DNA Methylation Reshapes Sex Development in Zebrafish
2021-12-03YanLiFengLiu
Yan Li, Feng Liu,*
1 State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
2 Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 100101 Beijing, China
3 University of Chinese Academy of Sciences, Beijing 100049, China
Sex determination is a complex biological process, through which the sex of an organism is established in a binary fate decision [1,2]. There are two main determining mechanisms:1) genotypic sex determination (GSD), whereby the individual’s sex is determined by its genotype; and 2) environmental sex determination (ESD), where the sex is driven by different external factors, such as temperature, pH, and social interactions [1].
Zebrafish sex determination can be affected by various factors. Recent studies have demonstrated that multiple sexrelated genes interact with each other as a network to dictate the gender, which is known as polygenic sex determination[3]. Many key candidate genes that determine the sexual development of zebrafish have been discovered, such as piwi[4], cyp19a1a [5], amh [6], dmrt1 [6,7], and wnt4a [8]. In addition,environmental factors such as temperature[9]and oxygen concentration [10] are also found to contribute to sex bias in zebrafish. In addition to genetic and environmental elements,there is an accumulating pile of evidence for involvement of epigenetic mechanisms in the regulation of sex differentiation in vertebrates [11–13]. However, the epigenetic regulation of sex determination and sexual plasticity remains largely elusive.
Recently,Wang et al.systematically analyzed the dynamics of DNA methylome and transcriptome during germline development and sex transition in zebrafish[14].They performed low-input whole-genome bisulfite sequencing (WGBS) and RNA-seq in germ cells at different stages during primordial germ cell (PGC) development and germ cell differentiation.Their data showed that there was no substantial change in the overall methylation level of early PGCs,which is in agreement with a previous study [15]. Furthermore, Skvortsova et al. demonstrated that the zebrafish PGC DNA methylome preserved paternal epigenetic imprinting during embryonic stages [15]. Importantly, Wang et al. found that the average methylation level of PGCs at 36 hours post fertilization(hpf), the time when they arrive at the genital ridge, began to decline and reached its lowest point at 9 days post fertilization(dpf), which is comparable to that of oocytes and ovaries.These results cast doubt on previous studies showing that PGCs maintain stable methylation patterns when migrating to the gonad at 9 dpf [16]. In addition, the methylation level in 35 dpf female zebrafish (35 dF) remained at a relatively low level, whereas the methylation level in 35 dpf male zebrafish (35 dM) was significantly increased, which is similar to the level in sperm.
Furthermore, the transcriptomic landscape was depicted and stage-specific genes and sex-specific genes were defined.Moreover,DNA methylation patterns on PGC signature gene promoters at 9 dpf are similar to that of oocyte/ovary. The genes with hypermethylated promoter can help PGCs to avoid premature differentiation at 9 dpf. It is proposed for the first time that zebrafish PGCs at 9 dpf have the dual potential to differentiate into male germ cells(MGCs)or female germ cells(FGCs).
Wang et al. then asked whether sex determination can be affected by changing DNA methylation level during gonadal development in zebrafish [14]. Given that treatment with DNA methyltransferase inhibitors 5-Aza-2′-deoxycytidine can feminize zebrafish[17],they treated zebrafish embryos with 5-Aza-2′-deoxycytidine and found that the treated embryos were more likely to develop into females.DNA demethylation in mammalian embryos can be regulated by the Tet proteins[18]. At the early stage of PGC development, there are over 10,000 demethylated differentially methylated regions(DMRs). They also found that blocking the tet3 expression can reduce the number of germ cells. Meanwhile, previous studies have reported that the number of germ cells in zebrafish can influence sex determination [19]. Their results suggest that Tet3 can regulate PGC development. However, it is unknown whether the deletion of Tet3 can change the level of methylation that further influences PGC determination.
The sexual identity of fish is variable, partly due to the cell type plasticity present in the gonads[1,2]. Some chemicals can reverse the sex of fish.Zhang et al.found that 2,4-dichlorophenol (2,4-DCP), a contaminant of aquatic environments, can induce feminization of zebrafish through inhibition of sox9a expression[20].It has been suggested that Aromasin can revert adult zebrafish from female to male [21]. After Aromasin treatment, the female sex cells begin to degenerate, accompanied by the gradual formation of male reproductive organs,while the sex-reversed fish can successfully fertilize and produce normal embryos. Wang et al. additionally used RNA-seq to determine the dynamic expression of genes during the sex transition from female to male [14]. The expression of female sexual developmental genes (cyp19a1a,foxl2,and esr1)was down-regulated and expression of genes associated with male and sperm production (sox9a, amh, and dmrt1) was upregulated.These results suggest that there are dynamic changes in the expression of key sex-related genes during sex transformation.
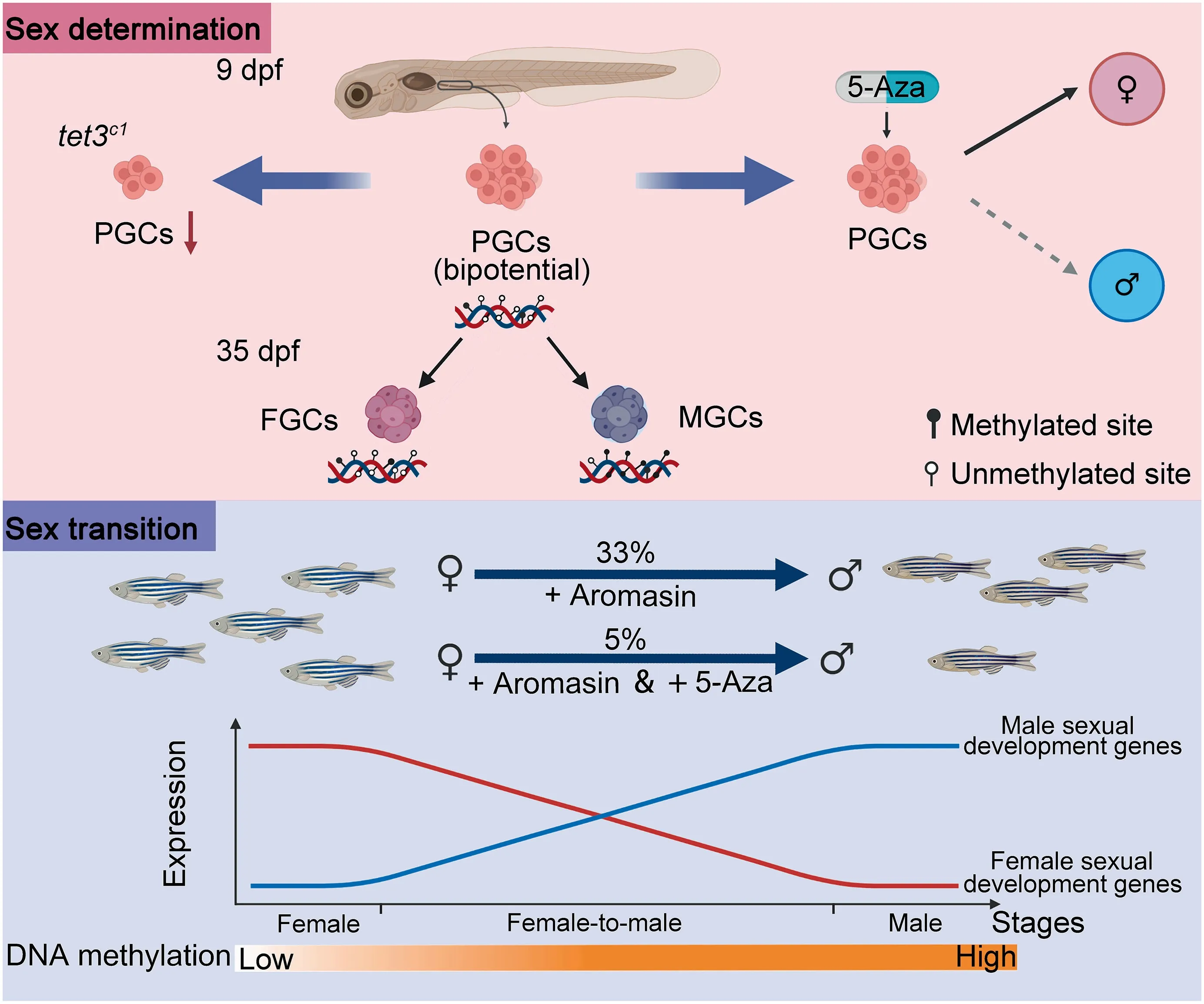
Figure 1 DNA methylation reprogramming during sex determination and transition in zebrafishSex determination and plasticity are regulated by epigenetic mechanisms in zebrafish.DNA methylation is crucial for sex determination in the larval stage (on the top). 5-Aza treatment can reduce the sex transition induced by Aromasin, suggesting that DNA methylation dynamics plays an essential role in sex reversal in the adult stage (at the bottom). PGC, primordial germ cell; FGC, female germ cell;MGC, male germ cell; dpf, day post fertilization.
Finally, Wang et al. provided evidence that similar DNA methylation reprograming occurs in the transition stage of Aromasin-induced female ovary to male testis [14]. In agreement with the observations in normal sexual development,the levels of methylation in the gonads of adult fish artificially converted from females to males are increased. It has been widely acknowledged that DNA methylation can affect gene expression by inhibiting gene transcription. Wang et al.found that there is a negative correlation between promoter methylation of sex determination genes and their expression. For instance, during sexual transition, there are an increase in the promoter DNA methylation level and a concomitant decrease in transcription of cyp19a1a, a gene associated with ovarian development in fish. On the contrary, a male-related gene dmrt1 has hypomethylated promoter and exhibits high expression. Moreover, methylation inhibitor treatment can partially inhibit the artificially induced conversion of zebrafish from female to male caused by Aromasin. Therefore, DNA methylation reprogramming is necessary during sex transformation in zebrafish.
Together, the reprogramming of DNA methylation can reshape germ cell development and gender transition, which is a new direction for regulating sex determination and transition in zebrafish (Figure 1). In this work, Wang et al. found that PGCs at 9 dpf possess dual potentials [14]. DNA methylation can regulate the development of PGCs to FGCs or MGCs. However, the authors did not identify the specific mechanism that regulates DNA methylation reprogramming during early PGC development. In addition, Tet3 defects can affect the number of PGCs.It is unclear whether DNA methylation is altered in PGCs, and whether sex determination is biased in tet3c1mutation. Recently, DNA methylation has been found to regulate the expression of key sex determination genes [22]. Changing the methylation level of key factors at specific sites is expected to reverse zebrafish sex determination.Moreover, chemicals that cause endocrine disorders can change sex in zebrafish. Aromasin-induced sex conversion at the adult stage is accompanied by DNA methylation reprogramming. It remains to be determined how DNA methylation concretely regulates zebrafish sex determination through specific gene loci.
Sex determination and differentiation are crucial to the formation of sexually reproducing organisms. Studies on sex determination mechanisms in fish is not only helpful to fishery production, but also to the evolutionary analysis of sex determination mechanisms.The sex determination process in fish is flexible and complex, mainly determined by intrinsic genetic factors and also influenced by the environment. However,the exact mechanisms of sex determination and sex differentiation in zebrafish are still unclear. The proposed role of DNA methylation reprogramming in this process may provide new insights into zebrafish sex determination mechanisms.
CRediT author statement
Yan Li: Writing - original draft. Feng Liu: Writing - review &editing. Both authors read and approved the finalmanuscript.
Competing interests
The authors declare no competing interests.
Acknowledgments
We thank our lab members for critical reading of this manuscript. This work was supported by grants from the National Key R&D Program of China (Grant No. 2018YFA0800200),the Strategic Priority Research Program of the Chinese Academy of Sciences, China (Grant No. XDA16010207),and the National Natural Science Foundation of China(Grant Nos. 31830061, 31425016, and 81530004), as well as the State Key Laboratory of Membrane Biology, China.
ORCID
0000-0003-4504-558X (Yan Li)
0000-0003-3228-0943 (Feng Liu)
杂志排行
Genomics,Proteomics & Bioinformatics的其它文章
- Genome-wide 5-Hydroxymethylcytosine Profiling Analysis Identifies MAP7D1 as A Novel Regulator of Lymph Node Metastasis in Breast Cancer
- The Role of DNA Methylation Reprogramming During Sex Determination and Transition in Zebrafish
- Glycoproteogenomics: Setting the Course for Next-generation Cancer Neoantigen Discovery for Cancer Vaccines
- Computational Screening of Phase-separating Proteins
- PM2RA: A Framework for Detecting and Quantifying Relationship Alterations in Microbial Community
- MACMIC Reveals A Dual Role of CTCF in Epigenetic Regulation of Cell Identity Genes
