Corydalis Rhizoma as a model for herb-derived trace metabolites exploration:A cross-mapping strategy involving multiple doses and samples
2021-07-20ChnjunYuFengyunWngXinyueLiuJiynMioSiqiTngQinJingXudongTngXioynGo
Chnjun Yu,Fengyun Wng,Xinyue Liu,Jiyn Mio,Siqi Tng,Qin Jing,Xudong Tng,Xioyn Go,*
aSchool of Chinese Materia Medica,Beijing University of Chinese Medicine,Beijing,102488,PR China
bGastroenterology Department,Xiyuan Hospital,China Academy of Chinese Medical Sciences,Beijing,100091,PR China
Keywords:
Alkaloid
Characteristic fragment Corydalis yanhusuo
In vivo metabolism
Metabolite
A B S T R A C T
Deciphering the metabolites of multiple components in herbal medicine has far-reaching significance for revealing pharmacodynamic ingredients.However,most chemical components of herbal medicine are secondary metabolites with low content whose in vivo metabolites are close to trace amounts,making it difficult to achieve comprehensive detection and identification.In this paper,an efficient strategy was proposed:herb-derived metabolites were predicted according to the structural characteristics and metabolic reactions of chemical constituents in Corydalis Rhizoma and chemical structure screening tables for metabolites were conducted.The fragmentation patterns were summarized from representative standards combining with specific cleavage behaviors to deduce structures of metabolites.Ion abundance plays an important role in compound identification,and high ion abundance can improve identification accuracy.The types of metabolites in different biological samples were very similar,but their ion abundance might be different.Therefore,for trace metabolites in biological samples,we used the following two methods to process:metabolites of high dose herbal extract were analyzed to characterize those of clinical dose herbal extracts in the same biological samples;cross-mapping of different biological samples was applied to identify trace metabolites based on the fact that a metabolite has different ion abundance in different biological samples.Compared with not using this strategy,44 more metabolites of clinical dose herbal extract were detected.This study improved the depth,breadth,and accuracy of current methods for herb-derived metabolites characterization.
1.Introduction
It is generally believed that the components of drugs absorbed into the body and related metabolites are the effective material basis for disease treatment[1-3].Compounds in herbal medicines that vary significantly in structural types and contents lead to more complex metabolome after metabolism in vivo.Moreover,the matrix of biological samples is complex,and some high-content or high-response endogenous substances may interact with drug metabolites,resulting in ion inhibition or enhancement,which interferes with the detection of herb-derived metabolites.All these factors pose a serious challenge for global detection and accurate identification of in vivo herb-derived metabolites.
In view of difficulties mentioned above in metabolic studies,more and more techniques have been employed to detect and characterize in vivo metabolites.With a view to improving detection sensitivity,some techniques based on mass spectrometry(MS)data such as mass defect filter[4],background subtraction[4,5],characteristic ions filtering and neutral loss filtering[6-8]were proposed.But these techniques might miss metabolites with inconspicuous chromatographic peaks whose MS/MS information is missing or submerged at the baseline.As a complement,multivariate statistical analysis[9]was applied to ensure the comprehensiveness of metabolite detection.The low sensitivity limited its application.
As for structure characterization accuracy,the structures of metabolites were only Confirmed by comparison with reference standards in previous reports.The other metabolites without standards were characterized by metabolic pathways and fragmentation patterns,and their structures were represented by the parent drugs and related metabolic reactions.
There is an urgent need for an effective method for detection and identification of metabolites.Therefore,in this study crossmapping of different doses and samples was used for trace metabolites detection.The chemical structure screening tables and characteristic fragments were combined to enhance the accuracy of metabolite structure identification.
The dry rhizoma of Corydalis yanhusuo W.T.Wang(Papaveraceae)(Yanhusuo in Chinese)has the effect of promoting blood circulation,promoting qi and relieving pain[10].Due to its strong physiological effects,more and more research on Yanhusuo has been carried out in recent years.Previous studies only showed metabolic pathways and metabolites of certain components like tetrahydropalmatine[11,12],corydaline[13,14],dehydrocorydalis[15]and palmatine[16].Few studies focused on the comprehensive metabolic pro file of Yanhusuo alkaloids,and metabolites were not fully discovered because of the inherent limitation of analytical methods[17].Through preliminary study of chemical constituents,we had a comprehensive understanding for Yanhusuo alkaloids[18].However,the pharmacodynamic constituents of Yanhusuo are still not clear.
Herein,Yanhusuo was selected as the model herb to establish a strategy for herb--derived metabolites characterization in this study.The strategy mainly included the following steps.First,representative authentic standards of homologous components contained in Yanhusuo were chosen,from which the fragmentation patterns were proposed.Second,the metabolic pathways and metabolic characteristic fragments(MCFs)of each family of compounds were acquired through metabolic analysis of standards.Third,the structure screening tables established on the basis of the structural characteristics and metabolic pathways of compounds in Yanhusuo were combined with fragmentation patterns and MS information to characterize structures of metabolites.Then,metabolites of highdose extract were used as pseudo-standard to identify those of clinical dose so as to identify metabolites with low ion abundance in the same biological samples.Finally,cross-mapping of different biological samples was applied to identify trace metabolites in different samples.
A total of 127 herb-derived metabolites were identified in biological samples after the clinical dose Yanhusuo administration.For the first time,the metabolite structures of four standards representing Yanhusuo alkaloids were clearly characterized,and metabolic pathways were proposed based on this.The strategy provided a reference for pharmacodynamic substances discovery in different biological samples and metabolism mechanism of TCMs.The specific work flow is shown in fig.1.
2.Materials and methods
2.1.Chemicals and reagents
Herbal materials of Yanhusuo were purchased from Beijing Tong Ren Tang Group Co.,Ltd.(Beijing,China)and identified by Professor Yaojun Yang,School of Chinese Materia Medica,Beijing University of Chinese Medicine.The voucher specimens were deposited in School of Chinese Materia Medica,Beijing University of Chinese Medicine, China. Protopine, tetrahydropalmatine, tetrahydroberberine and palmatine with purity of above 98% were obtained from Chengdu Biopurify Phytochemicals Ltd.(Chengdu,China).Liquid chromatography coupled with mass spectrometry(LC-MS)grade acetonitrile,methanol,and formic acid were purchased from Fisher Scientific(Fisher,Fair Lawn,NJ,USA).Distilled water was available from the A.S.Watson Group Ltd.(Hong Kong,China).
2.2.Animals
Male Sprague-Dawley rats((200±10)g)were purchased from Beijing Weitong Lihua Biotechnology Co.,Ltd.(Beijing,China).All the rats were kept under controlled environmental conditions(a 12 h light/12 h dark cycle,a temperature of(20±2)°C and humidity of(60±5)% with free access to food and water for three days before the experiments.All animal experiments were approved by the Animal Ethics Committee of Beijing University of Chinese Medicine(Beijing,China),and all procedures were performed in accordance with the Helsinki Declaration.
Then rats were randomly divided into 3 groups:the feces and urine group(n=12),the plasma group(n=12)and the bile group(n=14).The rats in the feces and urine group were divided into 6 groups and were orally administered four reference standards(2.00 mL),clinical dose(0.56 mL)and high dose(1.20mL)of Yanhusuo extract,respectively.The feces and urine samples of 12 rats were collected for 24h before and after administration,respectively.The urine samples were centrifuged at 12,000rpm for 15min and the supernatant was frozen in a refrigerator at-80°C.The feces samples were placed in a fume hood for drying.Then dried feces were crushed and stored in a centrifuge tube at-80°C.The other two groups used the same drugs and method of administration,except for the bile group,which had two more rats for blank control.Blood samples of 0.5 mL were collected via the ophthalmic veins into heparinized tubes at 0 and 5,10,30,45,60,120,180,240,360,480,600,720 min after the administration.The samples were centrifuged at 3,500rpm and 4°C for 15 min.Rats in the bile group were anesthetized by 10% chloral hydrate for bile duct intubation.The bile samples were collected for 0-12 h and 12-24h,and stored at-80°C.
2.3.Sample preparation
2.3.1.Sample preparation of Yanhusuo extract and standards
The standard solutions of four reference standards were prepared in 0.5% aqueous solution of sodium carboxymethyl cellulose at a concentration of 2mg/mL.Intragastrical administration dose of each standard solution was 20 mg/kg,i.e.,2mL per rat.
Herbal materials of Yanhusuo were powdered by a grinder and sieved through a 40-mesh sieve.About 20 g of powder was soaked in 200mL of distilled water for 1 h and decocted twice by boiling,each time for 30 min.Then,extracting solution was merged and drained with a water bath.The residue was dissolved in 40 mL of distilled water and Configured to be a concentration which was equivalent to 0.5 g of herbal materials(The oral clinical dosage of Yanhusuo in the Chinese Pharmacopoeia is 3-10g for per person.So,the clinical dosage of Yanhusuo is 1.4 g/kg,which is 0.28g per rat(0.56 mL).The high dosage was 3g/kg,which was 0.6g per rat(1.2 mL)).The extract was stored at-4°C.
2.3.2.Blood,urine and bile samples preparation
The frozen samples were dissolved at room temperature.200μL of plasma sample from each time point was mixed.An appropriate amount of mixed sample(100μL for plasma sample,1 mL for urine sample and 150μL for bile sample)was mixed with three times amount of methanol in polypropylene test tube.Then the mixture was vortex-mixed for 30 s and centrifuged at 12,000 rpm and 4°C for 10 min to remove the precipitated protein.The supernatants were dried with nitrogen.The residue was re-dissolved with 100μL of 70% methanol and then centrifuged at 12,000rpm and 4°C for 15 min.The supernatants were subjected to ultra-performance liquid chromatography coupled with quadrupole time of flight tandem mass spectrometry(UPLC-Q-TOF/MS)system.
2.3.3.Feces samples preparation
200mg feces were vortexed in 4 mL of methanol for 30 s and placed for1h.Then the suspensions were centrifuged at 12,000 rpm and 4°C for 15min.The supernatants were dried with nitrogen.The residue was re-dissolved with methanol,and then centrifuged at 12,000rpm and 4°C for 15min.The supernatants were subjected to UPLC-Q-TOF/MS system.
2.4.Instrumentation and conditions
The LC analysis was carried out on Waters ACQUITYTM UPLC IClass system(Waters,Milford,MA,USA)consisting of a binary solvent system,an autosampler and a column temperature controller.The system was operated under MassLynx V4.1 software(Waters,Milford,MA,USA).The chromatographic separation was carried out on an ACQUITY BEH C18column(2.1mm×100mm,1.7μm,Waters,Ireland).The mobile phase was composed of eluent A(0.1% formic acid in water)and eluent B(acetonitrile).The line gradient program was optimized as follows:0-2 min,1%-10%B;2-15min,10%-20%B;15-22 min,20%-30%B;22-27 min,30%-90% B;27-29 min,90% B;29-29.1min,90%-2% B;and 29.1-31min,2%-2%B.The column temperature was set at 40°C.The sample chamber temperature was set at 4°C.The mobile phase flow rate was set at 0.3 mL/min,and the injection volume was 2μL for each run.
The MS analysis was carried out by a Waters SYNAPT G2-SI MS system(Waters,Manchester,UK)equipped with electrospray ionization.The analysis was performed in the positive ion electrospray mode,and the source parameters were set as follows:the capillary voltage was 3.0 kV and the cone voltage was set at 40 V;the source temperature was 100°C,and the desolvation gas temperature was 400°C;the cone gas flow was 50 L/h,and the desolvation gas flow was 800L/h;the low collision energy was 6eV,and the high collision energy was 10-40 eV.The scan range was m/z 100-1200Da,and the 3D data were collected in the continuum mode.The leucine-enkephalin(m/z 556.2771 in positive ion mode)was invoked as the lock mass solution to ensure accurate mass measurement.The MS data were acquired by Waters MassLynx V4.1 software and processed by UNIFI™1.8 software(Waters,Milford,MA,USA).
3.Results
3.1.Fragmentation patterns of parent compounds
Generally,the parent drug and related metabolites have the same skeleton,and their fragmentation patterns are similar.In order to make better use of LC-MS technology to analyze metabolites,it is necessary to fully understand the fragmentation behaviors of parent compounds.
The Yanhusuo alkaloids can be mainly classified into the following four types: tetrahydroprotoberberine-type,protoberberine-type,protopine-type and aporphine-type.Tetrahydroberberine and tetrahydropalmatine easily underwent retrodiels alder(RDA)cleavage with complementary fragment ions in MS,since the C-ring was saturated(Figs.S1-S2).Among them,nitrogen-containing fragment ion had the highest abundance.The structure of protopine was similar to that of tetrahydroberberine,which was prone to RDA cleavage(Fig.S3).The fragment ion formed by further dehydration after RDA cleavage was the most abundant.The C-ring of palmatine alkaloid was not saturated,so it was difficult to experience RDA cleavage(Fig.S4).When the substituents contained two or more methoxy groups,the characteristic fragment ions were[M-CH3]+,[M-CH4]+,[M-CH4-CO]+,[M-2CH3]+and[M-CH3OH]+.Owing to the lack of the corresponding reference standards,the fragmentation pattern of aporphine-type alkaloids is summed up by related literature to determine the characteristic fragments[18,19].A weak bond existed in the amide group,so aporphine-type alkaloids very easily lost(CH3)2NH or CH3NH2substituents to produce the most abundant fragment of[M-45.0578]+or[M-31.0344]+as the characteristic ions(Fig.S5).

Fig.1.Summary diagram of proposed analytical strategy for identification of Yanhusuo alkaloids metabolites.

Fig.2.Proposed metabolic pathway of tetrahydroberberine(A),tetrahydropalmatine(B),palmatine(C),and protopine(D).
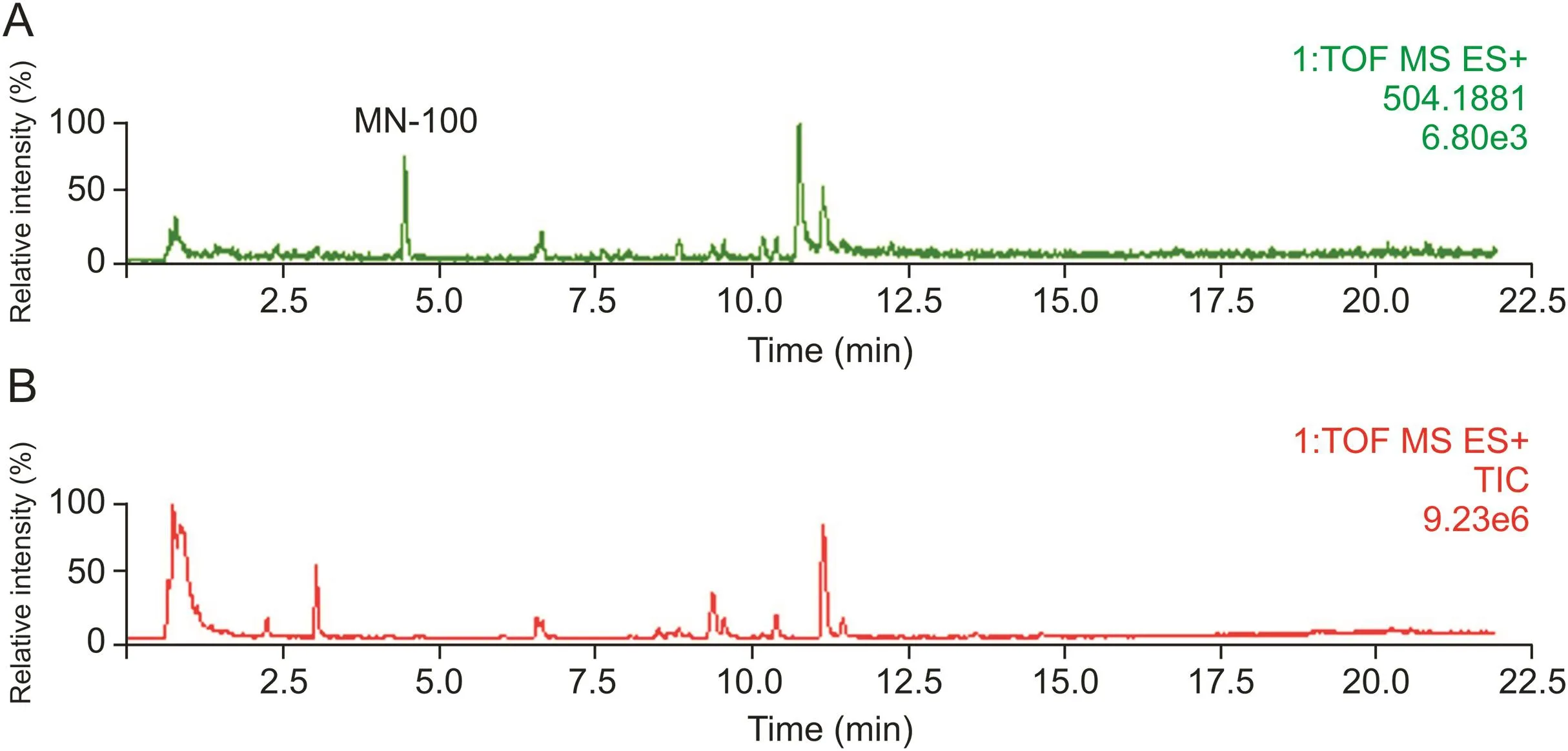
Fig.3.The chromatograms of clinical Yanhusuo extract administration plasma sample:(A)the extract ion chromatogram of m/z 504.1881,and(B)the total ion chromatogram of plasma sample.
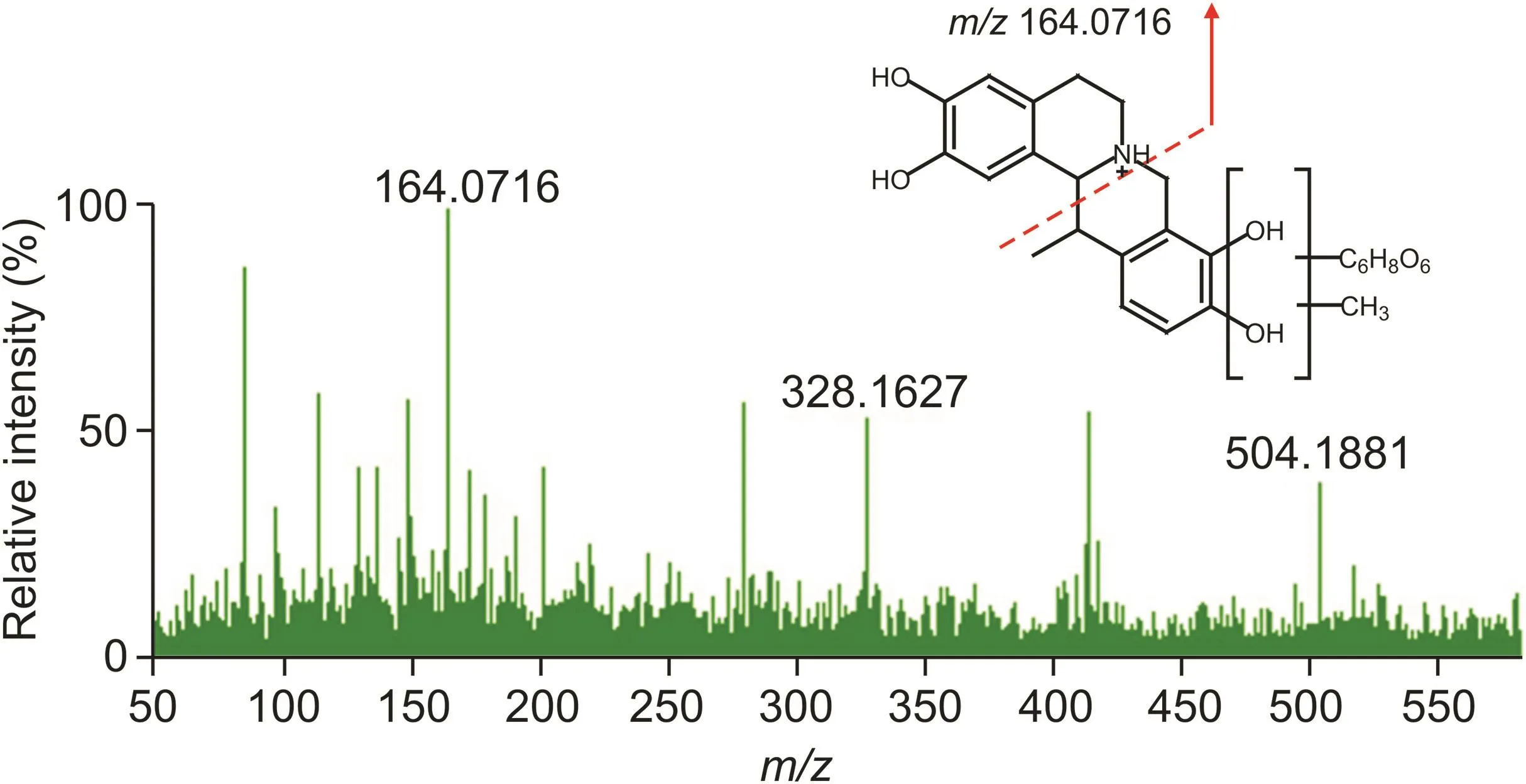
Fig.4.The MS/MS spectra and structure of MN-100.

Fig.5.The extract ion chromatograms(A)and MS/MS spectra(B)of m/z 518.2028 in plasma samples of clinical dose and high dose extracts.
3.2.Study on the metabolism of representative compounds
In order to study metabolites of various components in Corydalis comprehensively and systematically,we selected four representative standards,namely,tetrahydroberberine,tetrahydropalmatine,protopine,and palmatine.The metabolites of each compound were identified based on fragmentation patterns of parent compounds and metabolic pathways summarized from the literature.The metabolic pathways are shown in fig.2.As illustrated in this figure,the possible structures of all metabolites were clearly shown,which were not present in previous reports.Compared with those in the literature[11,12,16],new metabolic pathways such as dehydrogenation of tetrahydroberberine and new metabolites like M1-13,M3-7,etc.were also found in this study by the proposed strategy.All metabolites of four representative compounds are listed in Tables S1-S4.Based on the MS information and metabolic pathways summarized from standards,MCFs were proposed to characterize metabolites of Yanhusuo extract(Table 1).
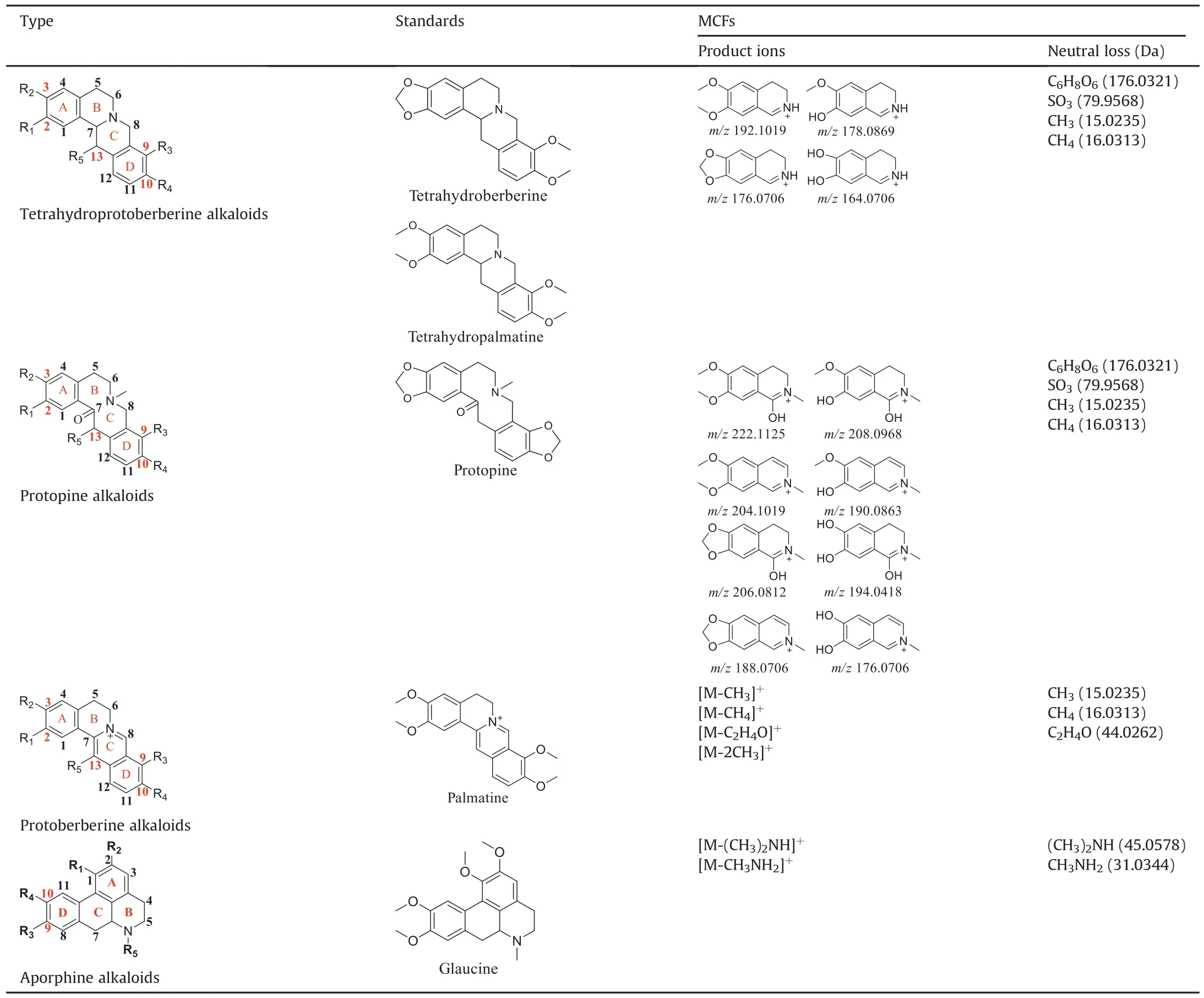
Table 1 The metabolic characteristic fragments(MCFs)of Yanhusuo-derived metabolites.
3.3.Establishment of chemical structure screening tables
Notably,it was found that the skeleton of Yanhusuo alkaloids did not change in metabolism.So,the chemical structure screening tables(Tables 2-4)were conducted according to the type and number of substitutes and in vivo metabolic pathways to offerreferences for structure identification.
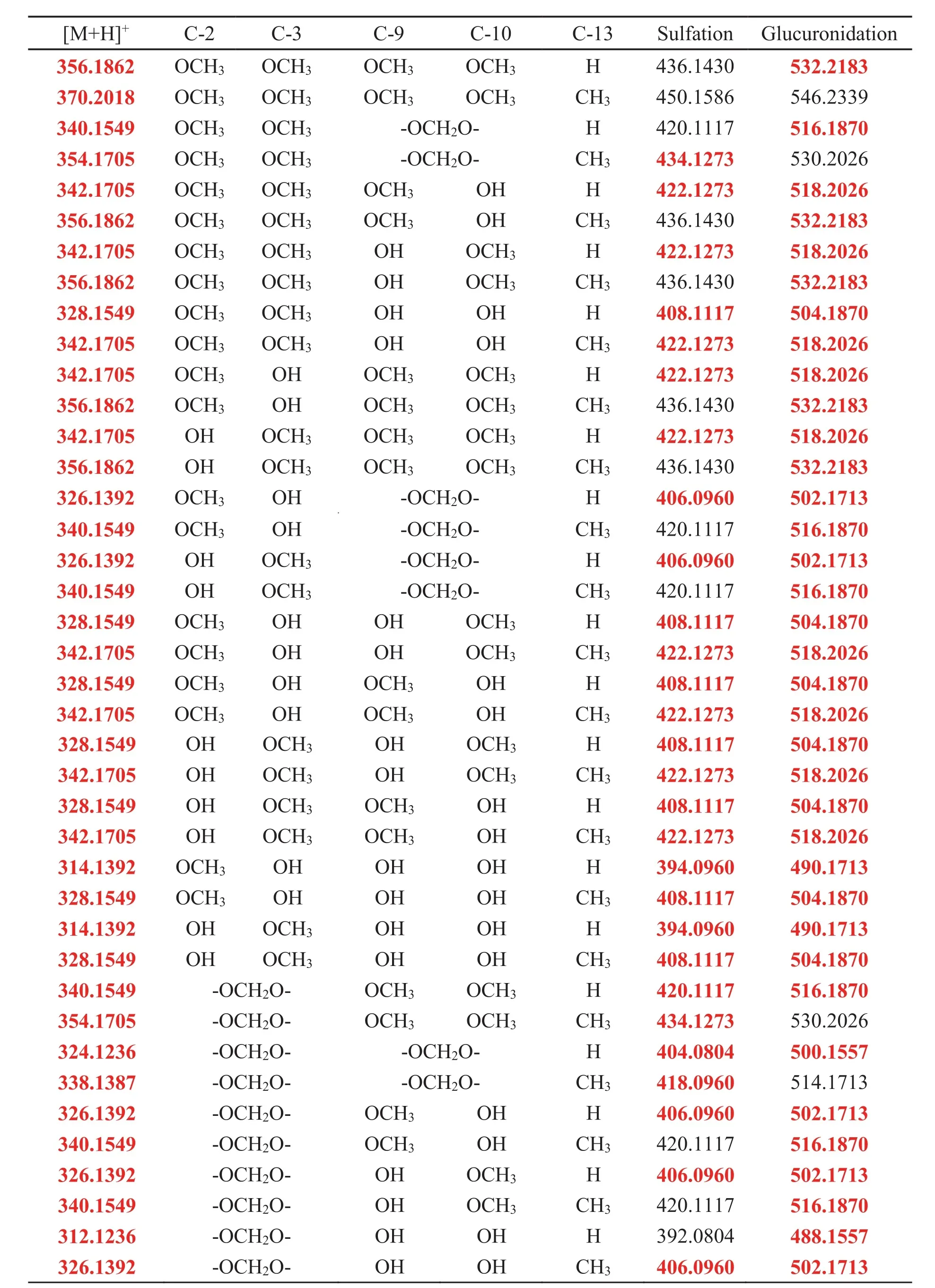
Table 2 The chemical structure screening table of tetrahydroprotoberberine alkaloids.

Table 3 The chemical structure screening table of protopine alkaloids.

Table 4 The chemical structure screening table of protoberberine alkaloids.
3.4.Global identification of Yanhusuo-derived compounds
3.4.1.Identification of metabolites detected in preliminary screening
Metabolites with relatively strong MS responses could be detected in preliminary screening based on metabolic characteristic fragments and metabolic pathways.The identification process was illustrated by taking metabolite MN-100 in plasma sample of clinical dose Yanhusuo extract as examples(Fig.3).The extract ion chromatograms(EIC)of MN-100 is shown in fig.3A,and Fig.3B is the total ion chromatogram of plasma sample.In positive mode,the quasi-molecular ion was m/z 504.1881[M+H]+and the chemical formula was C25H29NO10.The fragment ion m/z 328.1548[M+H]+suggested that the compound was a glucuronidated metabolite.The theoretical quasi-molecular ion m/z 504.1870 was retrieved in the chemical structure screening tables,corresponding to seven designed metabolite structures(Table 2).These seven metabolites were all tetrahydroprotoberberine-type alkaloids with two or more hydroxyl groups in the structure.The most abundant fragment ion m/z 164.0716 indicated the presence of two hydroxyl substitutions on the nitrogen-containing fragment ion produced by RDA cleavage.Therefore,we concluded that the compound had the same skeleton as tetrahydroberberine,with two hydroxyl substitutions at the C2-C3 position,a methyl substitution at the C13 position,and a glucuronic acid and a methoxy group at the C9-C10 position.The MS/MS spectra and structure of MN-100 are shown in fig.4.
3.4.2.Identification of metabolites with weak MS responses in the same biological samples from high dose to clinical dose
Some chromatographic peaks were missed in the preliminary screening due to low abundance,poor peak shapes,or inability to infer structures for lack of MS/MS information.In view of this,we proposed a method using metabolites of high dose Yanhusuo extract as pseudo-standards to identify those of clinical dose.The metabolite MN-106 in fig.5A was not detected in preliminary screening due to low abundance and poor peak shape.However,at the same retention time,the metabolite MH-119 in the high dose plasma sample was identified as methylated and glucuronidated tetrahydropalmatine.The EICs of m/z 518.2028 in plasma samples of clinical dose and high dose extract are shown in fig.5A.It can be seen from the figure that the response of the compound in sample of clinical dose extract was quite low.Partial amplification showed two peaks had the same chromatographic retention behavior.In the MS/MS spectra(Fig.5B),fragment ions in sample of clinical dose extract had a lower response value,almost submerged in the baseline,but it had the same fragmentation pattern as in sample of high dose,which indicated that MH-119 also existed in sample of clinical dose extract.
3.4.3.Identification of trace metabolites in the different samples by cross-mapping of different biological samples
The metabolites between the same biological samples obtained at different doses were almost completely coincident,so this method facilitated the identification of metabolites in the same samples.However,biological sample collection and data analysis of different administration doses required a large amount of repetitive work.What's more,high dose Yanhusuo extract did not enhance the MS response intensity of all metabolites.In the data analysis process,we found that there were many overlapping metabolites in different biological samples,while some samples,such as urine and bile,had many kinds of metabolites with high abundance.Thus,metabolites with strong abundance from one sample as pseudostandards were mapped to all the other three samples correspondingly by retention time and m/z value,so trace metabolites in the different samples could be identified.Taking the metabolites MN-28 and MN-29 in plasma samples of clinical dose Yanhusuo as an example,this strategy was illustrated.These two metabolites were not detected in high dose plasma samples during the preliminary screening.As shown in figs.6A and B,the MS responses of these two peaks in plasma samples with different doses were very weak,and one peak was even submerged in the baseline.The method from high dose to clinical dose could not identify them from plasma samples,while six metabolites with quasi-molecular ions of m/z 342.1715[M+H]+were detected in urine sample.In the preliminary screening results of urine sample,the peaks with retention time of 9.05 min and 9.83min were identified as hydrogenated tetrahydroberberine.Fragment ion at m/z178.0859[M+H-C10H12O2]+suggested that the structures of the compounds were similar to those of tetrahydroberberine and the hydrogenated reaction occurred at Aor B ring.The theoretical quasi-molecular ion m/z 342.1705 was searched in the chemical structure screening tables,and the possible structures were found to be three methoxyl groups and one hydroxyl group at C-2,C-3,C-9,and C-10 of the tetrahydroberberine skeleton.Therefore,hydrogenation occured on the A ring.The structures and MS/MS spectra are shown in fig.7.The ion of m/z 342.1715 was extracted from the plasma sample,as shown in figs.6A and B,to obtain four chromatographic peaks with the same m/z values at the same retention time as in urine(Fig.6C).The mass spectrum of metabolite MH-32 could be amplified locally to find the same fragment ions as those in urine.The other three chromatographic peaks lacked fragment ions but had the same quasi-molecular ions at the same retention time as metabolites in urine and were also identified as metabolites of hydrogenated tetrahydroberberine.The metabolites detected in rat biological samples after oral administration of clinical dose and high dose Yanhusuo extracts can be found in Tables S5-S6.Among them,127 herb-derived metabolites were identified in rat biological samples after oral administration of clinical dose.

Fig.6.The extract ion chromatograms of m/z 342.1715:(A)clinical dose plasma sample,(B)high dose plasma sample,and(C)clinical dose urine sample.
3.4.4.The relationship between structure and metabolism of Yanhusuo alkaloids
In the study of four standards,we found that the structures of Yanhusuo alkaloids were closely related to metabolism.The skeleton of alkaloids did not change during metabolizing.The structure contains many methoxyl groups.Therefore,demethylation was the most common and frequent metabolic reaction.It can be seen from Fig.8 that palmatine was the most susceptible one to have demethylation at methoxy substitution,followed by tetrahydropalmatine,tetrahydroberberine,and protopine,while the methylation reaction was reversed.The remaining hydroxyl groups were prone to have sulfation and glucuronidation.Both glucuronidation and sulfation reactions occurred only once and did not occur simultaneously.Only two metabolites were detected to be sulfated twice.The component containing methylenedioxy group would lose one molecule of CO,and the one that did not contain the substituent would not occur the cleavage,as shown in the case of tetrahydropalmatine and palmatine.Five-membered oxygen-containing ring and the carbonyl group of protopine alkaloids were easy to be hydrogenated.The former opened into a methoxyl group and a hydroxyl group,and the latter became a hydroxyl group.The hydroxyl group was unstable and dehydroxylation occurred easily.Dihydroxylation did not happen in other structural types.Protoberberine-type alkaloids did not undergo RDA cleavage due to the unsaturated C ring.So,the structure was difficult to determine and was very stable during the process of metabolism.Palmatine only underwent demethylation and hydroxylation reactions,and metabolites were mainly distributed in feces rather than in bile,suggesting that it might be metabolized by the intestinal tract rather than the liver.

Fig.7.The MS/MS spectra and structures of MN-28 and MN-29.
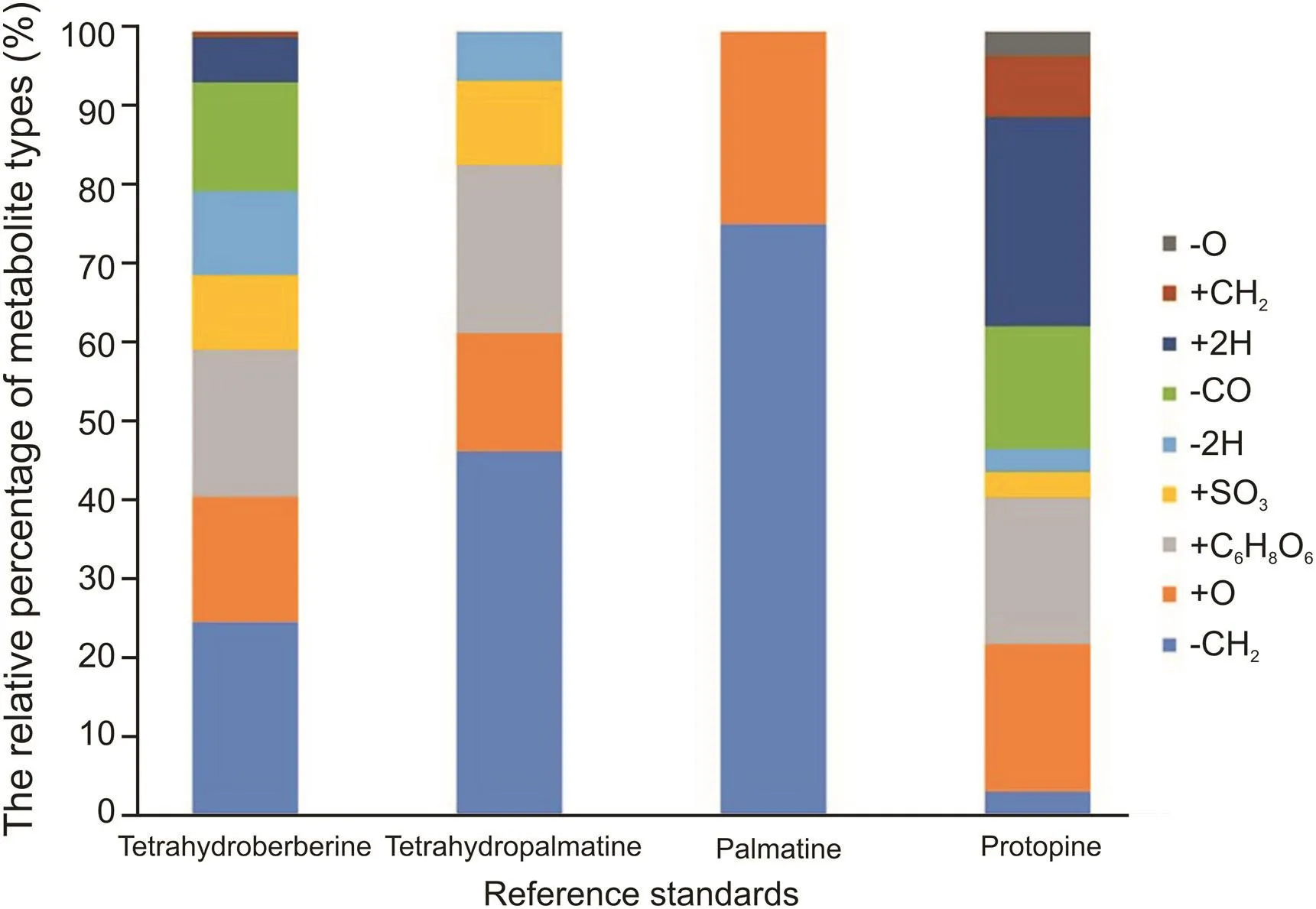
Fig.8.The relative percentage of metabolite types in four reference standards.
4.Discussion
The in vivo metabolic process of herbal medicine is very complicated,which makes it difficult to explain the mechanism.First,the structures of herbal medicine metabolites were diverse.Compared with previous chemical composition studies in vitro,we found that even though sulfated and glucuronidated metabolites were removed,the structure types of metabolites were more various than that of chemical constituents.In addition,there were still many metabolite isomers,some of which could be distinguished by characteristic fragment ions or ClogP values,but position-isomers or stereo-isomers could not be distinguished by LC-MS data,such as metabolites M4-17,M4-18,M4-19 and M4-20,indicating that more effective methods need to be developed for isomer identification.Second,the metabolic process was complex.Take M4-1 and M4-2 as examples.They were identified as the products of demethylation and dehydroxylation of protopine.This judgment only reflected the final result of element composition change rather than the process.The results of structural identification are shown in fig.S5.Protopine might experience more complex reactions to produce the metabolites.So,clarifying the structures of metabolites is of great importance for demonstrating the mechanism by which drugs work.Although this study proposed more realistic metabolic pathways based on the identification of metabolite structures,there were still many uncertainties.
5.Conclusions
In this study,127 metabolites were detected from rat biological samples of clinical dose of Yanhusuo extract using the crossmapping of different samples and doses,and the strategy from representative standards to herbal extracts and from high dose to clinical dose of herbal extracts.What's more,the chemical structure screening tables were combined with characteristic fragment ions to clarify the metabolite structures.More accurate metabolic pathways of four representative standards in rats were drawn,and the relationship between structure and metabolism of Yanhusuo alkaloids was demonstrated.Thus,this strategy had advantages in herb-derived metabolites research.It solved problems like the high response of biological samples matrix,low metabolite concentration,inconspicuous peak shape,and absence of MS/MS information,which caused barriers in structure identification of metabolites.Structural types of metabolites that are more abundant than in vitro chemical components and clear in vivo metabolic pathways will be a great help for finding bioactive components of Yanhusuo.The strategy proposed in this study will be a powerful tool in the discovery of pharmacodynamic substances and metabolic mechanism research.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.03.006.
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Molecular detection of SARS-CoV-2 being challenged by virus variation and asymptomatic infection
- The potential of miRNA-based therapeutics in severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)infection:A review
- Potential treatment with Chinese and Western medicine targeting NSP14 of SARS-CoV-2
- Accurate and sensitive determination of hydroxychloroquine sulfate used on COVID-19 patients in human urine,serum and saliva samples by GC-MS
- Fast saccharide mapping method for quality consistency evaluation of commercial xylooligosaccharides collected in China
- Dispersive liquid-liquid microextraction,an effective tool for the determination of synthetic cannabinoids in oral fluid by liquid chromatography-tandem mass spectrometry
