Molecular Evolution of Visual Opsin Genes during the Behavioral Shifts between Different Photic Environments in Geckos
2021-09-27YaoCAIYuefenFANYouxiaYUEPengLIJieYANandKaiyaZHOU
Yao CAI ,Yuefen FAN ,Youxia YUE ,Peng LI* ,Jie YAN* and Kaiya ZHOU
1Jiangsu Key Laboratory for Biodiversity and Biotechnology,College of Life Sciences,Nanjing Normal University,Nanjing 210023,Jiangsu,China
2School of Food Science,Nanjing Xiaozhuang University,Nanjing 211171,Jiangsu,China
Abstract Reptiles are the most morphologically and physiologically diverse tetrapods,with the squamates having the most diverse habitats.Lizard is an important model system for understanding the role of visual ecology,phylogeny and behavior on the structure of visual systems.In this study,we compared three opsin genes (RH2,LWS and SWS1) among 49 reptile species to detect positively selected genes as well as amino acid sites.Our results indicated that visual opsin genes have undergone divergent selection pressures in all lizards and RH2 and LWS suffered stronger positive selection than SWS1. Twelve positively selected sites were picked out for RH2 and LWS.Moreover,many diagnostic sites were found between geckos and non-gecko lizards,most of which were located near the positively selected sites and some of them have already been reported to be responsible for significant shifts of the wavelength of maximum absorption (λmax).The results indicated that the gecko lineage accelerated the evolution of these genes to adapt to the dim-light environment or nocturnality as well as the switch between nocturnality and diurnality.
Keywords Gecko,diurnality,molecular evolution,nocturnality,opsin gene
1.Introduction
Reptiles are the most morphologically and physiologically diverse tetrapods,which diverged from early tetrapods in the late Carboniferous period approximately 310-320 million years ago along with adaptive evolution (Hughes,2008).In particular,squamates represent the most diverse radiation of terrestrial vertebrates and occupy ecologically diverse habitat niches.Even though,most of them rely heavily on vision systems as their primary sensory modality which is closely related to the ability and means to interact with their environments (Fleishmanet al
.,2011).Vision in vertebrates is especially well studied,and studies of the evolution of their visual pigments have been able to both identify evolutionary changes,and to ascribe such changes to adaptive evolutionary processes (Hughes,2008).The vertebrate retina contains two types of photoreceptor:rods which are responsible for dim light and mediate scotopic vision,and cones acting in daylight,which mediate photopic vision and are necessary with color vision (Bowmaker and Hunt,2006).Comparative studies across all of the major vertebrates groups have detailed that there are one rod pigmentRH1
(rhodopsin) and four cone pigments encoded by distinct opsin gene families:LWS
/MWS
(long/middle-wavelength sensitive opsin gene) maximally sensitive in the red-green spectral region from 490-570 nm,RH2
(RH1
-like) sensitive in green from 480-535 nm,SWS2
(short-wavelength sensitive 2 opsin gene) sensitive in the blue from 410-490 nm andSWS1
(short-wavelength sensitive 1 opsin gene) sensitive in the violetultraviolet from 355-440 nm (Yokoyamaet al
.,2000).However,different lifestyles were in harmony with different habitat circumstance and various kinds of visual system.For instance,molecular evidence implies thatLWS
was inactivated on at least five cetaceans which are known to adapt to dim-light field dominated by blue light (Meredithet al
.,2013).Several groups including cetaceans,pinnipeds,bats,and mole rats,inhabit in low photopic environments,some of which have lost theirSWS1
opsins (Zhaoet al
.,2009).Furthermore,the significant divergent selection in batsSWS1
andLWS
was reported to be occurred in response to echolocation ability and foraging habitat,respectively (Gutierrezet al
.,2018).Among Squamata,dedicated fossorial snakes lost all visual opsins other thanRH1
,whereas all other snakes (including less dedicated burrowers) also have functionalSWS1
andLWS
opsins.In contrast,all lizards,even highly fossorial amphisbaenians express functionalLWS
,SWS1
,SWS2,
andRH1
genes,and most also expressRH2
(Simõeset al
.,2015).Lizards provide an important model system for understanding the role of visual ecology,phylogeny and behavior on the structure of visual systems in Squamata(Fleishman and Persons,2001;Loewet al
.,2002;Stuart-Foxet al
.,2007;Macedoniaet al
.,2009).This extensive group includes true chameleons,iguanid lizards,monitor lizards,geckos,and skinks.Some of them are diurnal,and some crepuscular or nocturnal members.Possible changes during transformation of the lifestyle in these reptiles from diurnal to nocturnal have induced some adaptive evolution (Bowmaker,2008).Therefore,it’s notable to explore the evolution of opsin genes owing to lifestyle transformation and understand molecular features and specializations in squamates.For example,Anolis carolinensis
,a diurnal lizard representive,possesses a complete set of opsin paralogs,however,a nocturnal gecko,Gekko japonicus
only has three functional opsins:SWS1
,LWS,
andRH2
(Liuet al
.,2015).This result from genomic sequence data is consistent with the inference of gecko opsin composition based on the wavelength of maximum absorption (λ
) values (Kojimaet al
.,1992,Loew,1994) and the experiments which failed to detect any hybridizing band using the chameleonRH1
andSWS2
cDNA clones as probes in the gecko genome (Shiet al
.,2001).With the loss ofRH1
,the question of which functional opsin is responsible for nocturnal vision in geckos needs to be solved.The amino acid replacement F89C ofRH2
was suggested to allow geckos to receive more light (Liuet al
.,2015).However,the evolutionary pattern of opsin they proposed was based on genome data of few species,and more genome data or molecular data from other related species are necessary to determine how particular changes occurred in these opsin genes.In this study,we aimed to further investigate adaptive evolution of gecko opsin genes during their behavior shift between different photic environments by comparing opsin genes among diverse lizard reptiles.2.Material and Methods
2.1.Taxon Sampling
Samples were collected by fieldwork or obtained by academic exchanges.Taxon sampling for surveys of opsin genes comprised 49 reptile species including 40 lizards(ingroups),6 snakes and 3 turtles (outgroups),27 of which were newly sequenced and others were downloaded from GenBank in this study (supplementary Table S1).Sample species includes seventeen geckos,six scincoid lizards;eleven Lacertoidea species;one Anguimorpha lizard;five iguanas;and six snakes.Samples used in the molecular analyses are detailed in supplementary Table S1.PCR and Sequence alignment
Genomic DNA was extracted from muscle tissues of each individual following standard phenol/chloroform extraction methods.We attempted to amplify each exon ofRH2
,LWS
andSWS1
visual opsin genes using primers newly designed according to the sequences fromGekko gecko
,Phelsuma madagascariensis
,andAnolis carolinensis
(supplementary Table S2-S4).All fragments were amplified in 50 μL PCR reactions:1 μL of each primer,10 μmol/L;1.8 μL DNA template,ca.200-300 ng;5 μL 10 × PCR buffer (Mg);4 μL dNTPs,2.5 mmol/L;0.5 μL TakaraTaq
(Takara Biomedical,Dalian,China) and 36.7 μL ddHO.The PCR condition was as follows:3 min at 94°C followed by 35 cycles of 30 s at 94 °C,30 s at 52-60 °C,90 s at 72°C,and a subsequent 10 min final extension step at 72 °C.PCR products were checked by electrophoresis on 1.5% agarose gels and purified with AxyPrep PCR Cleanup Kit (AxyGen MA,USA).Sequencing was carried out bidirectionally on an ABI PRISM 3730 DNA Sequence.The newly generated sequences were examined by comparison with the published ones by BLAST on NCBI,and then assembled by Lasergene v7.1 (DNASTAR Inc.,Madison,WI,USA).After manual editing,all sequences were aligned by MEGA7.0 (Kumaret al
.,2016).Totally 431 fragments with highquality were picked and concatenated to 80 opsin genes,and then analyzed together.The newly sequences were deposited in GenBank under accession numbers KU645203-KU645282(supplementary Table S1).2.2.Molecular evolution analysis
The CODEML program from the PAML 4.9 package (Yang,2007) was used to estimate the non-synonymous (d
) and synonymous (d
) substitution rates and their respective ratio (d
/d
orω
) for theRH2
,LWS
andSWS1
genes among squamates.Theω
indicates changes in selective pressures,whereω
=1,ω
< 1 andω
> 1 corresponds to neutral evolution,purifying and positive selection,respectively.A well-accepted squamate phylogenetic tree was used as a framework tree in our analysis for each gene (Rosleret al
.,2011,Pyronet al
.,2013,Zheng and Wiens,2016).Different datasets,dataset IA (all lizards),dataset IB (all squamates),dataset II (only geckos),and dataset III (non-gecko lizards) were used for PAML analyses to minimize the effects of sampling.To examine the probabilities of sites under positive selection in three opsin genes,we used site models M8a (β andω
=1)versus M8 (Swansonet al
.,2003;Wonget al
.,2004) implemented in the CODEML program of PAML 4.9 (Yang,2007).The nested models were compared using a likelihood ratio test(LRT) with aχ
distribution.Positively selected sites in the M8 were identified using a Bayes Empirical Bayes (BEB) analysis(Yanget al
.,2005) with posterior probabilities ≥ 0.80.Positively selected sites were also detected by fixed effects likelihood(FEL) performed in HYPHY (Pond and Frost,2005) (via the www.datamonkey.org web server),with the default settings of significance levels of 0.2.We then performed selective pressure detection using TreeSAAP v.3.2 (Woolleyet al
.,2003),which detected selection based on 31 physiochemical amino acid properties.All magnitude category 6-8 changes withP
≤0.05 were used as an index for the degree of radical amino acid substitutions and positive selection.To evaluate whether positive selection was restricted to specific squamate lineages,we used branch models (including free-ratio model and two-ratio model) (Yang,1998;Yang and Nielsen,1998) and branch-site model implemented in CODEML.The free-ratio model (M1) that assumes an independentω
ratio for each branch was compared to the null one-ratio model(M0) with the sameω
for all branches (Yang and Nielsen,1998).Two-ratio model and branch-site model require the foreground branches (lineages tested to be under positive selection) and background branches (rest of the lineages) to be defined a priori.Theω
ratios were estimated for two branch categories (geckos as foreground branches versus non-gecko lizards as background branches) in two-ratio model,and then also compared with the null M0 model.The modified branch-site model A withω
varying among sites and among lineages (Zhanget al
.,2005)was tested against the null hypothesis of no selection in any of the foreground or background branches.Each squamate lineage across the phylogeny was used as the foreground branch,respectively,where the remaining branches were treated as background branches for each gene.A false discovery rate (FDR) correction for multiple tests was applied to the LRTP
values for branch-site model analysis (Anisimova and Yang,2007).2.3.Sliding Window Analyses
To explore the further heterogeneous selection pressure atRH2
,LWS
andSWS1
opsin genes across squamate phylogeny,we constructed a sliding window analysis using the program SWAAP (Pride,2000)with window size at 30 bp and step size at 3 bp,which were repeated for several groups of interest.Sliding window analysis was performed to show variation inω
value (d
/d
) along each opsin genes in geckos,non-gecko lizards and all lizards.2.4.Mapping of positively selected sites onto opsin 3D structures
To gain insights into the functional significance of these putatively selected sites,we mapped these positive sites onto protein structures.The 3D structures of opsin genes under positive selection were predicted by using the homology modeling software provided on the I-TASSER server (Zhang,2008).The protein sequences of positively selected genes were derived from the common Tokay gecko (Gekko gecko
),which were obtained from the GenBank database.3.Results
3.1.Visual pigment complement
We assembled a dataset of opsin gene sequences for 49 squamates,comprising 17 Gekkota geckos,6 Scincoidea lizards,11 Lacertoidea lizards,1 Anguimorpha lizard,5 Iguania lizards,6 Alethinophidia snakes and 3 turtles (outgroups).We have successfully amplified three visual opsin genes (RH2
,LWS
,SWS1
) from genomic DNA of the lizards from 20 families.All sequences were assembled and concatenated correctly for the protein coding region ofRH2
,LWS
,andSWS1
,which contain 1065 bp (354 aa),1110 bp (369 aa),1044 bp (346 aa),respectively.The sequence alignments had no premature stop codons or frame-shift mutations,which indicated the presence of functional proteins in squamates.3.2.Selection on opsin genes
We found M8 that incorporated selection fitted the data better than the neutral model,M8a,at two opsin genes (RH2
andLWS
,P
< 0.05),suggesting that these genes were subjected to positive selection in squamates.However,forSWS1
,the LRT showed no significant difference between the models M8 and M8a implying no positive selection.Using M8,the most stringent model carried out in PAML,a small proportion of codons (0.84%-10.17%) were estimated to be under selection with averageω
values of 1.292-2.462 at the two positively selected genes in squamates (Table 1).3 (0.84%),36 (10.17%),and 3 (0.84%) positively selected sites were identified respectively by the BEB approach as having posterior probabilities ≥ 0.80 atRH2
in dataset IA (all lizards),dataset II (only gecko),and dataset III (non-gecko lizards).Meanwhile,6 (1.63%),7 (1.89%),and 8 (2.17%) positively selected sites were identified respectively atLWS
in dataset IA,dataset IB (all squamates),and dataset II.However,no positively selected sites were detected atLWS
in dataset III.When we used a significance threshold of 0.95 for posterior probabilities,the number of positively selected amino acids decreased to two,thirteen,and two for dataset IA,II,and III ofRH2
,while one,one,and two for dataset IA,IB,and II ofLWS
.FEL,performed in HYPHY (Pond and Frost,2005),was also used to test for selection in the three opsin genes.HYPHY can improve the estimation of theω
value by incorporating variation ind
whereasd
is fixed across sequences for all the PAML-basedanalysis.FEL analysis showed that significant signs of positive selection were detected at the three opsin genes with many more positively selection sites than that identified by M8 (Table 1).Combining the two different maximum likelihood (ML)methods,a total of 12 positively selected sites (9 atRH2
and 3 atLWS
) were picked out (sites in bold shown in Table 1).Sites that were identified to be under positive selection by two ML methods were regarded as robust candidates for sites under selection.We further employed a complementary proteinlevel approach implemented in TreeSAAP (Woolleyet al
.,2003)to evaluated destabilizing radical changes at each robust site.The result showed that 10 of 12 sites (83.88%) have at least three radical changes in properties (Table S5,Table S6)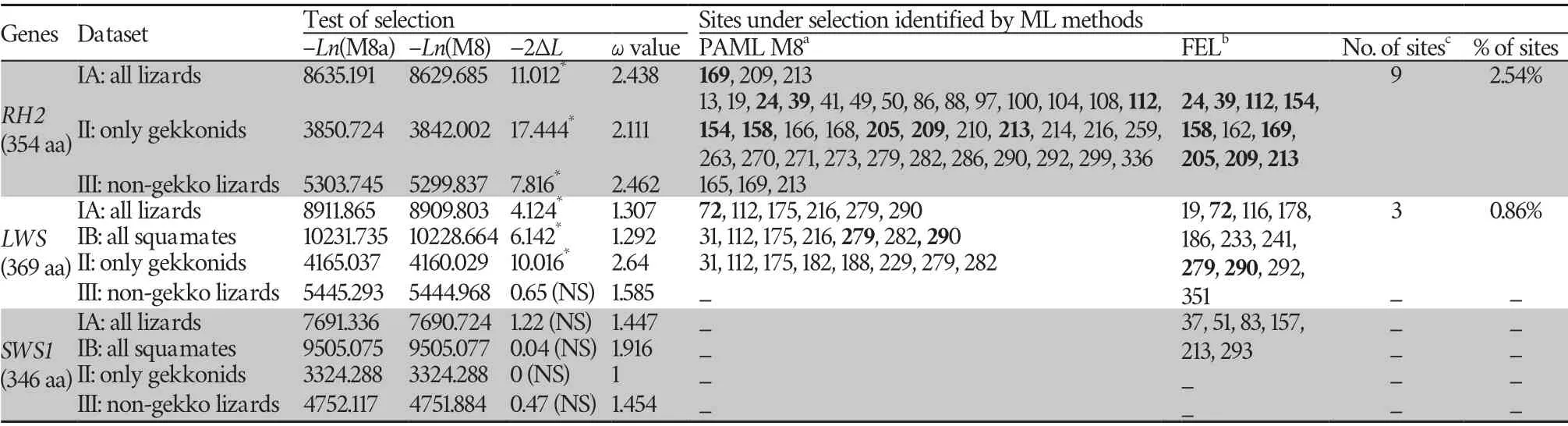
Table 1 Positively selected sited detected using two maximum likelihood (ML) methods across squamate phylogeny.
To evaluate whether positive selection is only limited to particular lineages at the three opsin genes,we first used branch models (including free-ratio and two-ratio models)that allow theω
ratio to vary among branches across the phylogeny.The LRT tests showed that relaxation of selective pressure was detected atRH2
(ω
=0.296,ω
=0.156) andLWS
genes (ω
=0.398,ω
=0.141;ω
=0.343,ω
=0.149) when lineage of geckos was treated as foreground branch and the remaining non-gecko lizards or other squamates were treated as background branches.On the contrary,theSWS1
(ω
=0.044) was implied to be under very strong purifying selection (data not shown).Using the stringent branch-site model,we found thatω
was greater than 1 atRH2
in four terminal branches leading toCnemaspis limi
,Phelsuma laticauda
,Gekko swinhonis
,andFylinia
sp.respectively,among which three were nested in the Gekkota clade (Figure 1,Table S5).As well atLWS
,evidences of positive selection were examined within lineage Gekkota.Theω
values were greater than 1 in the last common ancestor (LCA) of Gekkota,terminal branches leading toGonatodes annularis
,and leading toHemidactylus bowringii
(Figure 1,Table S5).In contrast,in branch-site model significant signs of positive selection were also examined atSWS1
in some ancestral branches(LCA of Gekkota,LCA of scincids,LCA of lacertids,LCA ofHemidactylus
,LCA ofPachydactylus vansoni
+Phelsuma
) and in one terminal branch leading toPhelsuma madagascariensis
(Figure 1,Table S5).Furthermore,each identified branch was detected some codons under positive selection.The results of branch-site models suggested more widespread positive selection across opsin genes in geckos than other squamates.This apparent difference can also be observed in the result of our sliding window analyses,with many peaks significantly greater than 1 for geckos,while the curves were uniformly below 1 for the negative control clades with only one exception(Figure 2).3.3.Spatial distribution of the positively selected residues on opsin 3D structures
A total of 12 radical amino acid changes subjected to positive selection identified by two ML methods were mapped onto the 3D structures of two opsin genes (nine atRH2
and 3 atLWS
).Opsins are G-protein coupled receptors (GPCRs),which contains seven α-helical transmembrane domains,an extracellular domain,and an intracellular domain to coupled (Terakita,2005).We found that 11 of the 12 (91.67%) positively selected sites were localized in the α-helical transmembrane domains (supplementary Figure S1).
Figure 1 Evidences of positive selection across Squamata phylogeny identified by branch-site model are shown.Three genes are marked with different colors,i.e.,RH2 (red),LWS (yellow),and SWS1 (green).The ω values greater than 1 for individual lineage according to free-ratio and two-ratio are shown.Amino acid changes identified using TreeSAAP across colored branch were indicated by blue vertical lines.The circles were on behalf of diurnal genera in geckos.All detailed results were listed in supplementary Table S5.

Figure 2 Sliding Window Analyses with window size at 30 bp and step size at 3 bp to show variation in ω value (dN/dS) along each opsin genes (A:RH2,B:LWS,C:SWS1) between only geckos (Red),non-gecko lizards (Green) and all lizards (Black).
3.4.Diagnostic amino acid sites between geckos and nongecko lizards
We also compared the amino acid sequences of the three opsin genes between geckos and non-gecko lizards to obtain the information of diagnostic sites.As shown in Table 2,when putting nocturnal and diurnal geckos together,16 amino acid residues of RH2 differed from those of non-gecko lizards (at residue 40,41,42,43,49,50,54,56,60,64,83,85,98,285,289,and 300,on a gray background in Table 2).However,residues 89 and 101 in diurnal geckos represented both statusfound in nocturnal geckos (89C,101A) and non-gecko lizards(89F,101G),indicating an intermediate status (shown on a dark blue background).Among all these amino acid residues,three sites (41,49,and 50,with a star in Table 2) were identified as positively selected sites by M8 model in PAML,and most of the rest are surrounding the positively selected sites.Moreover,certain changes at three sites (49,83,and 89,underlined in Table 2) have already been reported to cause significantλ
maxshifts in various visual pigments during vertebrate evolution(Yokoyamaet al
.,2007;Takenaka and Yokoyama,2007).Furthermore,the amino acid in codon position 89 has been reported to be an important functional determinant of rod and cone visual pigments (Yokoyama and Blow,2001),and the amino acid replacements F89C enables RH2 to have rod pigment-specific biochemical characteristics (Yokoyama and Blow,2001;Yokoyama and Tada,2010).F89C was found in all geckos included in this study except one diurnal species(Gonatodes annularis
).For LWS and SWS1,three and eight diagnostic sites can distinguish geckos and non-gecko lizards,respectively,and one to two sites have been reported to be important spectral tuning sites (Yokoyama and Radlwimmer,1998;Yokoyamaet al
.,2006).Among these sites at SWS1,in diurnal geckos both 85A (same to nocturnal gecko) and 85S(same to diurnal lizards) were detected.Besides,two sites which are shared between nocturnal gecko and diurnal lizards (298N and 321Y) were totally different in diurnal geckos (298H/Y and 321W,respectively) (on a light blue background in Table 2).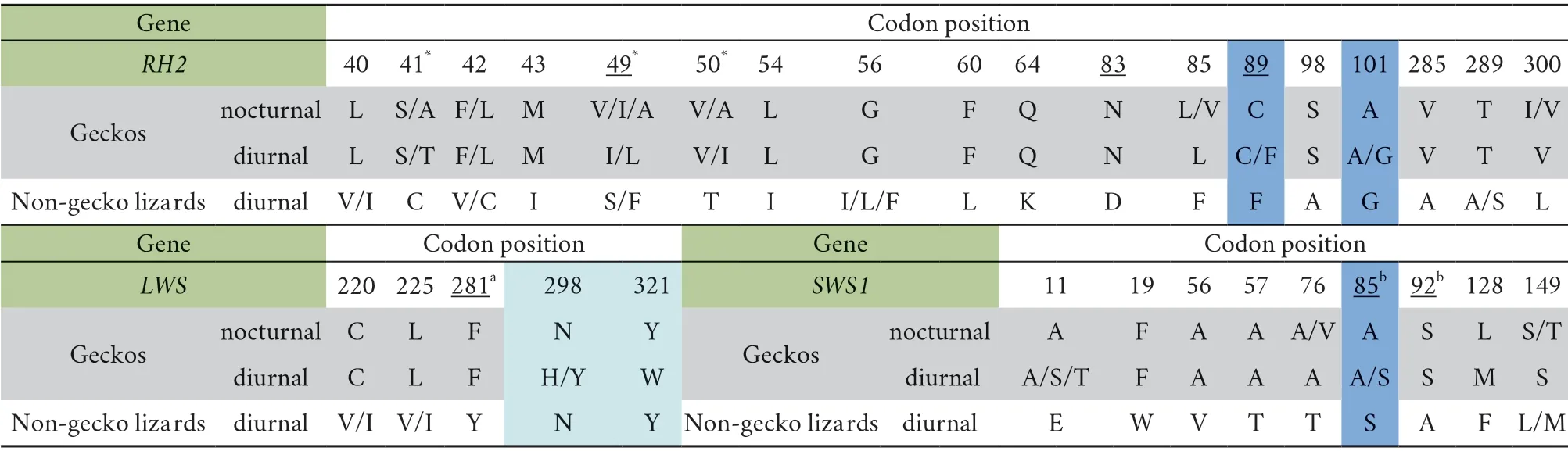
Table 2 Comparison of the diagnostic amino acid sites between the opsin genes of geckos and non-gecko lizards.
4.Discussion
Most vertebrates use a combination of rod and cone photoreceptors to enable vision in conditions ranging from starlight to direct sunlight.However,geckos are known to possess a mix of all-rod (nocturnal geckos) or all-cone retinas(diurnal geckos),which are all suggested to be evolutionary derived from cones by both morphological and molecular data(Roll,2000;Roll,2001b;Zhanget al
.,2006).Based on comparative retinal and photoreceptor morphology,Walls (1942) proposed a scenario for the evolution of photoreceptors in geckos that originated with the loss of rods in a diurnal ancestor of geckos,resulting in an all-cone retina.From this diurnal all-cone state,ancestral geckos transitioned to nocturnality and evolved an all-rod retina through photoreceptor transmutation.Some extant gecko lineages subsequently reverted back to diurnality and re-evolved all-cone retinas through transmutation of the “rods” back into cones (Schottet al
.,2019).Recent genome researches provided the evidence of aRH1
deficiency at the genome level in nocturnal geckos (Haraet al
.,2018;Liuet al
.,2015),and the loss ofRH1
occurred earlier than that ofSWS2
,which was in agreement with the hypothesis that the ancestors of modern geckos were diurnal lizards without rod opsin(Liuet al
.,2015).Moreover,the F89C amino acid substitution inRH2
gene shared between the Madagascar ground gecko and Japanese gecko suggested a possible functional switch or compensatory change in cone pigment of geckos that allow them to receive more light (Haraet al
.,2018;Liuet al
.,2015).However,the evolution pattern of opsin functional switch was proposed based on genome data of few species.More data from more related species are needed.As well known,most lizards are strictly diurnal like iguanids,chameleonids,agamids,scincids,lacertids,anguids,pygopodids,and varanids (Roll,2001a).However,as the largest extant lizard families,most geckos are nocturnal,with some lineages have been reversed back to diurnal.In this study,three opsin genes (RH1
,SWS1
,andLWS
) from nocturnal as well as diurnal geckos,and several lizard species were sampled and analyzed to further investigate the possibly important amino acid changes during evolution or adaptation.Site models (M8) identified 9 and 3 codons as robust sites under positive selection by at least two Maximum Likelihood methods forRH2
andLWS
respectively.However,model M8 failed to identify any positively selected sites forSWS1
opsin(Table 1),suggesting somewhat purifying selection onSWS1
opsin gene.In Branch-site model,ω
was found to be greater than 1 atRH2
in four terminal branches,among which three were nested in the Gekkota clade.Similarly atLWS
,evidences of positive selection were examined within lineage Gekkota(Figure 1).Moreover,significant signs of positive selection were also examined atSWS1
in some ancestral branches and some codons under positive selection were found which were not supported by the site model.Both of the results of branchsite models and the sliding window analyses supported more positive selection across opsin genes in gecko lineage than in non-gecko lizards.Among geckos,there are only 15 genera being diurnal (such asPhelsuma
,Sphaerodactylus
,andGonatodes
) that have purecone retinas as a consequence of a change of their habitat (Roll,2001a).Previous researches revealed difference in anatomical of the retina between nocturnal and diurnal geckos.They pointed that nocturnal geckos had photoreceptors with large rod-shaped outer segments and colored oil droplets in the inner segment whereas diurnal species had much smaller somewhat more cone-like outer segments and colorless oil droplets (Roll,2000;2001a).However,our results provided an additional molecular supplement of the opsin genes between nocturnal and diurnal geckos.In all our samples,three species (Gonatodes annularis,Phelsuma madagascariensis
,and Phelsuma laticatus
) on behalf of diurnal genera had undergone stronger adaptive selection than others geckos resulting in transformation of diurnal ambient light (Figure 1).Previous researches have suggested thatRH2
opsin amino acid site 89 is an important functional determinant of rod and cone visual pigments (Shiet al
.,2001),and the amino acid replacements F89C in geckos enablesRH2
to have rod pigment-specific biochemical characteristics to allow them to receive more light in dim-light environment (Kojimaet al
.,1992;Shiet al
.,2001;Yokoyama and Tada,2010;Liuet al
.,2015;Haraet al
.,2018).But these researches are based on the comparison of few opsin sequences.In our study,comparing the orthologousRH2
pigment among 39 species,we deduced that all geckos have amino acid site 89C exceptG.annularis
(diurnal) and all non-gecko lizards were 89F (Table 2).89 F inG.annularis
is likely to be the result of reversion due to the re-adaptation to daylight.Similar change was also found at site 85 inSWS1
,which was also a reported spectral tuning site.Furthermore,we also found sites like 298 and 321 inLWS
,where nocturnal gecko and diurnal lizards share the same residue;however diurnal geckos have different residues.These changes are likely to occur during the evolution of geckos from nocturnality to diurnality.In conclusion,our results indicate that opsin genes are under different selection pressures in all lizards,and gecko lineage has accelerated the evolution of these genes to adapt to the dim-light environment or nocturnality as well as the switch between nocturnality and diurnality.We compared amino acid sequences of three opsin genes (RH2,SWS1
,andLWS
)from representatives of many extant lizard families,which provide clues and evidences for further study of the molecular specialization and evolution to response to different lifestyle(nocturnal or diurnal) in geckos.Acknowledgements
We express special appreciation to B.SUN,Y.LIN and K.GUO for their assistance in sample collection.We are grateful to Y.GUO,K.LI,and Z.WANG for discussions and valuable comments.Financial support was provided by the National Natural Science Foundation of China(NSFC) (Grant No.31672269 and 31000949 to J.YAN,the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJA330001 to P.LI),the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD),and Top-Notch Academic Programs Project of Jiangsu Higher Education Institutions (TAPP,PPZY2015B117).Appendix
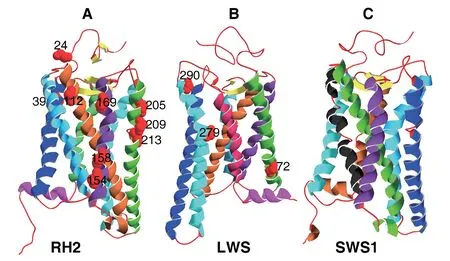
Figure S1 The predicted three dimensional structures (ribbon diagram) of RH2,LWS,and SWS1.α-helices,β-sheets,and loops are represented by spirals,arrows,and lines,and are coloured in different colours,yellow,and red,respectively.A:RH2;B:LWS;C:SWS1.The positive selection sites (24,39,112,154,158,169,205,209,213 for RH2;and 72,279,290 for LWS) are marked with red globes.

Table S1 Samples with taxon and Genbank accession number.Sequences newly generated for this study are indicated in bold.

Table S2 Primers forRH2 exons amplification from genomic DNA.

Table S3 Primers for LWS exons amplification from genomic DNA.

Table S4 Primers for SWS1 exons amplification from genomic DNA.
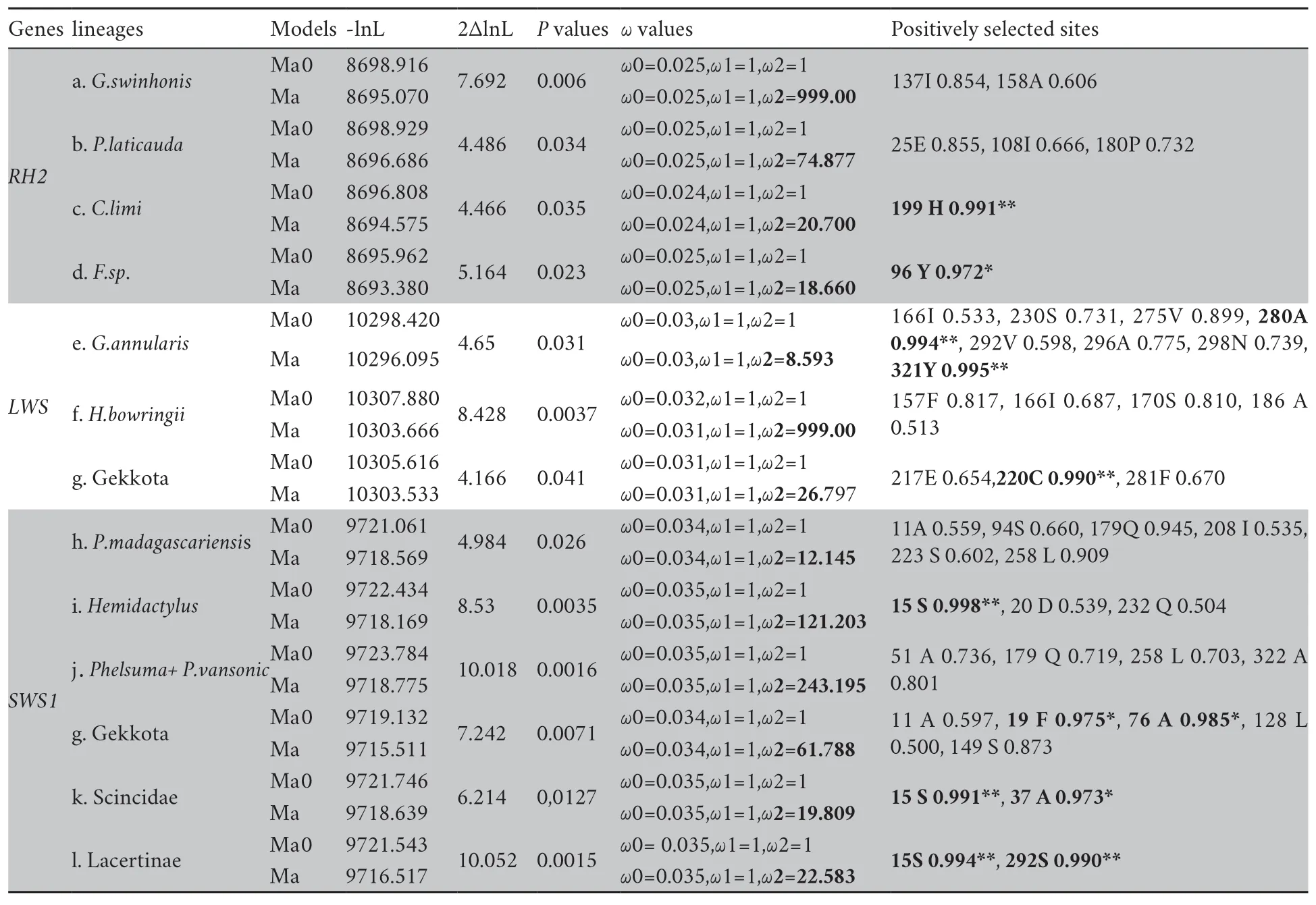
Table S5 Positive selection detected base on branch-site model in all squamates.
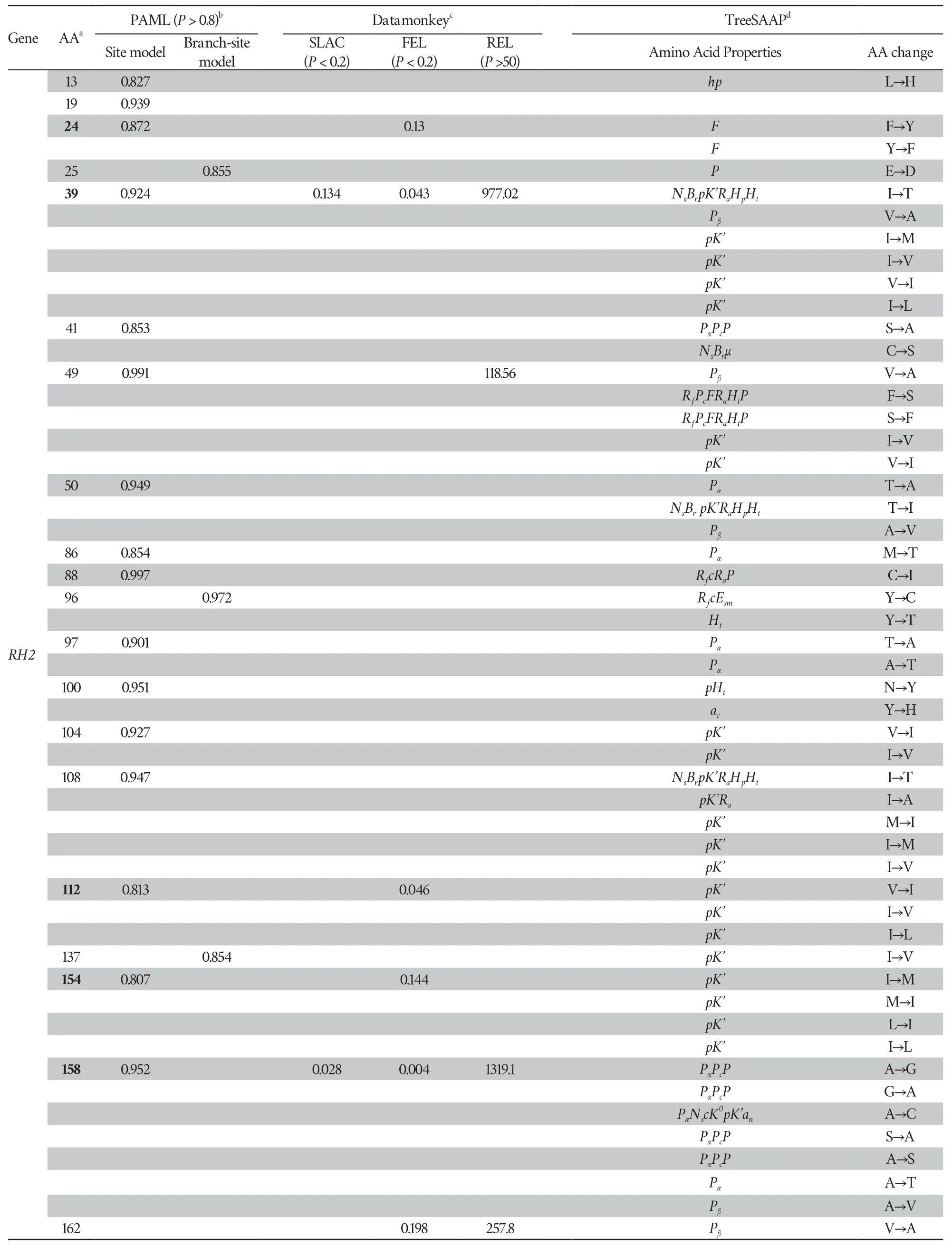
Table S6 Amino acid sites identified as positively selected sites detected using PAML,Datamonkey and TreeSAAP at RH2,LWS and SWS1 opsin genes.

(Continued Table S6)

(Continued Table S6)

(Continued Table S6)

(Continued Table S6)
杂志排行
Asian Herpetological Research的其它文章
- A New Species of Cyrtodactylus (Squamata:Gekkonidae) from the Karst Forests of Daweishan National Nature Reserve,Yunnan,China
- Disordered Translocation is Hastening Local Extinction of the Chinese Giant Salamander
- Effects of Life Histories on Genome Size Variation in Squamata
- Age Structure of Two Species of Odorous Frogs (Odorrana margaretae andOdorrana grahami)
- Complete Mitochondrial Genome of the Steppe Ribbon Racer (Psammophis lineolatus):The First Representative from the Snake Family Psammophiidae and its Phylogenetic Implications
- Transcriptome Sequencing Provides Evidence of Genetic Assimilation in a Toad-Headed Lizard at High Altitude
