Improved Nucleic Acid Spot Hybridization Technique for Detection of Potato Spindle Tuber Viroid (PSTVd)
2022-04-06QiuCailingLuDianqiuLiuDefuShiJiaoxuBaoLiuyuanMaZhonglianLiuLiFengZhenyueHuangXianminJiangRuiChenYueandWangShiminCollegeofAgronomyandLifeSciencesZhaotongUniversityZhaotong657000YunnanChina
Qiu Cai-ling , Lu Dian-qiu, Liu De-fu , Shi Jiao-xu , Bao Liu-yuan , Ma Zhong-lian , Liu Li ,Feng Zhen-yue Huang Xian-min Jiang Rui Chen Yue and Wang Shi-min College of Agronomy and Life Sciences, Zhaotong University, Zhaotong 657000, Yunnan, China
2 College of Agronomy and Biotechnology, State Cultivation Base of Crop Stress Biology for Southern Mountainous Land of Southwest University, Southwest University, Chongqing 400715, China
3 College of Potato Sciences, Zhaotong University, Zhaotong 657000, Yunnan, China
Abstract: Potato spindle tuber viroid (PSTVd) disease is one of the major diseases that threatens potato production. Therefore, an advanced, rapid and sensitive detection technology is needed to detect the disease for better control. In order to establish an easier nucleic acid spot hybridization (NASH) method, some studies were tried as the followings: (1) the pre-hybridization step of nucleic acid spot hybridization (NASH) was omitted compared with ordinary way; (2) RNA extraction (phenol extraction and Ames buffer extraction) methods were compared; (3) fixed RNA by UV lamp and oven compared with UV cross-linker; (4) hybridized the RNA in shaking incubator and so on. The results showed that RNA extracted by Ames buffer was more effective than by the phenol extraction method. Besides, the result of hybridization without pre-hybridization step was better than that with 1.5 h of pre-hybridization. The more important discovery was that the shaking incubator could replace the hybridization oven and the ordinary UV lamp could replace the UV cross-linker. After a long term repeated research and testing, a new hybridization system that could rapidly detect the PSTVd by improved NASH technique merely using common instruments and equipment was established.
Key words: potato spindle tuber viroid (PSTVd), nucleic acid spot hybridization (NASH), pre-hybridization, RNA extraction, detection technology
Introduction
Potato spindle tuber viroid (PSTVd) (Kasaiet al., 2013; Nie, 2012; Owenset al., 2012; Eiraset al., 2011) is a small, single-stranded, highly structured, circular non-coding RNA pathogen that lacks detectable messenger RNA (mRNA) activity and can replicate autonomously in susceptible plant species. The genome ranges from 245 to 401 nucleotides (nt) (Abraitieneet al., 2008; Wiesyket al., 2011) and the molecular is about 359 nt (Owenset al., 2003). The viroid is highly infectious (Nie, 2012; Takedaet al., 2011), and it can be easily transmitted by physical contact, pollen and seed or some insects such as green capsids, grasshoppers and potato beetles. Therefore, it appears to be widely transmitted. The PSTVd can lead to 60% maximum and 20% minimum yield loss, resulting in large economic losses (Soliman, 2012). In most countries, the PSTVd is a quarantine disease and China is no exception. In China, the disease occurs from south to north. In the main potato producing areas of Chinese northern, the PSTVd has done serious harm to potato production (Singh, 1971). Consequently, the PSTVd is a problem that requires prompt resolution (Solimanet al., 2010; Solimanet al., 2012). The viroid is difficult to be extincted by meristem tip culture. Until now, there is no specific preventative medicine. Effective control of infection is only depended on the early identification of the PSTVd (Janskyet al., 2009; Baiet al., 2010). Therefore, a rapid, sensitive and simple PSTVd detection technology is urgently needed to prevent the disease.
Currently, the most common detection techniques used for the PSTVd are as the followings: biological detection (indicator plant), return polyacrylamide electrophoresis (R-PAGE) (Kumaret al., 2013), reverse transcription polymerase chain reaction (RTPCR) and nucleic acid spot hybridization (NASH) (Noor and Krull, 2013).
The environment may have an interfering effect on biological detection and the technique has a long detection cycle. The R-PAGE has low sensitivity and poor reproducibility, while the RT-PCR is cumbersome, costly and proned to false positives. The NASH is a biological technique that is used to detect specific DNA or the RNA molecular sequences (target sequences).
Firstly, the DNA or the RNA is fixed and transferred into nitrocellulose or nylon membranes, then the DNA or the RNA probes are labeled with radioactive or nonradioactive and hybridized with the RNA that need to be detected. During the hybridization process, the probes bond with the complementary target sequence through hydrogen bonds. After washing away the unbound probes, the specific binding of the probes is detected by autoradiography or color reaction. The technology is highly sensitive, with the potential to detect a large amount of test sample. However, this method is quite cumbersome and requires specialized equipment (such as hybridization oven and the UV crosslinker), which mades the usage of this technique detection difficult in many laboratories. In order to facilitate this technology, some research work was done to make the NASH (on PSTVd detedtion) easy to be used.
Materials and Methods
Two kinds of RNA extraction methods
There were many kinds of the RNA extraction methods for the NASH, such as the RNeasy Mini kit, Trizol. However, phenol extraction (Fanson, 2000; Ma and Yang, 2011) and Ames buffer extraction (Vassilakoset al., 2012; Perez-Leightonet al., 2011) methods would be introduced and compared in this study.
Phenol extraction
Added 0.3 g sample into liquid nitrogen and grinded it into powder. Then, added 0.6 mL extraction buffer(0.1 mol • L-1Tris+0.1 mol • L-1NaCl+0.01 mol • L-1EDTANa2+2% sodium dodecyl sulfate (SDS), sterilized the solution at 121℃ for 20 min and adjusted pH to 9-9.5 with HCl), 0.6 mL water-saturated phenol and 0.6 mL chloroform, the sample was mixed evenly and transferred into a 2 mL eppendorf (EP) tube. It was centrifuged under 10 000 r • min-1for 15 min at 4℃ and extracted the supernatant (350 μL). Then, added three times volume cold ethanol and 1/3 volume NaAc into the supernatant, mixed them well and stored them at -20℃ at least for 1.5 h. After that, it was centrifugated under 10 000 r • min-1for 15 min at 4℃. Then, discarded the supernatant, washed the deposit for three times with 70% ethanol, discarded the ethanol and dried the product in air, and then added 100 μL double distilled water with DEPC and stored at -20℃.
Ames buffer extraction method
Ames buffer preparation: 160 mL distilled water+11.7 g NaCl+0.4 g MgCl2+8.21 g sodium acetate (NaAC)+ 40 mL ethanol+6 g SDS, dissolving NaCl firstly, then adding other salts, ethanol and SDS finally. Adjusted pH to 6.0 with HCl or NaOH.
Added 0.5 g sample into a sterilized mortar, added 0.75 mL Ames buffer to the sample and ground them into powder. Then, transferred the sample from mortar to 2 mL EP tube, incubated them at 37℃ for 15 min. Added 0.75 mL chloroform and mixed well into an emulsion condition. Centrifuged it briefly for separating the contents into aqueous (top) and chloroform (bottom) layers. Extracted the aqueous layer into a new tube.
Eight potato samples were extracted by above two mentioned methods and each sample was extracted twice correspondingly. The samples underwent detection by the NASH. The signal was detected by chemiluminescence method in order to compare the similarities and differences of test results and to determine which method was more suitable for NASH.
Pre-hybridization
In general, before the hybridization process, there was a pre-hybridization step by conventional NASH detection methods (Lianget al., 2011). In practice, the arrangements of detection time often caused complications. In this study, the following method was used to verify whether the pre-hybridization step was necessary. Both nylon A and B membranes had the same samples: number 1 to 5 samples were extracted by Ames buffer, number 6 to 8 samples were potato leaf juice which were directly dotted onto the membrane. All the samples were the PSTVd positive except sample 2.
Nylon A membrane was treated with conventional methods, pre-hybridized for 1.5 h combined with 1 μL probe and hybridized overnight. Nylon B membrane was immersed into hybridization solution containing 1 μL probe (without pre-hybridization), then it underwent overnight hybridization. Other steps were identical between A and B nylon membranes. Finally the A and B membrane results were compared to determine the necessity of pre-hybridization step.
Hybridization methods
Firstly using a water bath (without shaking) or shaking incubator instead of a hybridization oven, besides, using a UV lamp or an ordinary oven to fix the RNA instead of a UV crosslinker. Test Ⅰ (Fig. 3): the RNA was fixed on nylon+membrane by ordinary oven, and two hybridization ways were used separately (water bath and shaking incubator). The normal way (UV cross-linker fixed RNA, hybridization oven hybridized) was control. Test II: the RNA was fixed on nylon+membrane by UV light and oven, respectively.
Results
Comparison between two kinds of RNA extraction methods
The results demonstrated in Fig. 1 showed that the RNAs extracted by Ames buffer were more effective than by the phenol extraction method. The former method resulted in hybridization spots that were clear and demonstrated a uniform distribution, while the latter method resulted in less clear and more uneven spots. In addition, less time was required using the Ames buffer and the process was easier than the other one.
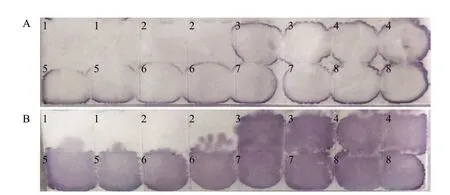
Fig. 1 Chemical staining results of NASH by two different methods of RNA extractionResults of hybridization without pre-hybridization step are better than that with 1.5 h of pre-hybridization method. Clearer spots are shown from nylon membrane B than A (Fig. 2). Moreover, hybridization without pre-hybridization is more easier and takes less time.
Pre-hybridization results
The results of hybridization without pre-hybridization step were better than that with 1.5 h of pre-hybridization. Clearer spots were got from B than that of A (Fig. 2). Moreover, the direct hybridization method was performed easier and took less time.
Comparison of different hybridization techniques
Fig. 3 showed the result of test Ⅰ. In this experiment, the ordinary oven could not fix the RNA perfectly, water bath (without shaking) could not replace the hybridization oven, while shaking incubator could. Shaking was the indispensable step of hybridization. Fig. 4 exhibited the result of test II. This test further demonstrated that an ordinary oven could not replace the UV cross-linker, but the ordinary UV lamp could. Several UV light treatments were used to fix the RNA and the results showed that there were no significant differences. Shaking incubators could replace the hybridization oven.

Fig. 2 Comparison of pre-hybridization and direct hybridization by chemiluminescenceA, 1.5 h of pre-hybridization followed by hybridization; B, Hybridization without pre-hybridization.
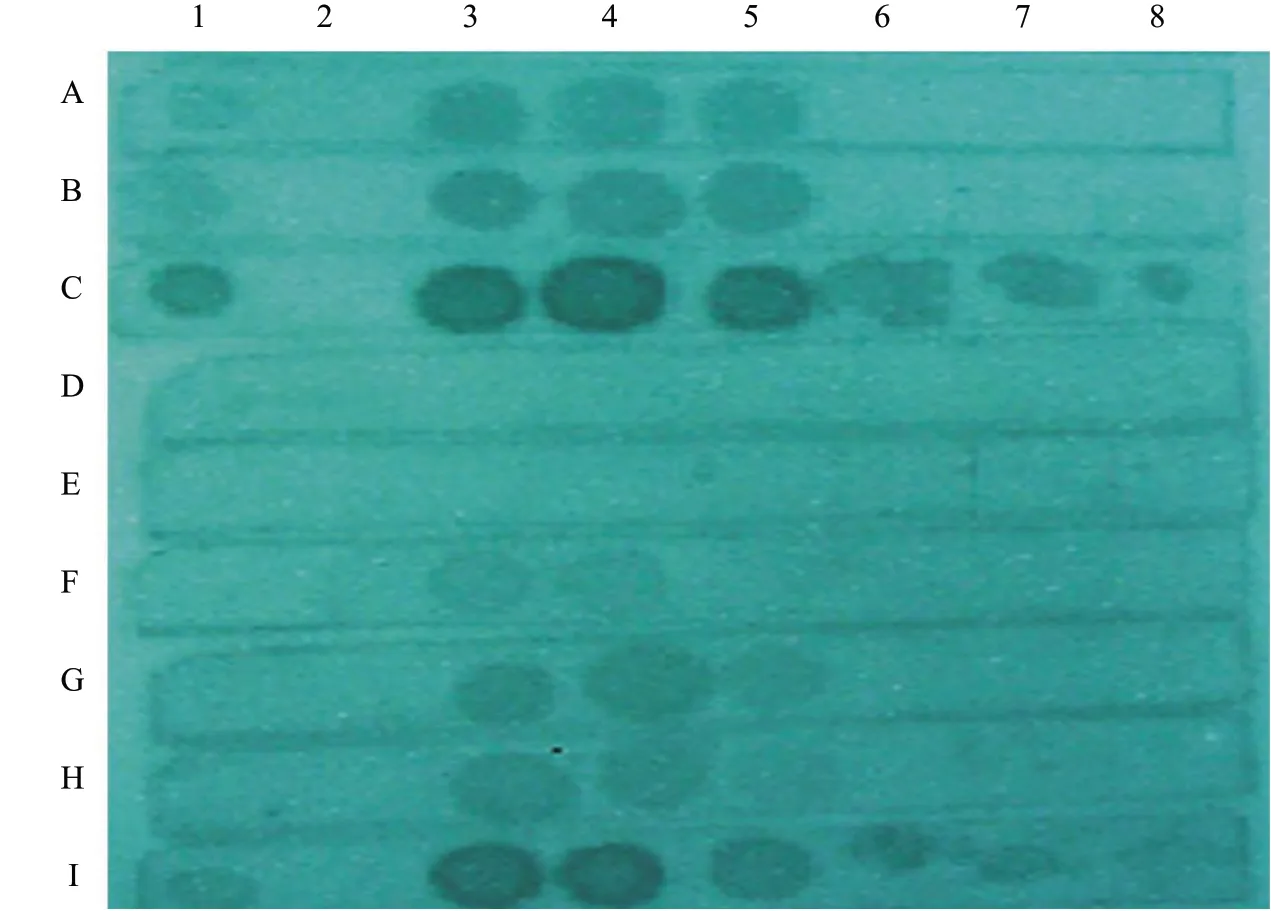
Fig. 3 Different hybridization results by different instruments using chemiluminescenceA, Oven heated at 80℃ for 1 h to fix RNA, shaking incubator hybridized; B, Oven heated at 80℃ for 2 h to fix RNA, shaking incubator hybridized; C, UV cross-linker fixed RNA, shaking incubator hybridized; D, Ordinary oven heated at 80℃ for 1 h to fix RNA, water bath hybridized; E, Ordinary oven heated at 80℃ for 2 h to fix RNA, water bath hybridized; F, UV cross-linker fixed RNA, water bath hybridized; G, Ordinary oven heated at 80℃ for 1 h to fix RNA, hybridization oven hybridized; H, Ordinary oven heated at 80℃ for 2 h to fix RNA, hybridization oven hybridized; I, UV crosslinker fixed RNA, hybridization oven hybridized. Except for sample 2, all the others are PSTVd positive samples, 2-5 are samples of Ames buffer extraction, and 6-8 are potato leaves juice.

Fig. 4 Result of different RNA fixed and hybridized methodsRNAs of A1-K2 point in nylon membrane are extracted by Ames buffer extraction method. The first sample is undiluted solution (643 ng • μL-1, not diluted), the second sample is diluted 10 times, the third sample is diluted 100 times and so on, the eighth sample is diluted 107 fold, all the ninth samples on each membrane are negative controls. All the first parts of nylon membranes (A1-K1) are hybridized in a hybridization chamber, while the second nylon membranes (A2-K2) are hybridized in a constant temperature shaking incubator. RNAs of K3 and K4 extracted by Trizol, dilution method as above, RNA fixed way as K1 and K2. K3 hybridized in a hybridization chamber, K4 hybridization incubator at a constant temperature shock. A, UV light irradiates the front and back for 5 min; B, UV light irradiates the front and back for 10 min; C, Bakes for 30 min at 80℃ and then UV irradiates the front and back for 5 min; D, Bakes for 30 min at 80℃ and then UV irradiates the front and back for 10 min; E, UV irradiates the front and back for 5 min and then bakes for 30 min at 80℃; F, UV irradiates the front and back for 10 min and then bakes for 30 min at 80℃; G, Only bakes for 30 min at 80℃; H, Only bakes for 60 min at 80℃; I, Only bakes for 120 min at 80℃; J, Only bakes for 180 min at 80℃; K, UV cross-linking irradiates the front and back each for 1 min, with an energy of 1 200 J.
Discussion
Nucleic acid extraction from plant tissues was the most laborious and time-consuming step in gene diagnosis (Nakaharaet al., 1999). Williams and Ronald (1994) and Nakaharaet al. (1999) had given us some rapid and simple nucleic acid extraction methods. However, another more suitable way for the NASH was explored in this study. Two kinds of the RNA extraction methods were compared in this study and the results demonstrated that Ames buffer extraction method which was efficient, easy to be utilized and was more suitable for the NASH. It also had the potential to save time and costs. Only 30 min were required for one sample extraction by use of this method, while the phenol extraction method took about one day. This process made it much easier to detect the PSTVd by the NASH technique.
It was reported (Salazaret al., 1988) that the RNA could be fixed by conventional ovens. However, this study obtained an opposite conclusion. Nylon membrane A-F used merely UV light to fix the RNA, acquired almost the same detection sensitivity of K which utilized UV cross-linking. While baked nylon membrane (G1-J2) at 80℃ had few effects, even extended the baking time, samples with 643 ng • μL-1RNA concentration could be detected and showed relatively shallow spots. Besides, the sensitivity was reduced more than 100 times. Therefore, the ordinary UV lights could fix the RNA better than the oven and the later could not be sed. All the first parts of nylon membranes (A1-K1) were hybridized in a hybridization chamber, while the second nylon membranes (A2-K2) were hybridized in a constant temperature shaking incubator. Whatever the RNA fixed method, the two kinds of hybridization results were the same. Therefore, there was no significant difference, when the hybridization oven was replaced by shaking incubator. From the K1 to the K4, it could be further confirmed that the two hybrid methods had basically same effect. Besides, the two RNA extraction methods for the NASH detection also made the same effect, while Ames buffer method was more simple, saving time and money, and more suitable for the NASH. In addition, the results were not satisfactory, when the fixed time was 30 min, 1 h or 2 h. In contrast, when the UV lamp on a clean workbench was used to fix the RNA, the results were more satisfactory.
This study showed that shaking incubator could replace the hybridization oven. Concurrently, shaking incubator is common in most laboratories. Therefore, most of them does not need to buy a hybridization oven to detect the PSTVd by the NASH technique.
Since clean bench and shaking incubator are common laboratory instruments, using the NASH technology to detect the PSTVd will be easy to apply in China or other countries. In conclusion, a new hybridization system that could rapidly detect the PSTVd by improved the NASH technique merely using common instruments and equipment was established.
Conclusions
When the PSTVd was detected by the NASH, the RNA extracted by Ames buffer was more effective, saving more time and easier than using of the phenol extraction method. Better results could be gotton without pre-hybridization. Moreover, the direct hybridization method was performed easier and took less time. For RNA fixing, the ordinary oven could not fix RNA perfectly and the UV light was necessary in fixing RNA. For incubation, shaking was an indispensable step of hybridization and water bath (without shaking) could not replace the hybridization oven, while shaking incubator could.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Analysis on Cloning and Expression of Promoter of Flowering Time GmFT2a in Soybean
- Effect of Allelic Variation of β-conglycinin α-subunit Locus cgy-2 on Soybean (Glycine max) Quality I. Evaluation of Free Amino Acid Content
- Nutrient Accumulation and Distribution in Cotton Promoted by Removal of Mulch Film
- Effects of Sowing Periods on Growth and Development, Yield and Quality of Maize in Cold Area
- Cloning of GsTPS9 Gene from Glycine soja and Study on Its Responses to Stresses
- Comparative Analysis of Hydrolytic Amino Acids in Human and Cow Milk of Different Lactation Periods
