Deletion of prominin-1 in mice results in disrupted photoreceptor outer segment protein homeostasis
2021-09-14YuShuXiaoJianLiangMinGaoJunRanSunYangLiuJieQiongChenXiaoHuanZhaoYiMinWangYuHongChenYuWeiWangXiaoLingWanXueTingLuoXiaoDongSun
Yu-Shu Xiao, Jian Liang,2, Min Gao, Jun-Ran Sun, Yang Liu, Jie-Qiong Chen,Xiao-Huan Zhao, Yi-Min Wang, Yu-Hong Chen, Yu-Wei Wang, Xiao-Ling Wan,2,Xue-Ting Luo,2, Xiao-Dong Sun,2,3,4
1Department of Ophthalmology, Shanghai General Hospital(Shanghai First People’s Hospital), Shanghai Jiao Tong University, School of Medicine, Shanghai 200080, China
2Shanghai Key Laboratory of Fundus Diseases, Shanghai 200080, China
3Shanghai Engineering Center for Visual Science and Photomedicine, Shanghai 200080, China
4National Clinical Research Center for Ophthalmic Diseases,Shanghai 200080, China
Abstract
● KEYWORDS: photoreceptor degeneration; Prominin-1;homeostasis; retinitis pigmentosa
INTRODUCTION
Inherited retinal degenerations (IRDs), a clinically and genetically heterogeneous group of diseases, primarily affect retinal photoreceptor or retinal pigment epithelium.Patients with IRDs suffer progressive vision loss, eventually leading to complete blindness. Currently, more than 300 genes have been identified as pathogenic genes (RetNet:https://sph.uth.edu/RetNet/). The emergence of cutting-edge treatments, such as gene therapy and stem cell transplantation,have brought hope to patients suffering from IRDs. However,the underlying mechanism how genetic defects contribute to IRDs in most of related genes is poorly understood, and this knowledge gap hinders the successful development and implementation of efficient therapy.
Prominin-1(also known asProm1, CD133, and AC133) is conserved across animal kingdom including human, mouse,zebrafish, drosophila, and caenorhabditis elegans, which is a transmembrane protein with 5-transmembrane domain,containing two large N-glycosylated extracellular loops,and a cytoplasmic tail[1]. It is usually identified as a surface antigen in human haematopoietic stem and progenitor cells[2-3].Prom1is selectively enriched in cholesterol based membrane microdomains and interacts with plasma membrane cholesterol[4-5]. Besides interaction with cholesterol,Prom1also involves in glucose metabolism and may affect cellular glucose through its impact on cytoskeleton[6].
Variants inProm1have been reported to be involved in wide spectrum of genetic disorders which can cause progressive degeneration of photoreceptor, including retinitis pigmentosa(RP)[7-11], cone-rod dystrophy[12-14], and Stargardt disease[15-16].This genetic overlap suggests basic similarities of the underlying pathogenic mechanisms and the common genetic pathways, despite different clinical phenotypes. Among these diseases, RP is the most common form of IRDs that affects about 2.5 million people worldwide[17]and the prevalence of RP has been reported approximately 1 in 4000[18]. For most of the RP cases, the degeneration of rods occur initially followed by subsequent loss of cones[19].
Previous studies have shown thatProm1distributed throughout the entire cone outer segment (OS) and basal part of rod OS in human, while located exclusively in the basal disks of cones and rods in the mouse[20].Prom1expression can be detected at birth, and deletion ofProm1will result in abnormal morphogenesis of photoreceptor OS[21]. In cervical loop epithelium stem cells,Prom1binds with arl13b and hdac6,and modulates ciliary dynamics through sonic hedgehog pathway[22]. Mutations in humanProm1can result in decreased ciliary length[23]. Furthermore, high concentration ofProm1was found in extracellular membrane vesicles, released by midbody and primary cilium[24]. These phenotypes and the potential localization ofProm1imply thatProm1may involve in the proper function of primary cilium. However, detailed function ofProm1in photoreceptor degeneration remains elusive.
The photoreceptor OS is a large specialized primary ciliumderived organelle, comprises of tightly stacked membrane discs[25], and filled with large amount of proteins necessary for the phototransduction cascade. The proteins in the OS are synthesized in the photoreceptor inner segment, and then trafficked to their default destination during OS morphogenesis and disks renewal[26-27]. The process is heavily depending on the integrity of cilium structure and proper function of ciliary protein, and the mutations impairing ciliary protein trafficking can cause photoreceptor degeneration[28-29]. The photoreceptor degeneration has been proven to be a key process of IRDs.
In this study, we generatedProm1knockout (Prom1-KO)mice by CRISPR/Cas9 technology. Using these models, we demonstrated thatProm1was required for maintaining normal homeostasis of OS proteins and deletion ofProm1can cause photoreceptor degeneration.
MATERIALS AND METHODS
Ethical ApprovalAll animal experiments were conducted with strict adherence to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.All animal experiments were approved by the Animal Research Committee at Shanghai Jiaotong University. All animal experimental procedures were approved by the Shanghai General Hospital Review Board.
CRISPR/Cas9 technology was used to generate Prom1-KO mice. Two target sites were chosen using the online tool (http://crispr.mit.edu/), both of which are located in the second exon of theProm1gene. After off-target rate prediction, target site with higher scores was picked. One knockout allele resulted in the deletion of 184 base pairs in the exon2 ofProm1gene.
After successful implementing the CRISPR/Cas9 strategy, the validated F0 mice were raised to adult and then crossed with wild-type (WT) mice to produce F1 generations. The F1 mice were further intercrossed to produce stable homozygotes F2.Homozygous males and females mated with each other to make F3 for research use.
Genotypes of all produced mice were confirmed by Sanger sequencing with the following primers:Prom1forward,5’-GCTTGGGTTCCACCTCGTTA-3’,Prom1reverse,5’-TTTGATACCCTCCTTAGGGCA-3’.
ElectroretinographyElectroretinography (ERG) was performed to assess retina function of Prom1-KO and agematched WT mice. The mice were pre-adapted overnight and all the procedures were performed in dim red light. After dark adaption, mice were anesthetized using a combination of ketamine and xylazine (16 mg and 80 mg/kg body weight respectively), pupils were dilated with 0.5% tropicamide. The response to a light flash stimulation (3.0 candela seconds/m2)was amplified. Cone-related ERGs were recorded 10min after complete light adaption. The baseline to maximum a-peak was recorded as a-wave amplitude. The b-wave amplitude was recorded as maximum a-peak to maximum b-peak. For each group, four Prom1-KO mice and four age-matched WT was tested.
Hematoxylin and Eosin Stain and Outer Nuclear Layer ThicknessFor hematoxylin and eosin (H&E) staining,mice were sacrificed and transcardially perfused with 4%paraformaldehyde (PFA) using a peristaltic pump (Masterflex L/S, USA). And then eyes were enucleated and fixed in 4%PFA for 4h before sinking in 30% sucrose solution overnight.The eyes were embedded in paraffin and cut into 4 μm slices.After deparaffinated, the sections were stained with H&E. The thickness of the outer nuclear layer (ONL) was measured at three evenly spaced locations on each side of the optic nerve headand analyzed by software Image J (NIH, USA).
Western Blot AnalysisThe whole retina dissected from WT or Prom1-KO mice were lysed in RIPA lysis buffer (P0013B,Beyotime) with inhibitors of protease and phosphatase (from Roche Inc). Then, the sample was sonicated and collected the supernatant after centrifuging. Protein quantification kit(BCA Assay; 23325, Thermo Fisher) was used to test the concentration of samples. Equal amount of sample (20 μg) was resolved by SDS-PAGE and transferred to PVDF membranes.Primary antibodies used for probing were shown in Table 1.An HRP-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies were used to bind with chemiluminescent substrates. The signal was detected by Amersham Imager 600(GE, USA).
Optical Coherence TomographyOptical coherence tomography (OCT) images were acquired from anesthetized mice using a Phoenix Research Lab systems (Phoenix, USA).Several B scans of retina was captured and averaged to produce final OCT images. Images tool was used to take ONL thickness measurement in OCT images.
Immunohistochemistry AnalysisThe eyes from Prom1-KO and WT mice were enucleated and fixed with 4% PFA in phosphate buffer saline (PBS) for 2h prior to removal of the lens and cornea. Then the eyes were soaked in 30% sucrose in PBS overnight for dehydration. Following dehydration, the eyes were placed in optimal cutting temperature and frozen in -80℃. Cryosection was performed by LEICA CM3050S into 10 μm sections. The sections were washed with PBS for three times and incubated with blocking buffer (PBS with 5% goat serum, 0.5% Tween20). After blocking, the retinal sections were incubated with primary antibody overnight at -4℃. Then, the sections were washed with PBS for three times before incubation with secondary antibodies for 2h in room temperature. The 4’,6-diamidino-2-phenylindole(DAPI) nuclear stain was used to label nuclei. The primary antibodies that were used as shown in Table 1. Besides those antibodies, Alexa Fluor 488 and Alexa Fluor 594 secondary antibodies and DAPI were also used in the experiments. The immunohistochemistry images were captured by confocal microscope (LEICA, USA).
RNA-seq AnalysisThe retinas from 10-day Prom1-KO and WT (5 per group) were dissected and soaked in liquid nitrogen. Samples were sent to Beijing Genomics Institution(BGI) China for further process. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and proteinprotein interaction analysis was generated through online bioinformatic software afforded by BGI China.
Statistical AnalysisStatistical significance was determined by Student’st-test using GraphPad Prism 7 software. Quantitative data are presented as the mean±SEM.
RESULTS
Generation of Prom1-KO Mice Using CRISPR/Cas9 TechnologyTo investigate the function ofProm1in vivo, we generated Prom1-KO mice by CRISPR/Cas9 technology. The mouseProm1cDNA and genomic sequences was downloaded from the NCBI database. Two target sites in exon2 ofProm1was selected (Figure 1A). Off-target rate prediction was performed by online tool (http://crispr.mit.edu/). The targetsite with higher score was picked. A frameshift deletion allele Mut was generated, which has the deletion of 184 base pairs(bp) in exon 2 ofProm1and confirmed the deletion by Sanger sequencing (Figure 1B). Mice carrying this deletion were backcrossed to C57BL/6J for three generations and then the offspring mice were intercrossed to generate homozygous Prom1-KO mice for further research. Prom1-KO mice were born at the expected Mendelian ratios and were healthy and fertile, with no apparent differences compared with control littermates. Immunoblot and immunofluorescence analysis(Figure 1C, 1D) using commercial antibody demonstrated the absence ofProm1. We have also compared the body weight of Prom1-KO and WT mice, and no significant changes observed.
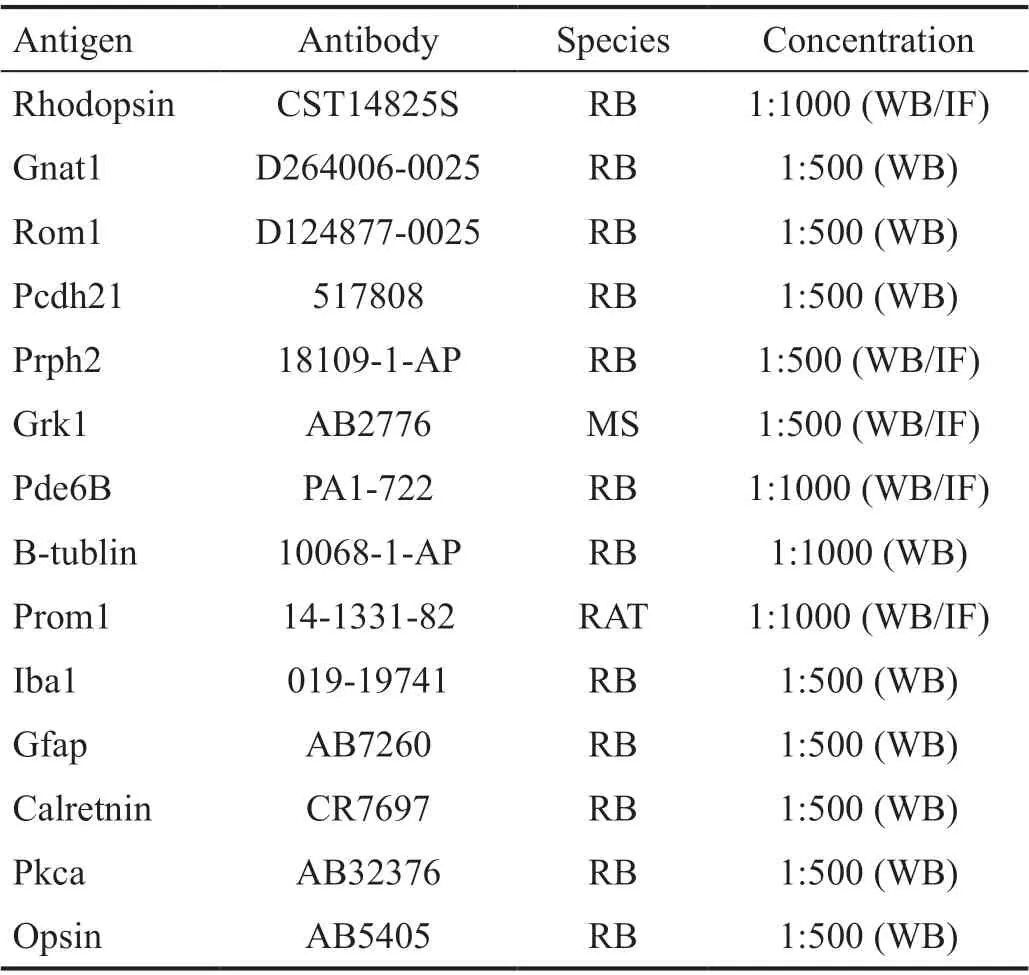
Table 1 Antibody used in the study
Progressive Photoreceptor Degeneration in Prom1-KO MiceTo investigate the severity of photoreceptor degeneration after Prom1-KO. We performed histological analysis and noninvasive OCT in Prom1-KO and WT mice. H&E staining was carried out on retinal cryosections. Retinal H&E images showed that the thickness of the ONL gradually decreased at 4,16, and 24 weeks of age (Figure 2A-2F). At 24-week Prom1-KO mice, the ONL thickness at the midperipheral retina was reduced by approximately 90% compared with WT (Figure 2C, 2F). OCT analysis showed similar results as H&E staining images. At 16 weeks of age, the average ONL thickness of Prom1-KO mice decreased by 47% (Figure 2G, 2I). At 24 weeks of age, further ONL thinning was observed in Prom1-KO mice (Figure 2H, 2J). At the midperipheral retina, the average ONL thickness was reduced by 84% compared with WT (Figure 2J). These results demonstrated that deletion ofProm1resulted in progressive photoreceptor degeneration.
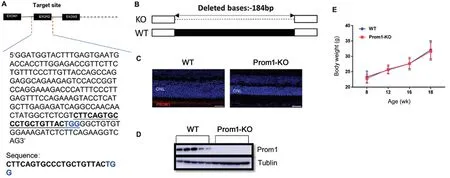
Figure 1 Generation of Prom1-KO mice A: Part of Prom1 exon2 sequence is shown. The gRNA target site is underlined. B: Sanger sequencing results comparing with WT (del 184bp); C: Immunofluorescence analysis of Prom1-KO and WT mice with commercial Prom1 antibody; D: Absence of Prom1 protein in Prom1-KO mice retina comparing with WT. β-tublin was used as a loading control. ONL: Outer nuclear layer. E: Body weight of Prom1-KO and WT mice. Data are presented as mean±SEM, n=6 for each group.

Figure 2 Prom1-KO mice show progressive photoreceptor degeneration A-C: Hematoxylin and eosin (H&E) staining of cryosections of the Prom1-KO and WT retinas at the ages of 4wk (A), 16wk (B), and 24wk (C). Scale bar, 20 μm. D-F: Quantification of ONL thickness of the Prom1-KO and WT retinas at the ages of 4wk (D), 16wk (E), and 24wk (F). G, H: Representative OCT images of Prom1-KO and WT retinas at the age of 16wk (G), and 24wk (H). I, J: Quantification of ONL thickness of Prom1-KO and WT mice retina at the ages of 16wk (I) and 24wk (J)through OCT images. n=4 for each group. Data are presented as mean±SEM. aP<0.05, bP<0.01. INL: Inner nuclear layer; ONL: Outer nuclear layer.
Reduced Photoreceptor Function in Prom1-KO MiceIn order to studying the photoreceptor function of mice withProm1deletion, we performed ERG in 8-week and 16-week Prom1-KO mice and age-matched WT mice under dark and light pre-adapting conditions. In the scotopic response tests, the mean a-wave and b-wave amplitudes of 8-week Prom1-KO mice were reduced by approximately 67% and 20%, respectively (Figure 3A-3D). With the progression of photoreceptor degeneration, the mean a-wave and b-wave amplitudes of 16-week Prom1-KO mice were reduced by approximately 88% and 83%, respectively (Figure 3E, 3G),indicating progressively impaired rod-cell function. Under photopic conditions, similar results were also observed, with a relatively flat wave compared with age-matched WT mice(Figure 3F, 3H), indicating that cone-cell function was also compromised due to degenerated photoreceptors.
Loss ofProm1Leads to Mistrafficking of Rod and Cone OpsinTo further study the pathological changes caused by Prom1-KO, we performed immunofluorescence analysis of retinal cryosections from WT and Prom1-KO mice. Rhodopsin is an indispensable protein for normal phototransduction,mislocalization of rhodopsin was shown to associated with retinal toxicity[30-31]. We observed mislocalized rhodopsin protein distributing in both the ONL and the inner nuclear layer (INL) of 2-week Prom1-KO mice retina (Figure 4A).However, no significant mislocalization of rhodopsin was observed in the 6-week Prom1-KO mice retina (Figure 4B).Interestingly, in the 24-week Prom1-KO mice retina that only one-layer ONL left, we observed overlapping rhodopsin with ONL (Figure 4C). Besides rhodopsin, we also stained cryosections with opsin and Alexa Fluor-488-tagged peanut agglutinin (PNA, which binds with cone matrix sheaths).Different with rhodopsin immunostaining results, there is no abnormal distribution of opsin protein in 2-week Prom1-KO mice (Figure 4D). While in 6-week Prom1-KO mice, as shown in Figure 4E, we observed mislocalized opsin in the ONL and the outer plexiform layer. Similar with rhodopsin immunostaining, overlapping opsin with ONL was observed in 24-week Prom1-KO mice retina (Figure 4F). In addition,the PNA-stained structure was significantly shorter in 8-week Prom1-KO mice (Figure 4G).
In order to investigate if the trafficking of other photoreceptor OS proteins is also impaired in the Prom1-KO mice, we performed immunofluorescence analysis on 8-week and 24-week Prom1-KO mice retina cryosection using antibodies against peripherin 2 (Prph2), Grk1, and Pde6β. Comparing with the WT, we found no evident mislocalization of these three proteins.
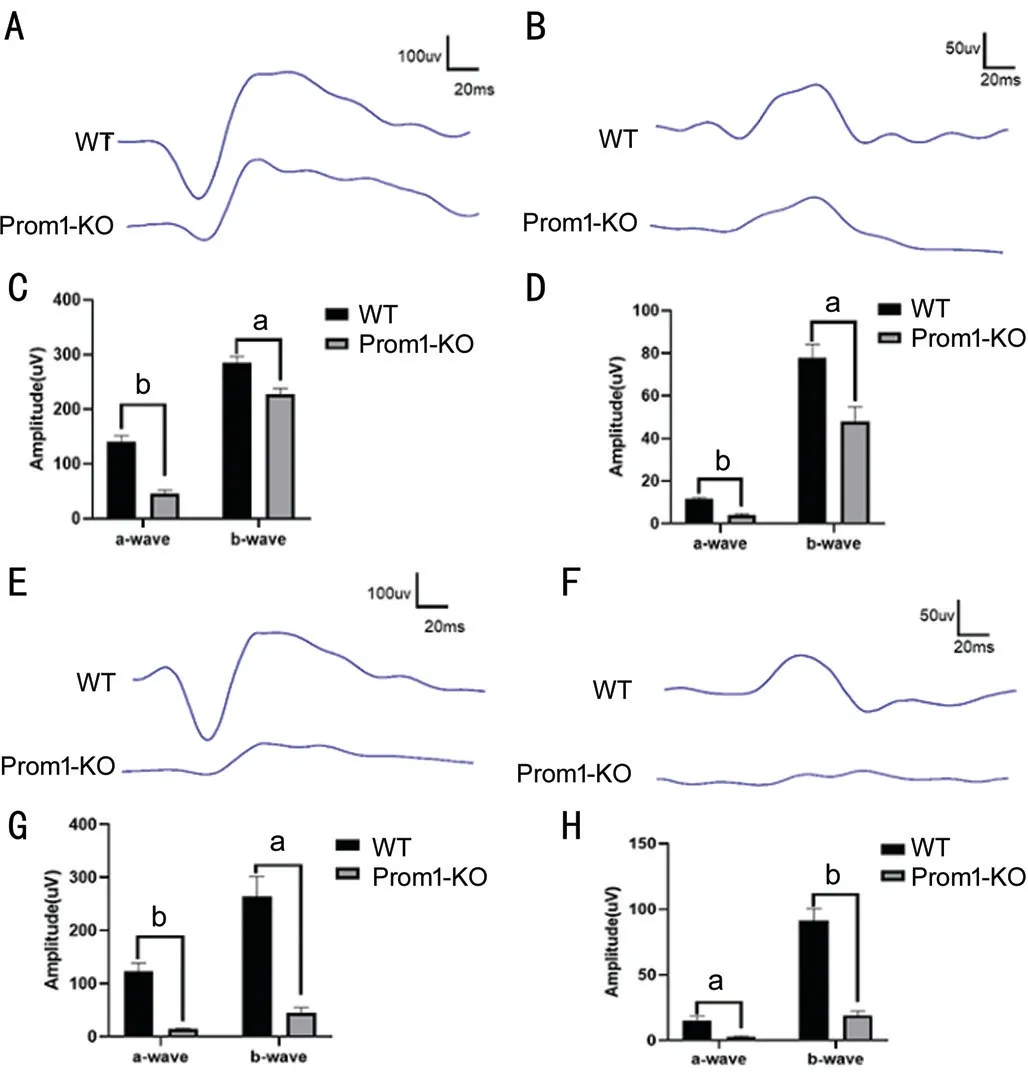
Figure 3 Prom1-KO mice have decreased scotopic and photopic ERG responses A, B: Representative scotopic and photopic ERG responses at 3.0 logcds/m2 for scotopic ERG (A) and 1.48 logcds/m2 for photopic ERG (B) from 8-week Prom1-KO and WT mice; C, D:Reduced scotopic (C) and photopic (D) ERG a-wave amplitudes,b-wave amplitudes of 8-week Prom1-KO mice compared with agematched WT; E, F: Representative scotopic and photopic ERG responses at 3.0 logcds/m2 for scotopic ERG (E) and 1.48 logcds/m2 for photopic ERG (F) from 16-week Prom1-KO and WT mice; G,H: Reduced scotopic (G) and photopic (H) ERG a-wave amplitudes,b-wave amplitudes of 16-week Prom1-KO mice compared with agematched WT. n=4 for each group. Data are presented as mean±SEM.aP<0.05, bP<0.01.
To investigate effect of Prom1-KO in other types of retina residential cell, we used the markers for microglial, Müller,bipolar, and horizontal cells (IBA1, GFAP, Pkcα, and calretinin respectively). As shown in Figure 4H, microglial was activated and migrated toward ONL in Prom1-KO mice retina. Dramatic increased expression of GFAP compared with WT (Figure 4I).Normal distribution and expression of bipolar and horizontal cells was found in Prom1-KO mice retina (Figure 4J, 4K).
These abovementioned results suggested thatProm1selectively participate in the trafficking of OS proteins and microglial and muller cells were involved in the Prom1-KO associated photoreceptor degeneration.
Reduction of Photoreceptor OS-Resident Proteins in the Absence ofProm1To investigate if the amount of photoreceptor OS proteins was also affected, we evaluated photoreceptor OS protein expression by semi-quantitative immunoblot analysis. We performed immunoblot analysis at 10-day mice retinas when there is no evident degeneration of photoreceptor OS. We observed the amount of photoreceptor OS proteins, including Prph2, retinal OS membrane protein 1(Rom1), Pde6β, rhodopsin, and protocadherin-21 (Pcdh21),was decreased by 22%-48% compared with WT (Figure 5A, 5B,Table 2). At 4-week mice retinas, semiquantitative immunoblot analysis yielded similar results (Figure 5C, 5D, Table 3). The extent of OS protein reduction varied between 47% and 66%, with Gnat1 and Grk1 being far more robustly reduced from 10 days of age (Tables 2 and 3).

Figure 4 Immunofluorescence staining analysis of the Prom1-KO mice retinas A-C: Retinal cryosections from 2-week (A), 6-week (B), and 24-week (C) old mice were labeled with rod specific markers rhodopsin (outer segment); D-F: Retinal cryosections from 2-week (D), 6-week(E), and 24-week (F) old mice were labeled with cone specific markers opsin (outer segment). G: Prom1-KO and WT mice retinas 8-week were immunostained with PNA marker; H, I: Staining for IBA1 (H) and GFAP (I) showed that microglial and Müller cells were activated in Prom1-KO mice retina; J, K: Staining for calretinin (J) and Pkcα (K) showed that amacrine and bipolar cells were relatively unaffected in Prom1-KO mice retina. Scale bars, 25 μm. INL: Inner nuclear layer; ONL: Outer nuclear layer.
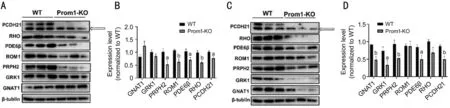
Figure 5 Reduced expression of outer segment proteins in Prom1-KO retinas A, C: Representative immunoblot images of Prom1-KO and WT at 10 days of age (A) and 4 weeks of age (C); B: Quantification of OS proteins in Prom1-KO and WT retinas at 10 days of age; D:Quantification of OS proteins in Prom1-KO and WT retinas at 4 weeks of age. The expression level of each protein was first normalized by its corresponding loading control, and then normalized by the mean value of the WT group. Data are presented as mean±SEM in the chart (B, D).aP<0.05, bP<0.01.

Figure 6 Transcriptomic analysis of 10-day Prom1-KO and WT mice retinas A: Heatmap of differentially expressed genes (DEGs); B, C:GO and KEGG enrichment analysis of DEGs; D: Protein-protein interaction analysis based on RNA-seq data.

Table 2 Quantification of OS proteins in Prom1-KO and WT retinas at 10 days of age mean±SEM
Taken together, the abovementioned results show that the expression of numerous OS proteins was reduced as a result of the Prom1-KO and the extent of this reduction vary among different proteins.
Changes in the Retinal Transcriptome upon Loss ofProm1To investigate the downstream molecular events brought on by the loss ofProm1, we analyzed the transcriptome of 10-dayProm1-KO and WT mice retina by RNA-seq (heatmap Figure 6A). GO analysis indicated that differentially expressed genes(DEGs) were closely related to biological process involved in regulation of lipid metabolic process and triglyceride metabolic process, and lipoprotein metabolic process (Figure 6B). KEGG pathway analysis indicated that these genes were strongly associated with the retinol metabolism pathway, the cholesterol pathway, and peroxisome proliforator-activated receptor (PPAR) signaling pathway (Figure 6C). Besides that,we also constructed a protein-protein interaction analysis based on the DEGs of Prom1-KO and WT groups (Figure 6D). However, we did not observe a strongly inter-connected network.
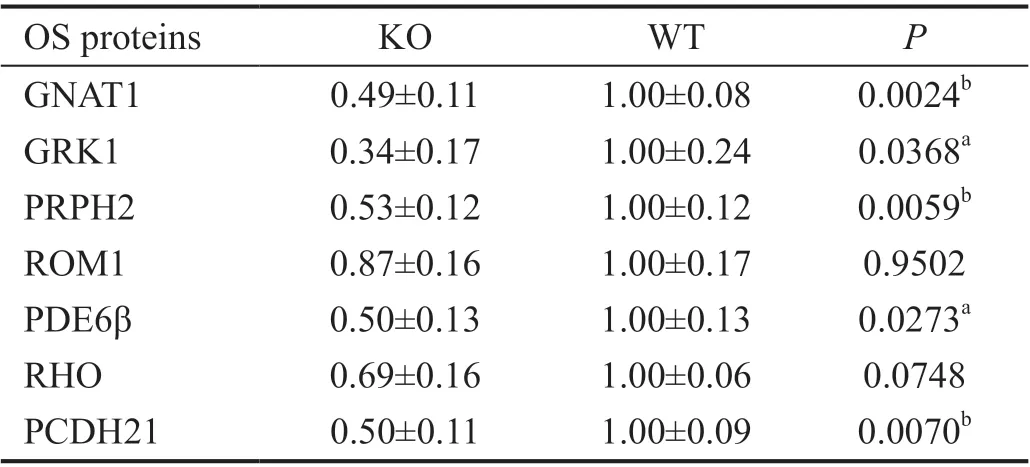
Table 3 Quantification of OS proteins in Prom1-KO and WT retinas at 4 weeks of age mean±SEM
These data imply that abnormal lipid metabolism may involve in the retinal degeneration process caused by Prom1-KO which is consistent with the known role of Prom1 in maintaining normal lipid regulation[32].
DISCUSSION
Prom1mutation is involved in various IRDs. Elucidating underlying pathophysiological mechanisms associated with genetic defects inProm1is a key to develop efficient therapeutic strategies for the related patients. In this study,we generated a Prom1-KO mice model by CRISPR/Cas9 technology. The Prom1-KO mice in this research exhibited retinal degeneration phenotype including decreased ONL thickness, compromised ERG amplitudes, and disrupted OS membrane discs, consistent with other Prom1-KO mice models used in previous study[21,33]. In our Prom1-KO mice, rhodopsin and cone opsin trafficking were impaired which resulted in mislocalization of these two essential phototransduction proteins. And we also found reduction of OS proteins in the absence ofProm1, suggesting thatProm1plays an important role in maintaining the normal amount of these proteins.
Mislocalization of OS proteins was treated as a typical sign of abnormal retinal protein trafficking. Proper localization of OS proteins required three crucial steps. First, protein-contained vesicles which was synthesized by Golgi fused with the ciliary base. Second, proteins are transported through a narrow transition zone to enter the axoneme. Last, the cargo proteins are transported from the axoneme to the default OS localization[34-35]. Mutations in genes involved in these steps have shown mislocalization of OS proteins[36-41]. Such as Ift172, which mediates bidirectional movement of cargo proteins along the axoneme, caused mislocalization of alpha subunit of transducin and rhodopsin after being knockout[37]. And Rpgr, which interacts with vital transition zone proteins including Cep290 and Nphp to regulate protein transportation in transition zone, caused mislocalization of rhodopsin after being knockout[41-42].However, the underlying mechanism of mislocalization of OS proteins followed by retinal degeneration is still elusive in such studies. In our study, we found mislocalization of opsin and rhodopsin. The initial mislocalization of rhodopsin was observed at 14-day Prom1-KO mice, while mislocalization of opsin occurred at 6 weeks of age. As the disease progressed,totally mislocalized rhodopsin and opsin were observed at the 24-week Prom1-KO mice. Interestingly, we also checked the distribution of Grk1, Pde6β, and Prph2, however, no evidence of mislocalization was observed even at 24 weeks of age for these three proteins. These results triggered our speculation that there were different OS protein trafficking systems for specific proteins and Prom1 was key part of one of these trafficking systems. But which step Prom1 was involved in and its multiple protein interactors participated in this process is still a subject that need to be addressed in future studies.
In our study, we also analyzed the expression level of seven OS proteins, all of which were associated with IRDs[43-48].Rhodopsin is the main structural component of photoreceptor OS disk. It worked with Grk1 and Pde6B, playing an important role in phototransduction cascade. Gnat1 and Pcdh21 were richly gathered in the OS disk membrane. Prph2 and Rom1,which can facilitate the folding of the OS discs, are crucial for OS biogenesis[49]. Reduction of these OS proteins was observed at 10 days of age when there was no evident sign of OS degeneration. Of the proteins with affected expression levels in Prom1-KO mice, rhodopsin is the main component of rod OS[50], and its protein level is positively related with OS volume. Our Tunel assay data showed there was no sign of photoreceptor apoptosis in Prom1-KO mice retina until age of 14d. This meant decreased OS volume started from 14 days of age. However, 39% reduction of rhodopsin was already observed in 10-day Prom1-KO mice. This result demonstrated the reduction would be associated with abnormal protein trafficking rather than decreased OS volume. At 4 weeks of age, robust reduction in the expression level of Gnat1 and Grk1 was observed, compared with relatively normal expression at 10 days of age. These results may imply that reduction of expression of these two proteins can be a consequence of decreased rhodopsin expression. Previous study using heterogeneous rhodopsin model which contained approximately 50% of the normal amount of rhodopsin[51-52],however, the Prph2 expression level was normal in their models, with shorter OS, and organized discs[51,53]. In our study, we observed significantly decreased Prph2 expression.The disparity of phenotypes between heterogenous rhodopsin mice and Prom1-KO mice further proved that the reduction of other OS proteins in the retina of Prom1-KO mice is not the consequence of reduction of rhodopsin, instead was a primary defect caused byProm1deficiency. Besides that, it was supposed the expression level of OS proteins decreased with continuous death of photoreceptor. However, for some yetunknown reasons, relatively unchanged or even increased OS proteins expression was reported in previous RP studies[28,54].Our study also showed the expression level of OS proteins were not strictly correlated with the OS volume. At 10 days of age, robust reduction of Rom1 expression was observed.But at 4 weeks of age, the expression level of Rom1 is quite closed between Prom1-KO and WT mice. We speculated that it was due to the following two reasons. First, mistrafficked Rom1 was not degraded, instead accumulated near the OS.Our Western-blot samples were collected from retina, so it was possible to detect the continuously accumulated Rom1.Second, there are compensatory mechanisms may restore the trafficking of Rom1 in the absence of Prom1.
Our GO and KEGG analysis data identified changes which are directly associated with lipid metabolism including regulation of lipid metabolic process, triglyceride metabolic process and the cholesterol pathway. And we also found changes of PPAR signaling pathway. When the PPARs were activated, it may down-regulate hepatic apolipoprotein C-III and increases lipoprotein lipase gene expression, and then regulate lipid metabolism in an indirect way[55-56]. Besides that, some studies had also showed the correlation between abnormal lipid level and RP[57-58]. Taken together, our RNA-seq data revealed dysregulated lipid metabolism was involved, at least partially,in the photoreceptor degeneration process caused by Prom1-KO. These identified up- or downregulated genes in response toProm1loss should prove quite meaningful for understanding the biological mechanism ofProm1function and for future studies.
In summary, we generated a Prom1-KO mice model by CRISPR/Cas9 technology. Progressive degeneration of photoreceptor and disrupted photoreceptor OS were observed.We demonstrated thatProm1was required for proper localization of rhodopsin and opsin in the retina, and played an essential role in maintaining normal amount of OS proteins.The abnormal lipid metabolism was involved in the process of retinal degeneration caused by Prom1-KO.
ACKNOWLEDGEMENTS
The authors want to give special thanks to Dr. Mengxi Shen(Bascom Palmer Eye Institute, USA) for her kind help in revising the article.
Foundations:Supported by the National Natural Science Foundation of China (No.81730026); the National Key R&D Program (No.2017YFA0105301; No.2019ZX09301113);the Science and Technology Commission of Shanghai Municipality (No.17411953000).
Conflicts of Interest: Xiao YS,None;Liang J,None;Gao M,None;Sun JR,None;Liu Y,None;Chen JQ,None;Zhao XH,None;Wang YM,None;Chen YH,None;Wang YW,
None;Wan XL,None;Luo XT,None;Sun XD,None.
杂志排行
International Journal of Ophthalmology的其它文章
- Five-year results of refractive outcomes and visionrelated quality of life after SMlLE for the correction of high myopia
- Role of glycolysis in retinal vascular endothelium, glia,pigment epithelium, and photoreceptor cells and as therapeutic targets for related retinal diseases
- Predictive value of retinal function by the Purkinje test in patients scheduled for cataract surgery in Kinshasa, DR Congo
- Simultaneous pars plana vitrectomy, panretinal photocoagulation, cryotherapy, and Ahmed valve implantation for neovascular glaucoma
- Displacement of the retina after idiopathic macular hole surgery with different internal limiting membrane peeling patterns
- Analogs of microgravity: the function of Schlemm’s canal, intraocular pressure and autonomic nervous during the head-down tilt test in healthy subjects
