印度碗状红菇
2021-11-08陈彬宋杰王倩梁俊峰
陈彬 宋杰 王倩 梁俊峰

摘 要:本研究报道一个中国红菇属新记录种——印度碗状红菇(Russula indocatillus)。文中提供了该物种详细的形态学描述、线条图以及系统发育分析,并且与近缘种进行了比较。印度碗状红菇的主要形态学特征为:棕黄色到赭黄色的菌盖中央,奶油黄色到淡黄色的菌盖边缘,孢子印白色到奶油色,担孢子近球形到宽椭圆形到椭圆形,表面具圆锥状或近圆柱状孤立的疣,单细胞的盖表囊状体,短而纤细,有分叉和分隔的菌盖表皮末端细胞。综合形态学特征和系统发育分析,表明印度碗状红菇属于红菇属异褶亚属劣味组(Russula subg. Heterphyllidia sect. Ingratae)。
关键词:红菇科;新纪录种;系统发育;分类学
中图分类号:Q949.329 文献标识码:A
Russula indocatillus, a New Record Species in China
CHEN Bin1, 2, SONG Jie1, WANG Qian1, LIANG Junfeng
1. Research Institute of Tropical Forestry, Chinese Academy of Forestry, Guangzhou, Guangdong 510520, China; 2. Nanjing Forestry University, Nanjing, Jiangsu 210037, China
Abstract: Russula indocatillus was reported as new species to China. A detailed morphological description, illustrations and phylogeny are provided, and comparisons with related species are made. It is morphologically characterized by a brownish orange to yellow ochre pileus center with butter yellow to pale yellow margin, white to cream spore print, subglobose to broadly ellipsoid to ellipsoid basidiospores with bluntly conical to subcylindrical isolated warts, always one-celled pileocystidia, and short, slender, furcated and septated terminal elements of pileipellis. The combination of detailed morphological features and phylogenetic analysis based on ITS-nrLSU-RPB2 sequences dataset indicated that the species belonged to Russula subg. Heterphyllidia sect. Ingratae.
Keywords: Russulaceae; new record species; phylogeny; taxonomy
DOI 10.3969/j.issn.1000-2561.2021.09.014
1 Introduction
The genus Russula Pers. (Russulaceae, Russulales, Basidiomycota) is one of the most highly diverse genera of mushroom-forming fungi and range of habitats from the tropics to polar ecosystems, which resulted in many complex and multilevel classifications[1-3]. It is the largest group of all seven genera in Russulaceae, which includes eight subgenera and at least 2000 members within the genus[3, 4-6]. Some species of Russula can be as food for humans in the world. According to the recent statistics of resource diversity of Chinese macrofungi, there are 78 edible species in China[7-9].
Russula subg. Heterphyllidia Romagn. sect. Ingratae (Quél.) Maire is mainly characterized by a fetid odor (sometimes spermatic, waxy, or bitter), pale to dark yellowish brown to dark greyish brown colors, tuberculate striate pileus margin, articulated and branched hair cuticles, acute to subacute lamellae, spores print white to cream, inamyloid supraphilar spot, and short-celled and branched hyphal extremities in the pileipellis[10-11]. Resently, many species belonged to sect. Ingratae have been reported from China[12-14], indicating that China has been a hot spot for the exploration of unknown Russula species. During a survey on the habitat diversity and geographic distribution of Chinese Russula, some interesting specimens were made. Then they were identified as R. indocatillus A. Ghosh, K. Das & R.P. Bhatt, a species originally described from India, based on morphological features and phylogenetic analysis, which is a taxon new to China.
2 Materials and methods
2.1 Morphological study
The basidiomata were photographed under daylight in the field. Specimens were dried at 45-55 ℃ and deposited in the Herbarium of Research Institute of Tropical Forestry, Chinese Academy of Forestry (RITF). Terminology for descriptive terms follows Vellinga and Noordeloos (1988)[15]. Color designations were from Kornerup and Wanscher (1981)[16]. Macroscopic characteristics and ecological aspects were based on detailed notes and the photographs of fresh materials. Spores were observed and measured in Melzers reagent. After pretreatment of dried materials in 5% KOH, the observations and measurements of other microscopic features were made in Congo red with ZEISS Imager M2. Pileipellis was examined in Cresyl blue to verify the presence of ortho- or metachromatic reactions[17]. Cystidia were examined in Sulfovanillin (SV) solution. Structure and ornamentation of basidiospore were illustrated using scanning electron microscope (SEM-JEOL JSM-6510). The abbreviation (n/m/p) indicates measurements were made on n basidiospores from m fruit bodies of p specimens. In the dimension notation of basidiospores (a) b-c (d), b-c is the range containing 95% of the measured values for length or width with the extremes (a and d) of all measurements. Q value indicates length/width ratio of the basidiospores excluding ornamentation with bold Q denoting the average Q of all basidiospores ± standard deviation.
2.2 Molecular study
The total genomic DNA was isolated by an improved CTAB protocol[18] from dry herbarium. We amplified and sequenced the three following loci: 600 base pairs of the ITS region of rDNA using the primers ITS1 and ITS4[19], 900–1400 base pairs of the nuclear ribosome large subunit (nrLSU) using the primers LROR and LR5[20]; 700 base pairs of the second largest subunit of the RNA polymerase II (RPB2) using the primers bRPB2-6f and fRPB2-7cr[21-22]. The PCR products were subjected to automated DNA sequencing on an ABI 3730 DNA analyzer and an ABI BigDye 3.1 terminator cycle sequencing kit (Shanghai Sangon Biological Engineering Technology and Services CO., Ltd, Shanghai, China). The sequences newly generated were submitted to GenBank database (Tab. 1).
2.3 Phylogenetic analysis
Reference sequences were selected for phylogenetic analysis based on NCBI blast search results and published molecular analyses, and then combined with the newly generated sequence. NCBI accession numbers for the sequences used in the phylogenetic analysis are listed in Tab. 1. A preliminary multiple sequence alignment was performed using MAFFT 7.0 (http://mafft.cbrc.jp/ alignment/server/), with manually checking and adjusting when necessary in order to obtain reliable results in BioEdit[23]. The final sequence alignment was submitted to TreeBASE (http://purl.org/phylo/ treebase/phylows/study/TB2:S27534).
Both maximum likelihood (ML) and Bayesian analysis (BA) were employed. Maximum likelihood of the phylogenetic relationships were conducted with RAxML-HPC2 on XSEDE (8.2.12) through the Cipres Science Gateway (www.phylo.org). Bootstrap analyses were done to affirm consistency of results with 1000 replicates. The GTR model of nucleotide substitution and GAMMA rate heterogeneity parameters were implemented. Bootstrap support (BS)≥70% was regarded as significance. BA analysis were performed with XSEDE (3.2.7a) through the Cipres Science Gateway (www.phylo.org) under the GTR model. Analyses were run with 4 chains for a total of 50000000 generations and trees were sampled every 1000 generations, and first 25% of the trees were discarded as the burn-in phase of each analysis. Bayesian Posterior Probabilities (PP) values were obtained from the 50% majority-rule consensus trees and nodes with PP≥0.95 were considered as significantly supported.
3 Results
3.1 Phylogeny
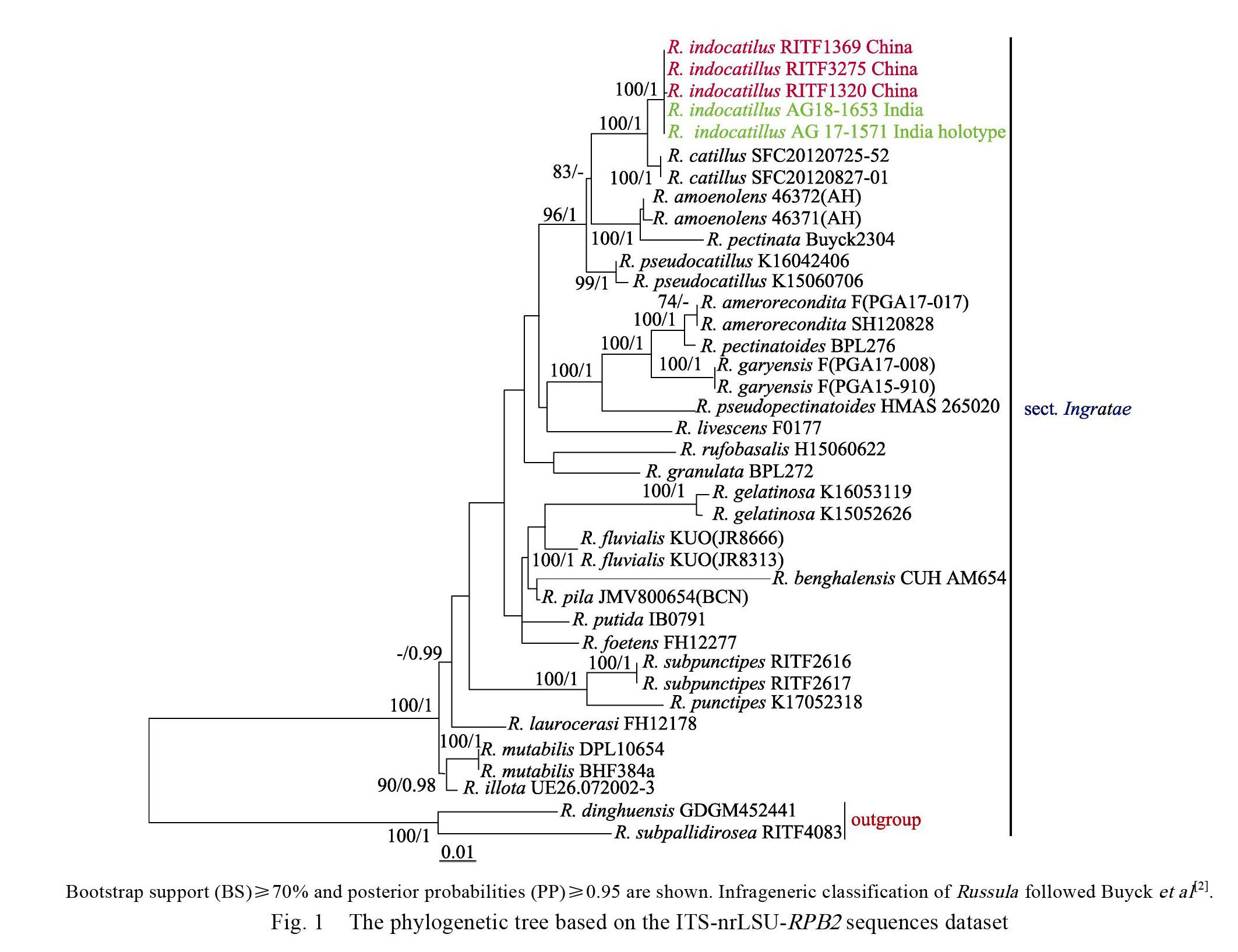
The final ITS-nrLSU-RPB2 sequences dataset was analysed by ML and BA. Both the ML and BA analyses result in similar tree topologies and only the best ML tree is shown in Fig. 1. BA analysis posterior probabilities are also showed along the branches.
The phylogenetic analysis showed that sect. Ingratae was a strongly supported monophyletic group by BS (100%) and PP (1) (Fig. 1). Our phylogeny indicates the Chinese collections and the holotype of Indian R. indocatillus formed an independent lineage within sect. Ingratae with high statistical support (BS=100%, PP=1) (Fig. 1), which is sister to the lineage consisting of South Korean species R. catillus Lee with 100% bootstrap support and 1.00 posterior probabilities (Fig. 1).
3.2 Taxonomy
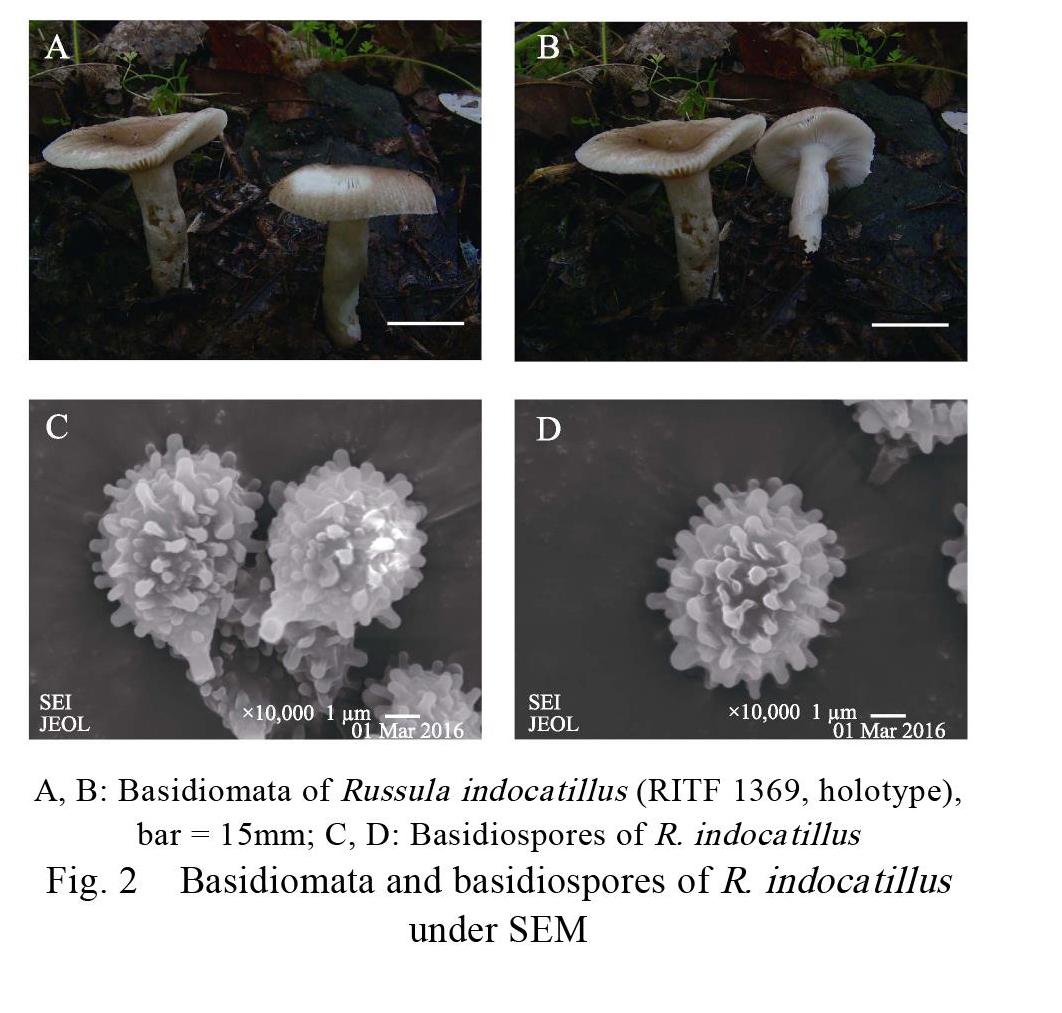
Russula indocatillus A. Ghosh, K. Das & R.P. Bhatt Figs. 2, 3 Basidiomata small to medium-size. Pileus 40–52 mm in diam., first hemispheric, then planoconvex, applanate with a slightly depressed center when mature; margin incurved when young, relatively flat at maturity, stripes 5–10 mm from edge inward; surface smooth, glabrous, viscid when wet, not peeling readily; brownish orange (6C7) to yellow ochre (6D7) at the center, margin first butter yellow (4A5) to apricot (5B6), turning pale (2A2) to pale yellow (2A3) after maturation, unchanging in color when bruised. Lamellae adnate, 3–4 mm in height, 15–20 pieces per centimeter at edge, not forked, slightly interveined, satin white (2A1) to yellowish white (3A2), unchanging after bruising, not brittle when toughing, lamellulae absent. Stipe 22–36 mm×8–12 mm, central to subcentral, subcylindrical to cylindrical, slightly curved towards center, slightly attenuate towards the base, surface dry, rugulose longitudinally, pithy and hollow when mature, pale yellow (3A3) to grayish yellow (5B5), unchanging when bruised. Context 2–3 mm thick at pileus center, white to cream, without color changing when bruised. Taste slightly bitter or slightly spicy. Oder indistinct. Spore print white to cream.
Basidiospores [60/3/3] (5) 5.4–6.8 (7.5)×(4) 4.5–5.2 (5.6) μm, [Q=1.10–1.32, Q=1.18±0.11], subglobose to broadly ellipsoid to ellipsoid; ornamentation amyloid, warts bluntly conical to subcylindrical, isolated or connected with irregular lines or ridge, not forming a reticulum, measuring 0.3– 0.55 (1.0) μm high; suprahilar spot large. Basidia (27) 32.8–44.8 (47)×(6.2) 7.4–9.6 (11) μm, mostly 4-spored, sometimes 2- and 3-spored, subclavate to clavate, thin-walled, sterigmata 3–6×1.5–2.3 μm. Lamellar trama mainly composed of numerous nested spherocytes (20–50×17–43 μm) surrounded by connective hyphae (1.5–4.5 μm wide). Pleurocystidia (46) 51–64 (69)×(7) 7.6–9.4 (11) μm, clavate to subclavate to subcylindrical, apically obtuse or mucronate to moniliform, granular or crystalline contents turning to blackish grey in SV. Cheilocystidia (42) 45.8–66 (73)×6–10 μm, mostly similar to pleurocystidia, clavate to subcylindrical, apically obtuse to mucronate, granular or crystalline contents blackish grey in SV. Pileipellis orthochromatic in Cresyl blue, 120–160 μm thick, distinctly two layered, not sharply delimited from the underlying sphaerocysts of the context, composed of suprapellis and subpellis; suprapellis 90–120 μm thick, consisting of thin- walled, often ramifying, septate, erect or repent hyphae and pileocystidia, terminal elements 12–36×2.4–6 μm, apically obtuse or attenuate; subpellis 30–40 μm thick, consisting of horizontally oriented hyphae (2–6 μm diam.); pileocystidia 25–45×4–7 μm, always one-celled, cylindrical to narrowly clavate, apically mucronate or obtuse, usually with an appendage, granular or crystalline contents turning to blackish grey in SV.
Specimens examined: China, Hubei Province, Shennongjia Forestry District, Hongping Town, 31°4017.86N; 110°2640.53E, 2560 m asl., in mixed hardwood forests, dominated by Fagus, 14 July 2012, Yuan117 (RITF1369). Hubei Province, Yichang City, Xingshan County, Nangyang Town, Longmenkou Forest Farm, mixed forests, 31°1839N; 110°4020E, 260 m asl., 11 July 2012, leg. Yuan54 (RITF1320). Yunnan Province, Kunming City, Yeya Lake, mixed forests, dominated by Quercus, 25°0734N; 102°5142E, 2126m asl., 27 July 2014, leg. LHJ14072713 (RITF3275).
4 Discussion
Our samples share a remarkable resemblance with the description of Ghosh et al (2020) [29]. Meanwhile, the Chinese collections clustered (BS= 100%, PP=1) with the holotype of Indian R. indocatillus and share 100% ITS sequence identity, suggesting that they are conspecific. Based on the macro-and micro-morphological character, as well as the significant support for its phylogenetic placement (Fig. 1), R. indocatillus was placed in Russula subg. Heterphyllidia sect. Ingratae.
The phylogenetic result indicated R. indocatillus is sister to South Korean species R. catillus, however, they can be easily distinguished for R. catillus has brittle and less lamellae, whitish stipe slightly inflate towards to the base, larger basidia (41.6–48.9×9.3–11.7 μm), smaller cheilocystidia (43.2–59.7×6.9–11.2 μm) and thicker pileipellis (110–210 μm), furthermore, R. catillus grows in the areas with higher latitudes and greater longitudes and is symbiosis with oaks[26].
In addition, the recently reported four new species (R. gelatinosa Y. Song & L.H. Qiu, R. rufobasalis Y. Song & L.H. Qiu, R. pseudocatillus F. Yuan & Y. Song and R. subpunctipes J. Song) and a new recorded species (R. punctipes Singer) of subg. Heterophyllidia sect. Ingratae from China are worthy of attention[12-14]. Comparing to R. indocatillus, R. gelatinosa has a surface gelatinized, a reddish brown to ochre to dark brown pileus, whitish lamellae with reddish hue, a longer stipe (7–9.5× 1.2–1.7 μm), larger basidiospores [(7.6) 8.2–9.0– 9.8×(7) 7.5–8.4–9.1 (9.5) μm] with ornamentation composing of high wings and short warts, and large basidia [(44) 48–67.5 (69)×(10) 12–19 (20.5) μm][12]; R. rufobasalis possess whitish to rusty tinged lamellae, a reddish stipe base, and a thicker pileipellis (360–500 μm)[12]; R. pseudocatillus has larger basidiospores [7.0–7.9–8.6 (9.2)×(5.1) 5.5–6.1–6.6 (6.7) μm] with conical to cylindrical isolated warts never forming a reticulum, shorter pleurocystidia (32–37.5×9.5–11.5 μm) and pileocystidia (13–33× 3–6 μm) [13]; R. subpunctipes possess sulfur yellow to honey yellow pileus with a margin cracking into patches, henna lamellar edge, henna warts on the hollow stipe, faint fragrant odor, larger basidiospores [8.7–10.5 (–10.7)×8.3–9.5 μm] with ornamentation composed of large wings and short isolated warts, larger basidia (65–72×11–14 μm) and pleurocystidia (110–132×9–25 μm) [14]; R. punctipes has a dull yellow to brownish stipe with ochraceous brown to dark brown punctuations, strongly fetid odour, larger basidiospores [(7.1) 7.9–9.3–10.3 (11)×(6.0) 7.2–8.7–10.1 (10.3) μm] with ornamentation composed of wings (up to 3 μm high) and short warts, wider basidia [(32)33.5–48.5 (54) × (10.5) 11–18.5 μm] and cheilocystidia (30.5–62.5× 9–18.5) [12].
References
[1] Knudsen H, Borgen T. Russulaceae in Greenland[M]//Arctic and alpine mycology 1. Seattle and London: University of Washington Press, 1982: 1-559.
[2] Buyck B, Zoller S, Hofstetter V. Walking the thin line… ten years later: the dilemma of above- versus below-ground features to support phylogenies in the Russulaceae (Basidiomycota)[J]. Fungal Diversity, 2018, 89: 267-292.
[3] Looney B P, Meidl P, Piatek M J, et al. Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates[J]. New Phytologist, 2018, 218: 54-65.
[4] Buyck B, Wang X H, Adam?íková K, et al. One step closer to unravelling the origin of Russula: subgenus Glutinosae subg. nov[J]. Mycosphere, 2020, 11: 285-304.
[5] Adam?ík S, Looney, B, Caboň M, et al. The quest for a globally comprehensible Russula language[J]. Fungal Diversity, 2019, 99(1): 369-449.
[6] Wijayawardene N N, Hyde K D, Al-Ani L K T, et al. Outline of Fungi and fungus-like taxa[J]. Mycosphere, 2020, 11(1): 1060-1456.
[7] Wang X H, Yang Z L, Li Y C, et al. Russula griseocarnosa sp. nov. (Russulaceae, Russulales), a commercially important edible mushroom in tropical China: mycorrhiza, phylogenetic position, and taxonomy[J]. Nova Hedwigia, 2009, 88: 269-282.
[8] Wu F, Zhou L W, Yang Z L, et al. Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species[J]. Fungal Diversity, 2019, 98: 1-76.
[9] 王向华. 红菇科可食真菌的若干分类问题[J]. 菌物学报, 2020, 39(9): 1617-1639.
Wang X H. Taxonomic comments on edible species of Russulaceae[J]. Mycosystema, 2020, 39(9): 1617-1639.
[10] Singer R. The Agaricales in modern taxonomy[M]. 4rd ed. K?nigstein: Koeltz Scientific Books, 1986: 1-981.
[11] Romagnesi H. Statuts et noms nouveaux pour les taxa infragénériques dans le genre Russula[J]. Documentation Mycologique, 1987, 18: 39-40.
[12] Song Y, Li J W, Buyck B, et al. Russula verrucospora sp. nov. and R. xanthovirens sp. nov., two novel species of Russula (Russulaceae) from southern China[J]. Cryptogamie Mycologie, 2018, 39(1): 129-142.
[13] Yuan F, Song Y, Buyck B, et al. Russula viridicinnamomea F. Yuan & Y. Song, sp. nov. and R. pseudocatillus F. Yuan & Y. Song, sp. nov., two new species from southern China[J]. Cryptogamie, Mycologie, 2019, 40(4): 45-56.
[14] Song J, Chen B, Liang J F, et al. Morphology and phylogeny reveal Russula subpunctipes sp. nov., from southern China[J]. Phytotaxa, 2020, 459(1): 16-24.
[15] Vellinga E C, Noordeloos M E. Glossary[M]//Noordeloos M E, Kuyper T W, Vellinga E C. Flora Agaricina Neerlandica 5. Rotterdam: Balkema Publishers, 1988: 6-11.
[16] Kornerup A, Wanscher J H. Taschenlexikon der farben[M]. 3rd ed. G?ttingen: Muster-Schmidt Verlag, 1981.
[17] Buyck B. Valeur taxonomique du bleu de crésyl pour le genre Russula[J]. Bulletin de la Société Mycologique de France, 1989, 105: 1-6.
[18] 周玲玲,梁俊峰. 大型真菌 DNA 提取方法的改进[J]. 广东林业科技, 2011, 27(1) : 13- 16.
Zhou, L L, Liang, J F. An improved protocol for extraction of DNA from macrofungi[J]. Guangdong Forest Science Technology, 2011, 27(1): 13-16.
[19] White T J, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies[M]// Innis M A, Gelfand D H, Sninsky J J, et al. PCR protocols, a guide to methods and applications. San Diego: Academic Press, 1990: 315-321.
[20] Vilgalys R, Hester M. Rapid genetic identification and mapping enzymatically amplified ribosomal DNA from several Cryptococcus species[J]. Journal of Bacteriology, 1990, 172: 4238-4246.
[21] Liu Y J, Hall B D. Body plan evolution of ascomycetes, as inferred from an RNA polymerase II, phylogeny[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 13: 4507-4512.
[22] Matheny P B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales)[J]. Molecular Phylogenetics and Evolution, 2005, 35: 1-20.
[23] Hall T A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT[C]// Nucleic Acids Symposium Series, 1999, 41: 95-98.
[24] Vidal J M, Alvarado P, Loizides M, et al. A phylogenetic and taxonomic revision of sequestrate Russulaceae in Mediterranean and temperate Europe[J]. Persoonia, 2019, 42: 127-185.
[25] Yuan H S, Lu X, Dai Y C, et al. Fungal diversity notes 1277-1386: taxonomic and phylogenetic contributions to fungal taxa[J]. Fungal Diversity, 2020, 104: 1-266.
[26] Lee H, Park M S, Jung P E, et al. Re-evaluation of the taxonomy and diversity of Russula section Foetentinaei (Russulales, Basidiomycota) in Korea[J]. Mycoscience, 2017, 58: 351-360.
[27] Zhang J B, Li J W, Li F, et al. Russula dinghuensis sp. nov. and R. subpallidirosea sp. nov., two new species from southern china supported by morphological and molecular evidence[J]. Cryptogamie Mycologie, 2017, 38 (2): 191-203.
[28] Looney B P, Ryberg M, Hampe F, et al. Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi[J]. Molecular Ecology, 2016, 25(2): 630-647.
[29] Buyck B, Hofstetter V, Eberhardt U, et al. Walking the thin line between Russula and Lactarius: the dilemma of Russula subsect. Ochricompactae[J]. Fungal Diversity, 2008, 28: 15-40.
[30] Ghosh A, Das K, Bhatt R P, et al. Two new species of Genus Russula from Western Himalaya with morphological details and phylogenetic estimations[J]. Nova Hedwigia, 2020, 111: 115-130.
[31] 谢雪丹, 刘培贵, 于富强. 云南松幼苗上红菇类菌根真菌的物种多样性及其菌根形态[J]. 云南植物研究, 2010, 32(3): 211-220.
Xie X D, Liu P G, Yu F Q. Species diversity of russuloid mycorrhizae forming fungi on Pinus yunnanensis seedlings and the mycorrhizal morphology[J]. Acta Botanica Yunnanica, 2010, 32: 211-220.
[32] Looney B P. Molecular annotation of type specimens of Russula species described by W.A. Murrill from the southeast United States[J]. Mycotaxon, 2014, 129(2): 255-268.
[33] Haelewaters D, Dirks A C, Kappler L A, et al. A preliminary checklist of fungi at the Boston Harbor Islands[J]. Northeastern Naturalist, 2018, 25(S9): 45-77.
[34] Miller S L, Buyck B. Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications[J]. Mycological Research, 2002, 106(3): 259-276.
[35] Li G J, Zhao D, Li S F, et al. Russula chiui and Russula pseudopectinatoides, two new species from southwestern China supported by morphological and molecular evidence[J]. Mycological Progress, 2015, 14: 1-14.
[36] Song J, Liang J F, Mehrabi-Koushki M, et al. Fungal systematics and evolution 5[J]. Sydowia, 2019, 71: 141-245.
責任编辑:崔丽虹
