型微卫星中的占比
Note: The percentage represents the proportion of different types of SSRs

图4 斑点叉尾全基因组微卫星重复类型重复数分布
3.2 斑点叉尾各碱基类型微卫星不同类别分布特征分析



四碱基、五碱基的数量较多的前2种类别分别是AAAT、AAAC和ATAAT、AAAAC,六碱基的数量相对于前5种重复类型比较少,数量最多的2种类别是TGACTA和ATAGTC。与前3种碱基类型一样,四碱基、五碱基和六碱基也表现出明显的A/T碱基优势。有研究表明,DNA序列中G/C含量越高,微卫星分布越少。对于这个现象,倪守胜等(2018)通过分析虾夷扇贝()基因组的微卫星分布特征,得出DNA复制滑动机制和重组机制使得A/T重复类型的产生几率更高的结论。Schorderet等(1992)认为,由于基因组中CpG甲基化,胞嘧啶C容易在脱氨基作用下突变为胸腺嘧啶T。这可能是二碱基中CG含量较少、TG (AC类型)含量较多的原因。然而Stallings (1992)的研究则表明,无论是否有CpG甲基化过程,基因组中的CG重复类别都是偏小的,他提出CpG结构是一种有害结构。
3.3 斑点叉尾各碱基类型拷贝数特征分析
BACHTROG D, AGIS M, IMHOF M,Microsatellite variability differs between dinucleotide repeat motifs: Evidence from. Molecular Biology and Evolution, 2000, 17(9): 1277–1285
BAI C C, LIU S F, ZHANG Z M. Characteristic analysis of microsatellite DNA in the genome of Gobiidae. Progress in Fishery Sciences, 2016, 37(5): 9–15 [白翠翠, 柳淑芳, 庄志猛. 虾虎鱼科(Gobiidae)基因组微卫星DNA的分布特征. 渔业科学进展, 2016, 37(5): 9–15]
ELLEGREN H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nature Genetics, 2000, 24(4): 400–402
GAO H, KONG J. Distribution characteristics and biological function of tandem repeat sequences in the genomes of different organisms. Zoological Research, 2005, 26(5): 555– 564 [高焕, 孔杰. 串联重复序列的物种差异及其生物功能. 动物学研究, 2005, 26(5): 555–564]
HANCOCK J M. Simple sequences and the expanding genome. BioEssays, 1996, 18(5): 421–425

HUANG J, DU L M, LI Y Z,Distribution regularities of microsatellites in thegenome. Sichuan Journal of Zoology, 2012, 31(3): 358–363 [黄杰, 杜联明, 李玉芝, 等. 红原鸡全基因组中微卫星分布规律研究. 四川动物, 2012, 31(3): 358–363]
HUANG J, ZHOU Y, LIU Y Z,Characteristics of microsatellites ingenome sequences using 454 GS FLX. Sichuan Journal of Zoology, 2015, 34(1): 8–14 [黄杰, 周瑜, 刘与之, 等. 基于454 GS FLX高通量测序的四川山鹧鸪基因组微卫星特征分析. 四川动物, 2015, 34(1): 8–14]
KATTI M V, RANJEKAR P K, GUPTA V S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Molecular Biology and Evolution, 2001, 18(7): 1161–1167
KOVTUN I V, GOELLNER G, MCMURRAY C T. Structural features of trinucleotide repeats associated with DNA expansion. Biochemistry and Cell Biology, 2001, 79(3): 325–336
LEOPOLDINO A M, PENSA S D. The mutational spectrum of human autosomal tetranucleotide microsatellites. Human Mutation, 2003, 21(1): 71–79
LI W J, LI Y Z, DU L M,Comparative analysis of microsatellite sequences distribution in the genome of giant panda and polar bear. Sichuan Journal of Zoology, 2014, 33(6): 874–878 [李午佼, 李玉芝, 杜联明, 等. 大熊猫和北极熊基因组微卫星分布特征比较分析. 四川动物, 2014, 33(6): 874–878]
LIU Z J, LI P, ARGUE B J,Inheritance of RAPD markers in channel catfish () × blue catfish () and their F1, F2and backcross hybrids. Animal Genetics, 1998, 29: 58–62
LIU Z J, LI P, ARGUE B,Random amplified polymorphic DNA markers: Usefulness for gene mapping and analysis of genetic variation of catfish. Aquaculture, 1999, 174(1): 59– 68
LU T, WANG C, DU C,Distribution regularity of microsatellites ingenome. Sichuan Journal of Zoology, 2017, 36(4): 420–424 [卢婷, 王晨, 杜超, 等. 林麝全基因组微卫星分布规律研究. 四川动物, 2017, 36(4): 420–424]
MICKETT K, MORTON C, FENG J,Assessing genetic diversity of domestic populations of channel catfish () in Alabama using AFLP markers. Aquaculture, 2003, 228(1/2/3/4): 91–105
MORGANTE M, HANAFEY M, POWELL W. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nature Genetics, 2001, 30: 194–200
NARASIMHAMOORTHY B, SAHA M C, SWALLER T,Genetic diversity in switch grass collections assessed by EST-SSR markers. Bioenergy Research, 2008, 1(2): 136–146
NI S S, YANG Y, LIU S F,Microsatellite analysis ofusing next-generation sequencing method. Progress in Fishery Sciences, 2018, 39(1): 107–113 [倪守胜, 杨钰, 柳淑芳, 等. 基于高通量测序的虾夷扇贝基因组微卫星特征分析. 渔业科学进展, 2018, 39(1): 107–113]
NIE H, CAO S S, ZHAO M L,Comparative analysis of microsatellite distributions in genomes ofandSichuanJournal of Zoology, 2017, 36(6): 639–648 [聂虎, 曹莎莎, 赵明朗, 等. 红尾蚺和原矛头蝮基因组微卫星分布特征比较分析. 四川动物, 2017, 36(6): 639–648]
PEARSON C E, SINDEN R R. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry, 1996, 35(15): 5041–5053
QI W H, JIANG X M, XIAO G S,Distribution regularities of microsatellites in the pig genome. Animal Husbandry and Veterinary Medicine, 2014, 46(8): 9–13 [戚文华, 蒋雪梅, 肖国生, 等. 猪全基因组中微卫星分布规律. 畜牧与兽医, 2014, 46(8): 9–13]
QI W H, JIANG X M, XIAO G S,Seeking and bioinformatics analysis of microsallite sequence in the genomes of cow and sheep. Acta Veterinaria et Zootechnica Sinica, 2013, 44(11): 1724–1733 [戚文华, 蒋雪梅, 肖国生, 等. 牛和绵羊全基因组微卫星序列的搜索及其生物信息学分析. 畜牧兽医学报, 2013, 44(11): 1724–1733]
QI W H, YAN C C, XIAO G S,Distribution regularities and bioinformatics analysis of microsatellite in the whole genomes of goat and Tibetan antelope. Journal of Sichuan University (Natural Science), 2016, 53(4): 937–944 [戚文华, 严超超, 肖国生, 等. 山羊和藏羚羊全基因组微卫星分布规律及其生物信息学分析. 四川大学学报(自然科学版), 2016, 53(4): 937–944]
SCHLÖTTERER C. Genome evolution: Are microsatellites really simple sequences. Current Biology, 1998, 8(4): 132– 134
SCHORDERET D F, GARTER S M. Analysis of CpG suppression in methylated and nonmethylated species. Proceedings of the National Academy of Sciences of USA, 1992, 89(3): 957–961
SHEN X, YANG G, LIU Y,Construction of genetic linkage maps of guppy () based on AFLP and microsatellite DNA markers. Aquaculture, 2007, 271(1): 178–187
SIMMONS M, MICKETT K, KUCUKTAS H,Comparison of domestic and wild channel catfish () populations provides no evidence for genetic impact. Aquaculture, 2006, 252(2/3/4): 133–146
STALLINGS R L. CpG suppression in vertebrate genomes does not account for the rarity of (CpG)n microsatellite repeats. Genomics, 1992, 13(3): 890–891
SUBRAMANIAN S, MISHRA R K, SINGH L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biology, 2003, 4(2): 1–10
TONG X L, DAI F Y, LI B,Microsatellite repeats in mouse: Abundance, distribution and density. Current Zoology, 2006, 52(1): 138–152 [童晓玲, 代方银, 李斌, 等. 小鼠基因组中的微卫星重复序列的数量、分布和密度. 动物学报, 2006, 52(1): 138–152]
TU F Y, LIU J, HAN W J,Analysis of microsatellite distribution characteristics in the entire genome ofChinese Journal of Wildlife, 2018, 39(2): 400–404 [涂飞云,刘俊,韩卫杰,等. 食蟹猴全基因组微卫星分布特征分析. 野生动物学报, 2018, 39(2): 400–404]
TU F Y, LIU X H, DU L M,Distribution characteristics of microsatellites in the rat genome. Acta Agriculturae Universitatis Jiangxiensis, 2015, 37(4): 708–711 [涂飞云, 刘晓华,杜联明, 等. 大鼠全基因组微卫星分布特征研究. 江西农业大学学报,2015, 37(4): 708–711]
WANG C, DU L M, LI P,Distribution patterns of microsatellites in the genome of the German cockroach (). Acta Entomologica Sinica, 2015, 58(10): 1037–1045 [王晨, 杜联明,李鹏,等. 德国小蠊全基因组中微卫星分布规律.昆虫学报,2015,58(10): 1037–1045]
WANG Y Y, LIU X X, DONG K Z,Distribution difference of microsatellite in 7 domestic animals genomes. China Animal Husbandry and Veterinary Medicine, 2015, 42(9): 2418–2426 [王月月, 刘雪雪, 董坤哲,等.7种家养动物全基因组微卫星分布的差异研究. 中国畜牧兽医, 2015, 42(9): 2418–2426]
XIA J, LIU F, ZHU Z,A consensus linkage map of the grass carp () based on microsatellites and SNPs. BMC Genomic, 2010, 11(1): 135– 151

XU J J, ZHENG X, LI J,Distribution characteristics of whole genome microsatellite ofGenomics and Applied Biology, 2020, 39(12): 5488–5498 [徐杰杰, 郑翔, 李杰, 等. 黄颡鱼()全基因组微卫星分布特征分析. 基因组学与应用生物学, 2020, 39(12): 5488–5498]
XU J J, ZHENG X, ZHANG X Y,Analysis of distribution characteristics of microsatellites in the four genomes of puffer fish. Genomics and Applied Biology, 2021, 40(4): 1441–1451 [徐杰杰, 郑翔, 张鑫宇, 等. 4种河鲀全基因组微卫星分布特征分析研究. 基因组学与应用生物学, 2021, 40(4): 1441–1451]
ZHANG L L, WEI Z M, LIAN Z M,Abundance of microsatellites in the entire genome and EST ofChinese Bulletin of Entomology, 2008, 45(1): 38–42 [张琳琳, 魏朝明, 廉振民, 等. 赤拟谷盗全基因组和EST中微卫星的丰度. 昆虫知识, 2008, 45(1): 38–42]

ZHENG Y, JIN G L, WU W R. Relationship between sequence completeness and polymorphism of microsatellites. Genomics and Applied Biology, 2012, 31(6): 587–591 [郑燕, 金谷雷, 吴为人. 微卫星序列完整性与多态性的关系. 基因组学与应用生物学, 2012, 31(6): 587–591]
Analysis of Microsatellite Distribution Characteristics in the Channel Catfish () Genome
TANG Rongye, SU Mengyuan, YANG Wenshan, XU Jiejie, WANG Tao①, YIN Shaowu①
(College of Marine Science and Engineering, Nanjing Normal University Nanjing, Jiangsu Province Engineering Research Center for Aquatic Animals Breeding and Green Efficient Aquacultural Technology, Nanjing, Jiangsu 210023, China)
To understand the distribution of perfect microsatellites in the genome of the channel catfish (), we used MISA, a bioinformatics software package, to search and analyze the microsatellites. A total of 510256 perfect microsatellites were isolated from 29 chromosomes of, with a total length of 11036941 bp. The chromosome containing the largest number of microsatellites was chromosome 2 (25284), followed by chromosomes 3, 1, and 5. Chromosome 29 had the lowest number of microsatellites (11591). The length of each chromosome was significantly correlated with the number of microsatellites it contained (SPSS,= 0.98,< 0.01). The highest relative abundance of microsatellites was found on chromosome 27 (785.03 ind./Mb), and the lowest was on chromosome 11 (615.89 ind./Mb). Among the six repeat types, mononucleotides were the most frequent, accounting for 45.31% of the total, followed by dinucleotides (38.53%), trinucleotides (8.73%), tetranucleotides (6.93%), pentanucleotides (0.46%), and hexanucleotides (0.04%). The predominantly repeated microsatellite sequences in thegenome were A, AC, AG, AT, AAT, AAAT, C, AAC, AAAC, and AAG, showing an obvious inclination towards A and T bases. The results of this study provide a reference for the further study ofgenome characteristics and contribute basic data for future investigations into molecular marker-assisted breeding and genetic information assessment of.
; Whole genome; Microsatellites; Distribution characteristics
WANG Tao, E-mail: seawater88@126.com; YIN Shaowu, E-mail: yinshaowu@163.com
S917.4
A
2095-9869(2022)02-0089-09
10.19663/j.issn2095-9869.20210126002
* 江苏省农业重大新品种创制项目(PZCZ201742)、江苏省重点研发计划(现代农业)重点项目(BE2017377)、江苏省农业科技自主创新资金[CX(19)2034]和南京师范大学大学生创新创业训练计划项目共同资助[This work was supported by Agricultural Major New Variety Creation Project of Jiangsu Province (PZCZ201742), Key Research and Development Program of Jiangsu Province (BE2017377), Jiangsu Agricultural Science and Technology Innovation Fund [CX(19)2034], and Innovation and Entrepreneurship Training Program for College Students in Nanjing Normal University].
唐荣叶,E-mail: 2508049280@qq.com
王 涛,副教授,E-mail: seawater88@126.com;尹绍武,教授,E-mail: yinshaowu@163.com
2021-01-26,
2021-02-23

TANG R Y, SU M Y, YANG W S, XU J J, WANG T, YIN S W. Analysis of microsatellite distribution characteristics in the channel catfish () genome. Progress in Fishery Sciences, 2022, 43(2): 89–97
(编辑 冯小花)
