Evaluation of autophagy-related genes and lncRNAs signature for prognositic prediction in thyroid carcinoma via bioinformatics analysis
2021-06-04ShanQiGuoYingJieJiaDengHaoXiaoJiangLi
Shan-Qi Guo, Ying-Jie Jia, Deng Hao*, Xiao-Jiang Li*
Evaluation of autophagy-related genes and lncRNAs signature for prognositic prediction in thyroid carcinoma via bioinformatics analysis
Shan-Qi Guo1, Ying-Jie Jia1, Deng Hao2*, Xiao-Jiang Li1*
1Department of Oncology, First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin, 300381, China.2Tianjin Key Laboratory of Translational Research of TCM Prescription and Syndrome, First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, Tianjin, 300381, China.
Autophagy plays a significant role in the pathogenesis and prognosis of thyroid carcinoma. The role of autophagy-related genes and long non-coding RNAs, as well as the risk model of thyroid carcinoma patients were investigated to predict clinical outcome of thyroid carcinoma. Different expression of autophagy-related genes and long non-coding RNAs in thyroid carcinoma patients was identified in The Cancer Genome Atlas database. Functional enrichment analysis and gene set enrichment analysis was used to hint the mechanism that autophagy might act in thyroid carcinoma. Univariate and multivariate Cox regression analyses were performed for screening the prognostic autophagy-related genes and long non-coding RNAs to construct prognostic related risk model. thyroid carcinoma patients were divided into the low-risk and high-risk groups. The overall survival time was both shorter in the high-risk groups than that in the low-risk groups. As for autophagy-related genes prognostic risk model, age and autophagy-related genes risk score are independent prognostic factors that affect the survival of thyroid carcinoma.andexpression was closely related to pathological stage and T status,expression was closely related to M status, age and gender. While autophagy-associated long non-coding RNA related prognostic risk model consequently demonstrated that the long non-coding RNA risk score could significantly predict the survival rate of thyroid carcinoma patients with areas under the curve of 0.972. gene set enrichment analysis presented that a total of 16 gene sets including 10 up-regulated and 6 down-regulated gene sets were significantly enriched. The autophagy-related genes and long non-coding RNAs based prognostic risk models are a reliable forecasting tool for thyroid carcinoma patients.
Autophagy-related gene, Long non-coding RNA, Prognosis, Thyroid carcinoma, The Cancer Genome Atlas
Background
Autophagy, named as type II programmed cell death, has become a significant role in the course of self-digestion, where damaged molecules or cell organs are degraded and recycled in many biological and pathological processes [1]. Previous studies indicated that autophagy act as the double-edged sword that can both promote and reduces the survival of malignant cells during the development of cancer [2]. In precancerous lesions or the prior stage of carcinoma, autophagy induces the degradation of damaged proteins or cell organs to decrease cell damage and chromosomal instability, which aims to inhibit the progress of cancer [3, 4]. Autophagy helps tumor cells to adapt to stress state and promote tumor progression during tumor formation [5, 6]. Thyroid cancer is the most common endocrine cancer. The incidence rate has been steadily increasing in recent decades. This can be interpreted by the increasing use of imaging modality to improve the detection rate of small and asymptomatic cancer [7, 8]. Although thyroid carcinoma (THCA) has a good prognosis, unfortunately, some thyroid cancers may be resistant to chemoradiotherapy through apoptosis escape, and the proportion of patients with metastasis is still high[9]. Autophagy involved in the thyroid carcinoma cells’ pathogenesis, differentiation, proliferation and participated in the process of THCA progression, recurrence, metastasis as well as the response to therapies [10−12]. Long non-coding RNAs (lncRNAs) have many functions, including RNA splicing, RNA decay, microRNA regulation, protein folding and so on [13]. More and more evidences indicated that lncRNAs could participate the transcription and post-transcription of autophagy related genes to regulate autophagy [14−16]. As we already known, the relationship between autophagy and thyroid carcinoma has been verified. Nevertheless, the relationship between autophagy and prognosis of thyroid cancer is rarely studied by large-scale gene expression characteristics.
In the current research, we executed gene expression microarray data to conclude autophagy-related gene expression characteristics and lncRNA expression signature, further establish a prognostic model as a risk score for overall survival. Of great significance, these autophagy-related models may have potential prognostic value for patients with thyroid cancer, and become potential therapeutic targets for predicting autophagy and forecasting the prognosis of thyroid cancer patients.
Methods
Data acquisition
Autophagy-related genes came from The Human Autophagy Database (HADb, http://www.autophagy.lu/index.html). RNA-sequencing data of ARGs and the clinical records of the thyroid carcinoma cohort were downloaded and extracted from TCGA database (https://tcga-data.nci.nih.gov/tcga/), which contains 510 thyroid carcinoma tissues and 58 adjacent non-tumor tissues. All the mRNA levels were normalized by log2 transformation. Pearson correlation method was used to calculate the correlation between lncRNAs and ARGs.The lncRNAs with correlation coefficient |R2| > 0.3 and< 0.05 were considered as autophagy-related lncRNAs.
Differentially expressed ARGs and enrichment analysis.
Limma software package was applied to calculate the differential expression of ARGs in thyroid cancer and non-tumor tissues. The threshold value was |log 2 fold change (FC)| > 2, and the correctedvalue was < 0.05. Then, we employed gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of genes and genomes (KEGG) enrichment analysis to identify the main biological characteristics of differentially expressed ARGs. The visualization of GO and KEGG enrichment maps of annotation analysis results were executed by R language using "ggplot 2", "goplot", "cluster profiler" and "pathview" packages. Moreover, gene set enrichment analysis (GSEA) was also used to explain the level of gene expression.
Construction of risk models based on autophagy related genes and lncRNAs
Objective to investigate the expression of ARGs and lncRNAs of thyroid carcinoma, we conducted the univariate Cox regression analysis and multivariate Cox regression analysis. Based on multivariate Cox regression, “glmnet” software package was used to establish the prognostic risk models with OS related prediction formula. Kaplan-Meier method was used for survival analysis. The high-risk groups and low-risk groups were compared through median risk score. The logistic transformation of hazard ratio (HR) in multivariate Cox regression analysis was conducted to calculate risk scores of the linear combination of lncRNAs expression levels weighted by regression coefficients.
The risk score for each patient was computed using the following formulation: βgene1× exprgene1+ βgene2× exprgene2+ ··· ··· ··· +βgenen× exprgenen. The survival receiver-operator characteristic software package in R was served to evaluate the predictive value of the prognostic model by areas under the curve (AUC) of the receiver-operator characteristic curve.
Statistical analysis
Data analysis were performed by R 3.6.3 (https://www.r-project.org/) and GraphPad Prism 8 ((GraphPad Software Inc., SanDiego, CA, USA). Cytoscape software (version 3.7.2; The Cytoscape Consortium, San Diego, CA, USA) was employed for constructing the autophagy-lncRNA co-expression network. Pearson correlation analyses and Cox regression analyses were performed using SPSS statistical software (version 23; IBM, Armonk, New York, USA). Survival status was the basis of univariate COX regression analysis. GSEA (version 4.0.3; http://www.broadinstitute.org/gsea/index.jsp) was employed to discriminate functional annotations in two sets. Statistical significance was set as the threshold of two tailed< 0.05.
Results
Conclude differentially expressed ARGs
The RNA sequences and clinical data of 510 thyroid cancer tissue samples and 58 non-tumor samples were extracted from TCGA database. Of which, 509 THCA patients had RNA-seq data and clinical follow-up information. Altogether 232 ARGs were acquired from the Human Autophagy Database (HADb, http://autophagy.lu/clustering/index.html). Considering the thresholds of |log 2 fold change (FC)| > 1, fdr < 0.05, twenty-six ARGs were differentially expressed, including 15 up-regulated (and) and 11 down-regulated ARGs () were obtained (Figure 1A). The volcano map and box map were observed to display the expression profile of ARGs in thyroid cancer and non-tumor tissues (Figure 1B and Figure 1C).
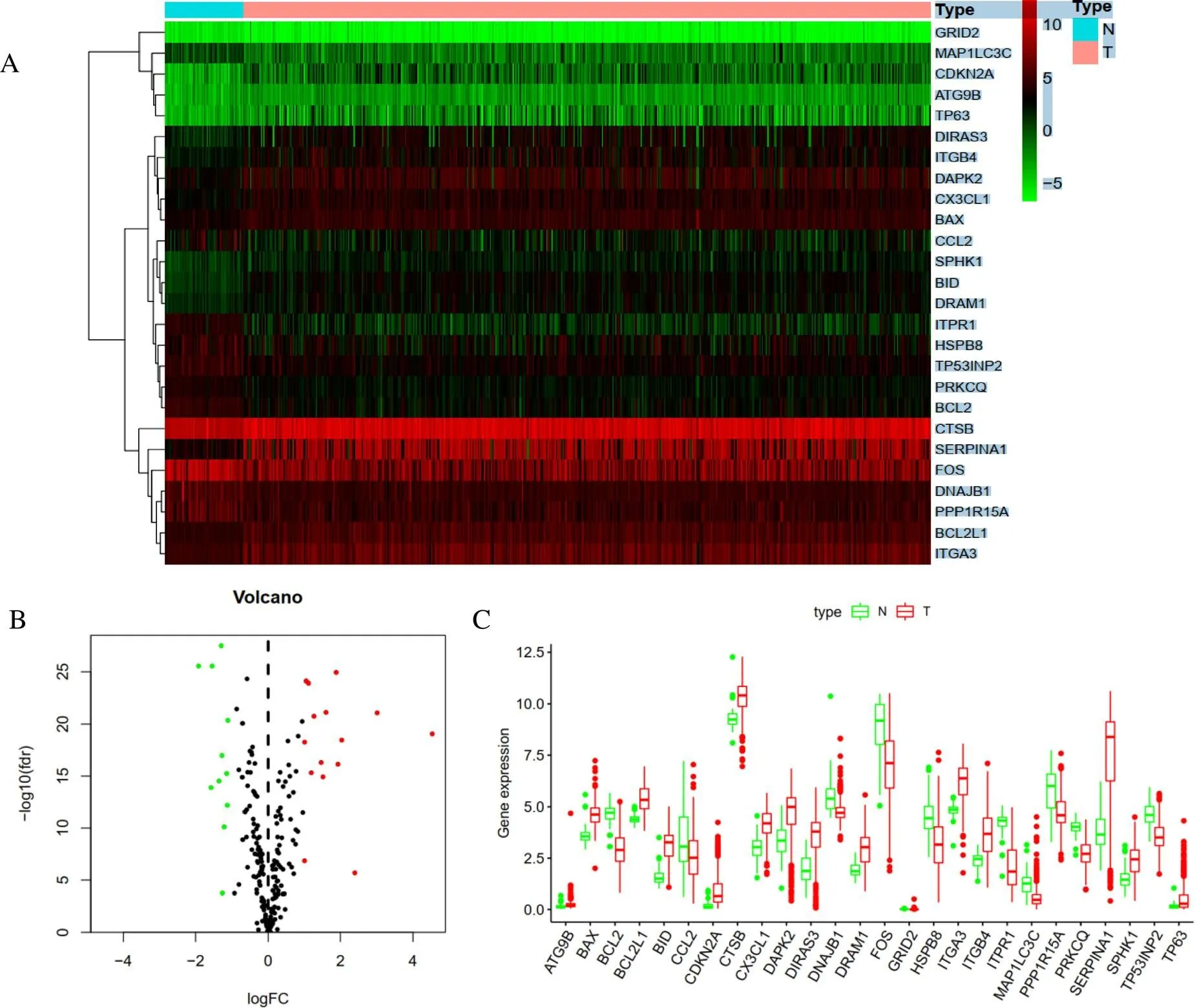
Figure 1 Differentially expressed autophagy-related genes between thyroid carcinoma and normal thyroid tissues. A, Heatmap of differentially expressed autophagy-related genes. B, The volcano plot for the 232 autophagy-related genes. Red indicates high expression, green indicates low expression. Black interprets that those genes showed no difference between hyroid carcinoma and normal thyroid tissues.C, The expression patterns of 26 differentially expressed autophagy-related genes in thyroid carcinoma and paired non-tumor samples. Red and green respectively indicate tumor tissues and normal tissues.
Analysis of GO and KEGG pathway enrichment
Go and KEGG enrichment analyses were executed according to the differential level of ARGs. We notice that the top terms in biological processes were autophagy, processes using autophagy and neuronal death (Figure 2A). The bubble diagram stated the relationship between ARGs and GO enrichment analyses (Figure 2B). In addition, the enrichment analysis of KEGG pathway in the differential expression of ARGs showed that Pathways in Autophagy-animal, Apoptosis, p53 signaling pathway etc. were notably associated with THCA (Figure 3A). The heatmap of the relation between ARGs and pathways were also showed (Figure 3B). The Z-score of enrichment pathway was less than zero, indicating that most cancer pathways were more likely to decrease.
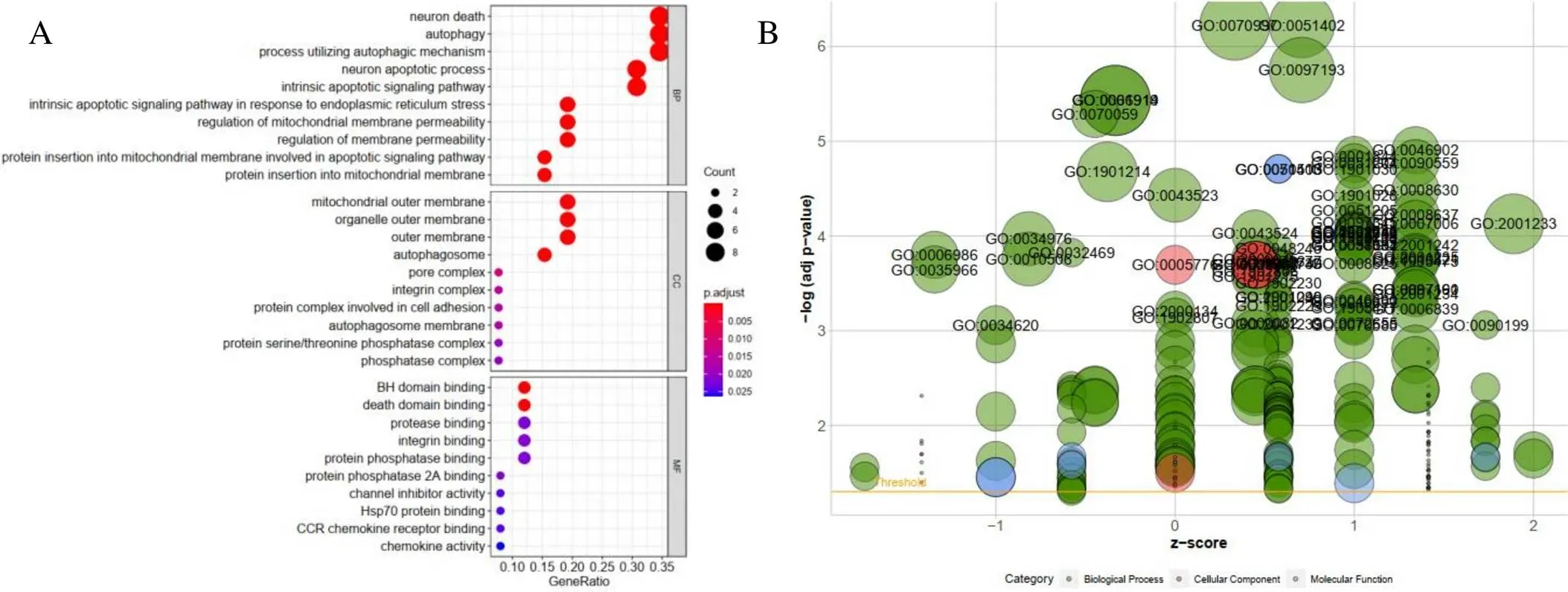
Figure 2 GO enrichment analysis of differentially expressed autophagy-related genes. A, Functional annotation of biological process, cellular component and molecular function category. The size of the circle represents the degree of enrichment (the bigger the more significant).B, The bubble plot of GO. The z-score is assigned to the x-axis, and the negative logarithm of the-value to the y-axis, as in the barplot (the higher the more significant). The size of the displayed circles is proportional to the number of genes assigned to the term. Greed circles correspond to the biological process, red indicates the cellular component, and blue shows the molecular function category.
Figure 3 KEGG enrichment analysis of differentially expressed autophagy-related genes. A, The outer circle shows a scatter plot for each term of the logFC of the differentially expressed autophagy-related genes. B, Heatmap of the relationship between autophagy-related genes and KEGG enrichment.
Prognosis prediction model construction
Then we further evaluated the differentially expressed ARGs and OS expression profiles. In univariate Cox regression analysis, 22 prognostic ARGs were markedly connected with OS (Figure 4). Multivariate Cox regression analysis was executed and 7 ARGs includingandwere finally identified to develop the prognostic risk model (Table 1). The formula of risk score is as follows: risk score = (0.9690 × expression value of) + (2.6643 × expression value of) + (−1.2237 × expression value of) + (1.9772 × expression value of) + (−1.8935× expression value of) + (0.5726 × expression value of) + (2.1638 × expression value of). Note that the coefficients ofandwere negative, it was suggested thatandwas negatively correlated with survival time of patients with thyroid cancer. In order to determine the performance of risk score in forecasting the clinical prognosis of patients with thyroid cancer, the Kaplan-Meier diagram was drawn to analyze the correlationship among survival time, high-risk group and low-risk group (Figure 5A). In addition, the distribution of patients in different risk groups (Figure 5B), the total THCA patients in TCGA dataset Figure 5C) and the expression profiles of seven key genes (Figure 5D) were also shown.
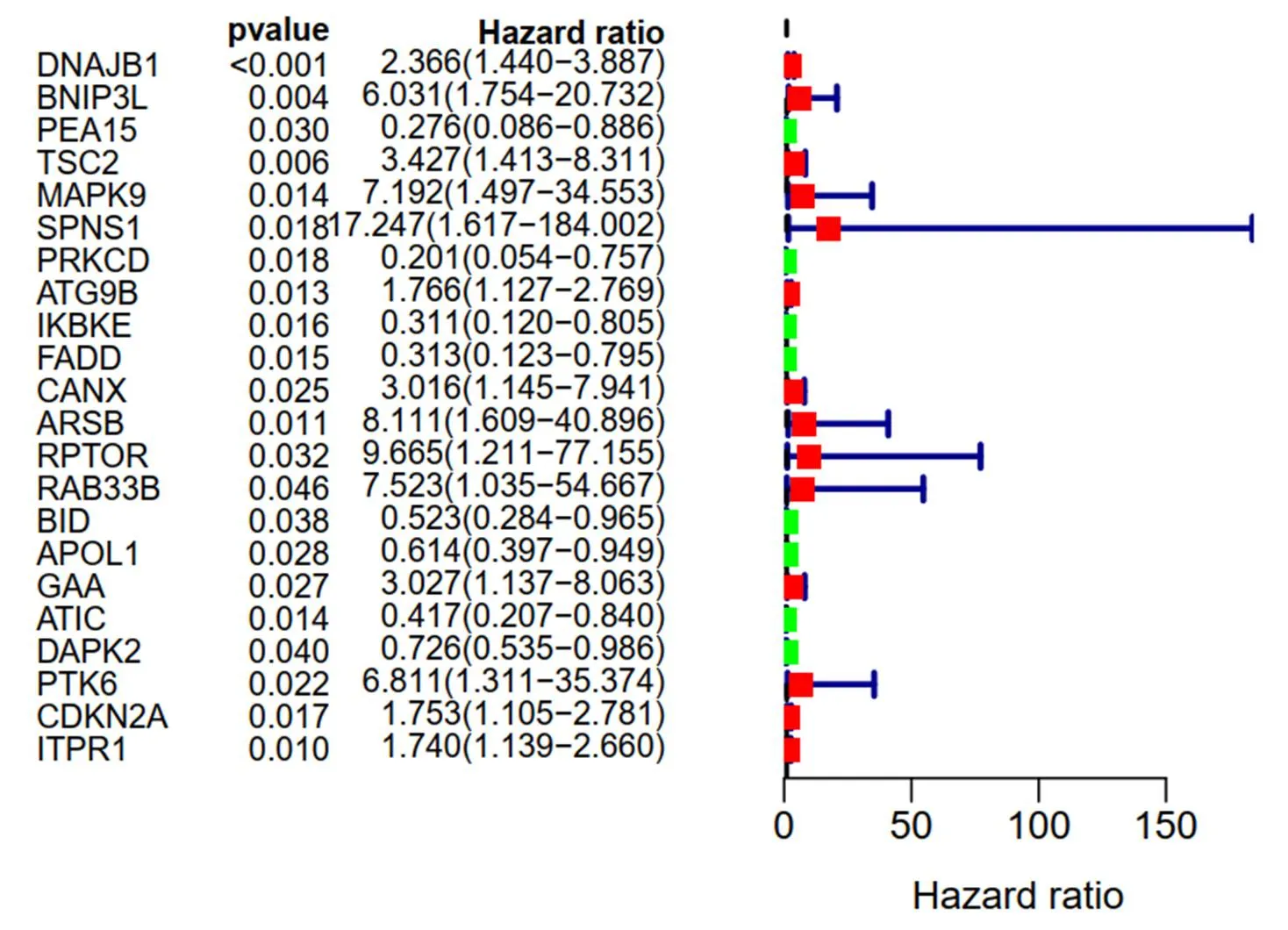
Figure 4 Result of prognosis-related autophagy-related genes in the univariate Cox regression analysis

Table 1 Result of prognosis-related autophagy-related genes with multivariate Cox regression analysis
Abbreviation: HR, hazard ratio.

Figure5 Overall survival-related autophagy-related genes prognostic model of thyroid carcinoma patients. A, Kaplan–Meier plot represents that patients in the high-risk group had significantly shorter overall survival than those in the low-risk group. B, The prognostic model distribution of thyroid carcinoma patients. C, The overall survival of patients in the The Cancer Genome Atlas dataset. D, The heatmap of thyroid carcinoma patients with the expression situation of seven key autophagy-related genes
Prognostic prediction by integrating risk score and clinical features
Univariate analysis and multivariate Cox regression analysis based on TCGA data set were used to find that age, gender, stage, t, N, m and risk score could be used as complementary prognostic factors. Univariate analysis manifested that age, stage, pathological T stage and risk score were the prognostic factors (Figure 6A). Age (< 0.001) and risk score (= 0.027) were independent prognostic factors for survival of thyroid cancer (Figure 6B). In addition, Figure 7 showed the clinicopathological significance of OS-related prognostic risk model in THCA patients. The expression ofgene in T1-2 was higher than that in T3−4 (= 0.002), and that in Ⅰ-Ⅱ was higher than that in Ⅲ−Ⅳ (= 0.005).gene expression showed the same distribution trend asexpression. Theexpression of T3-4 and Ⅲ-Ⅳ was significantly lower than that of T1-2 (= 0.001) and Ⅰ-Ⅱ (= 0.003). The difference ofexpression was closely related to age and gender. Patients of thyroid cancer > 30 years old and < 60 years old was higher than that of below 30 years old and over 60 years old (= 0.005). In addition, the expression ofin women was significantly higher than that in men (= 0.008), while the expression ofwas higher in patients without metastasis (= 0.013). The predictive model value of risk score had significant difference in age (= 0.017) and survival status (= 0.0026). No histological grade was reported in all patients with thyroid cancer in this study. Therefore, the relationship between histological grading and predictive models has not been analyzed.
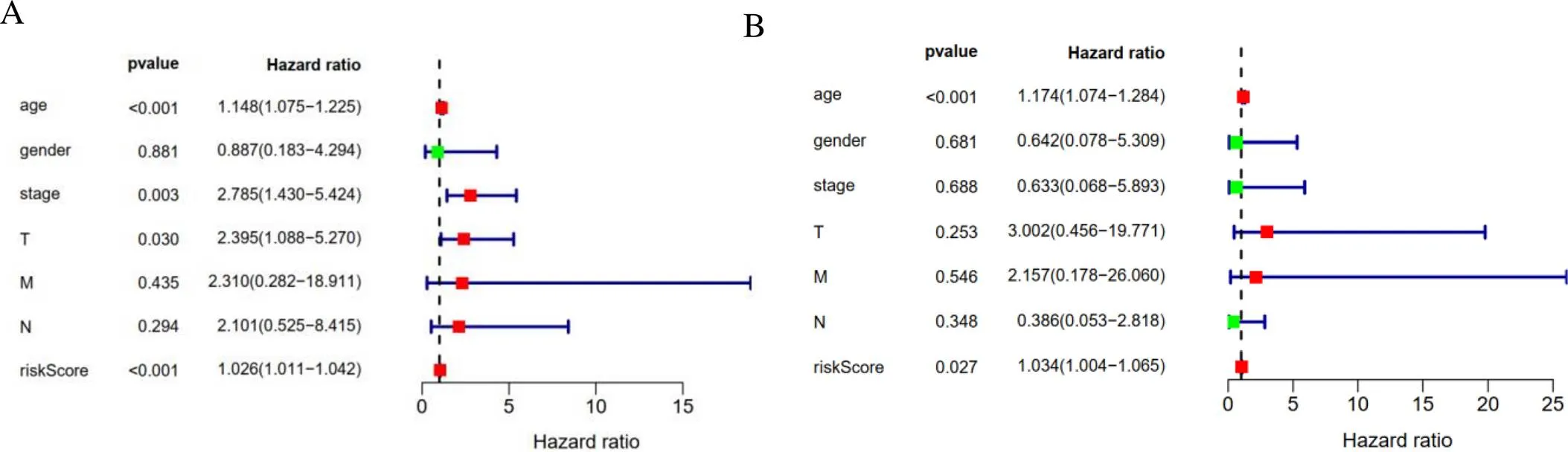
Figure 6 The results of autophagy-related genes prognostic risk model. A, Univariate analysis results. B, Multivariate analysis results.
Figure 7 The clinical pathological significance of Overall Survival-related prognostic risk model in thyroid carcinoma patients.
Co-expression network of ARGs and autophagy relevant lncRNAs
We extracted 14142 lncRNAs of the transcriptome data of 509 patients with thyroid cancer collected from TCGA dataset. Then, mRNA and lncRNAs were classified according to whether RNA could encode proteins. Using HADb database, we finally constructed the ARGs–lncRNA co-expression network to verified autophagy-related lncRNAs. Therefore, 1298 lncRNAs were indicated (|R2| > 0.3 and< 0.01) as autophagy-related lncRNAs. Then, 1298 autophagy-associated lncRNAs were screened as prognostic related lncRNAs by univariate Cox regression analysis (= 0.05). The results showed that 32 lncRNAs have prognostic value of thyroid cancer patients. Multivariate Cox regression analysis showed that 10 lncRNAs including FAM201A, LINC02454, AL162231.2, AC004918.3, AC011297.1, TONSL-AS1, AC008063.1, AC092279.1, CRNDE and LINC00900 have independent prognostic value in patients with thyroid cancer. Figure 8 showed a prognostic incremental RNA network with co-expression autophagy genes in thyroid cancer.
Prognostic risk model based on autophagy related lncRNAs
Multivariate Cox regression analysis were used to certified the above 10 lncRNAs, and the prognostic risk model was constructed. Of which four lncRNAs were favorable factors for thyroid cancer (TONSL-AS1, AC008063.1, AC092279.1and LINC00900) and six lncRNAs were unfavorable prognostic factors for thyroid cancer (FAM201A, LINC02454, AL162231.2, AC004918.3, AC011297.1, TONSL-AS1, AC008063.1, AC092279.1 and CRNDE) (Figure 9). The risk score was used to establish OS-related prediction model. Risk score= (0.908135113×expression value of FAM201A) + (0.600044489×expression value of LINC02454) + (0.679625648×expression value of AL162231.2) + (2.438695251×expression value of AC004918.3) + (0.171382272×expression value of AC092279.1) + (-0.781308235× expression value of TONSL-AS1) + (-1.720538412×expression value of AC008063.1) + (-3.758022369×expression value of AC092279.1) + (0.450466158×expression value of CRNDE) + (-2.382996609×expression value of LINC00900). In order to further improve the exactitude of the model in predicting OS of thyroid cancer patients, we used Akaike Information Criterion forward and backward regression method to achieve the optimal state.
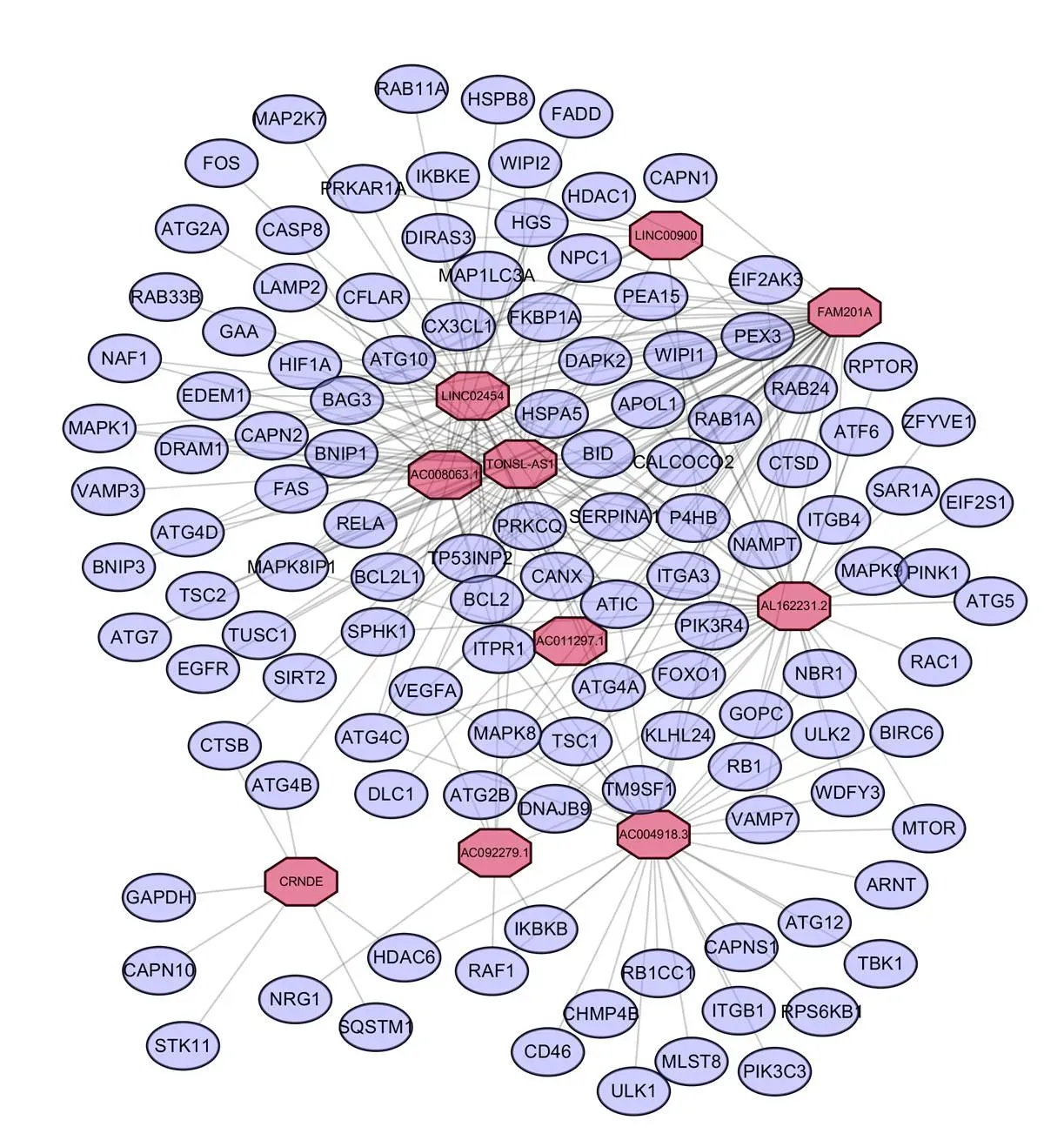
Figure 8 Network of prognostic long non-coding RNAs with co-expressed autophagy-related genes in thyroid carcinoma. In the centric position, red nodes indicate lncRNAs and the purple indicates autophagy genes. The coexpression network is visualized by CYTOSCAPE 3.7.2 software.
Figure 9 Kaplan–Meier survival curves for the 10 independent prognostic long non-coding RNAs for thyroid carcinoma in thyroid carcinoma dataset.
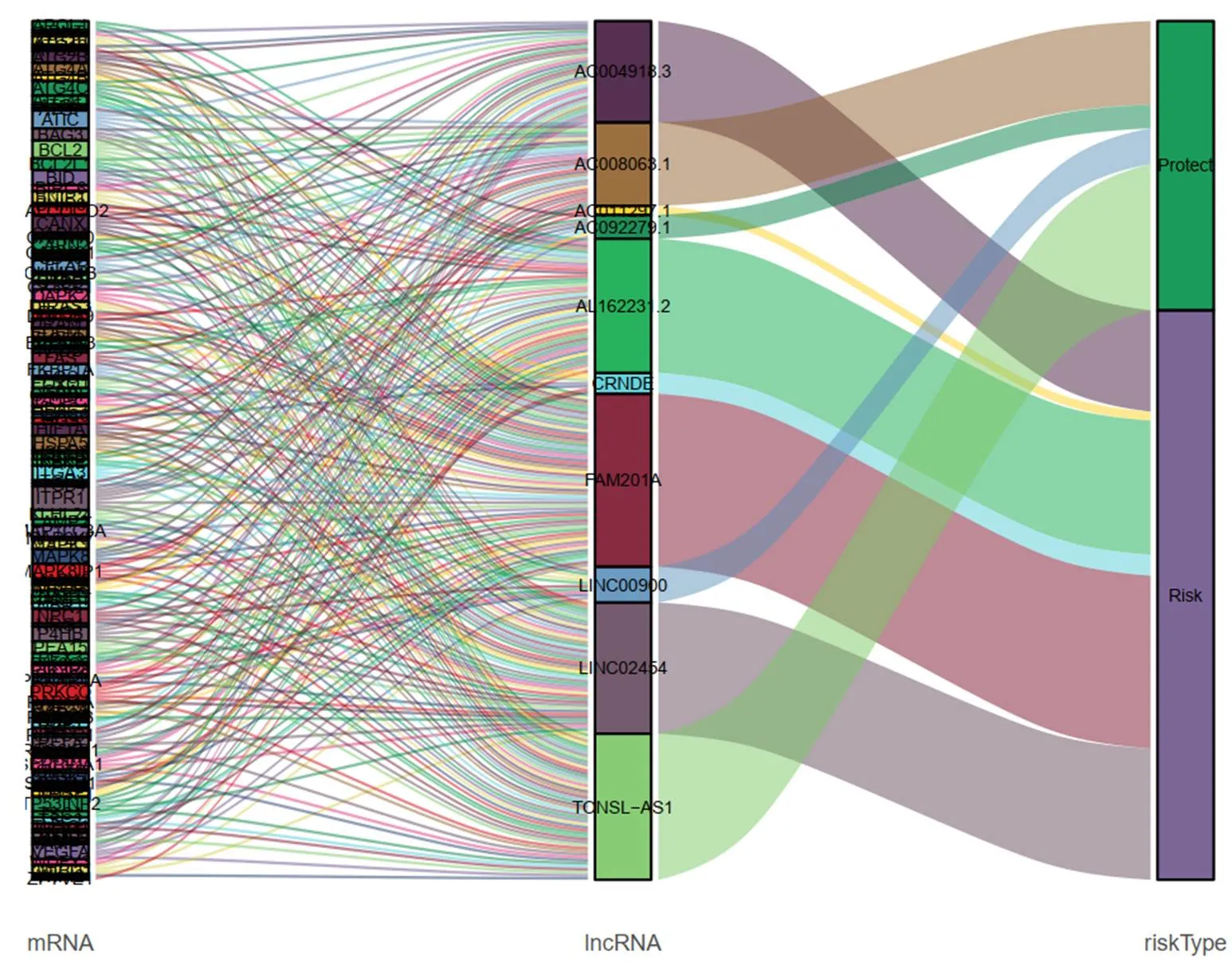
Figure 10 Sankey diagram illustrates the flow distribution of mRNAs, long non-coding RNAs in high-risk group and low-risk group.
Identification the prognostic features based on autophagy-related lncRNAs
The thyroid carcinoma patients were divided into low-risk group and high-risk group by median risk score. Sankey diagram also revealed the flow distribution of mRNAs, lncRNAs and the risk type (Figure 10). Results manifested that the risk score could predict the survival of thyroid carcinoma patients (Figure 11A). The receiver-operator characteristic curves of OS-related predictive signature were demonstrated in Figure 11B, with AUC of 0.972. Additionally, Figures 11C–E showed the number of patients with different risk, the survival related prediction model distribution of patients and the heatmap of the ten lncRNAs expression characteristics. In addition, univariate and multivariate Cox regression analyses were further explored whether risk score was an independent predictor of prognosis of thyroid cancer patients (Figure 12). Therefore, the HR of 1.003 indicated that risk score was helpful to predict the survival rate of THCA patients through the analysis of pathological stage, t, N, m, etc. Table 2 showed the correlation between risk scores and prognostic characteristics. Further functional annotation was performed by GSEA. The results showed that the differentially expressed genes between the two groups were enriched in autophagy-related and tumor related pathways. In all genomes, 94/178 gene sets were up-regulated in high phenotype genomes and 84/178 were up-regulated in low phenotype genomes. Among all the gene sets, 94/178 gene sets are up-regulated in high phenotype, and 84/178 gene sets are upregulated in low phenotype. The results showed that a total of 16 gene sets (including 10 up-regulated and 6 down regulated gen sets) were observably enriched when p value was < 0.05 (Table 3). GSEA of TCGA-THCA data showed that genes related to amino acid metabolism, energy metabolism and RNA polymerization in THCA were up-regulated (Figure 13).
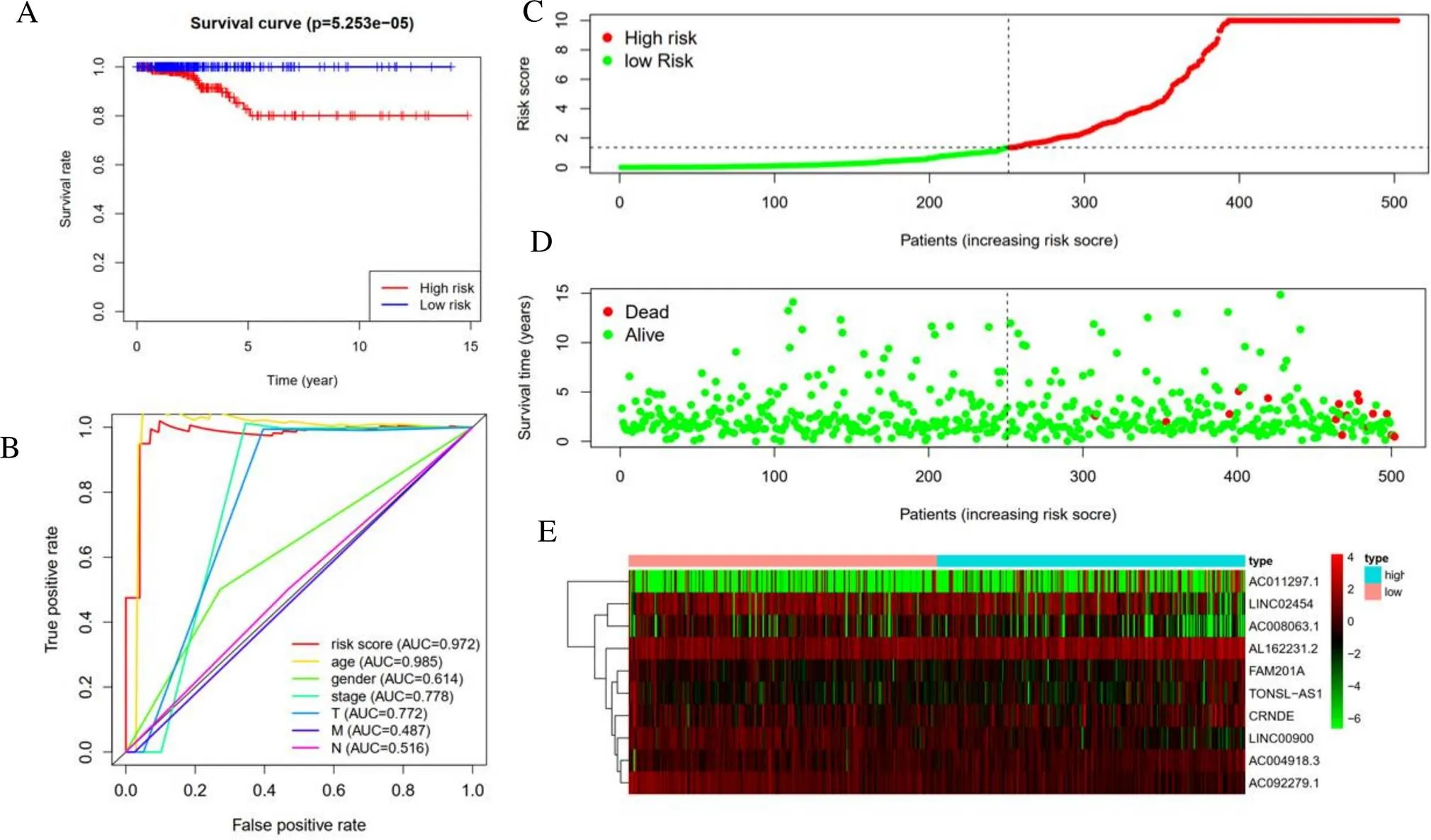
Figure 11 Overall Survival-related long non-coding RNAs prognostic model of thyroid carcinoma patients. A, Kaplan–Meier plot represents that patients in the high-risk group had significantly shorter overall survival than those in the low-risk group. B, Receiver-operator characteristic curve of Overall Survival-related prognostic risk model. C, The prognostic model distribution of thyroid carcinoma patients. D, The overall survival of patients in the The Cancer Genome Atlas dataset. E, The heatmap of thyroid carcinoma patients with the expression situation of seven key long non-coding RNAs
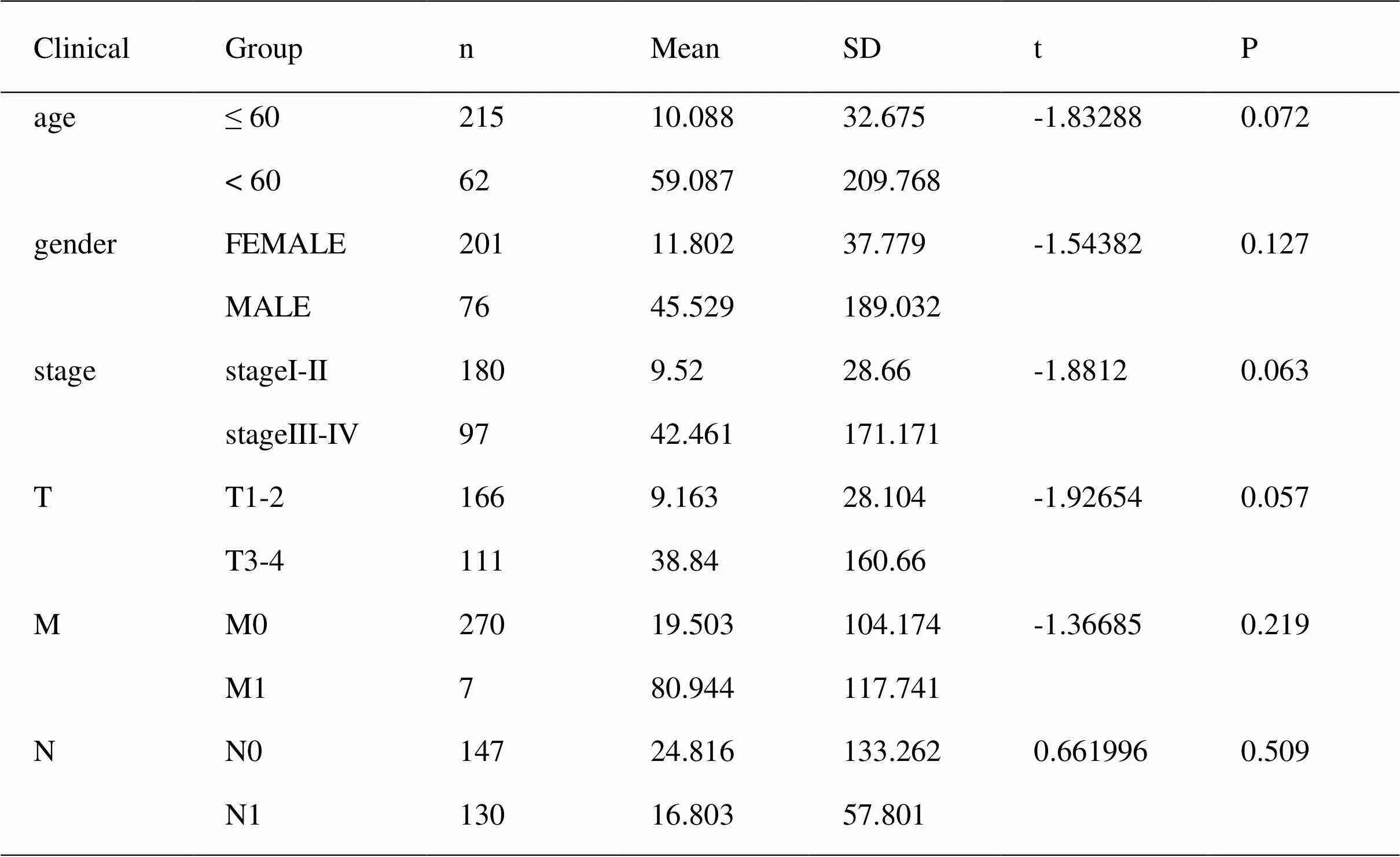
Table 2 clinical impact of risk score signature of thyroid carcinoma patients
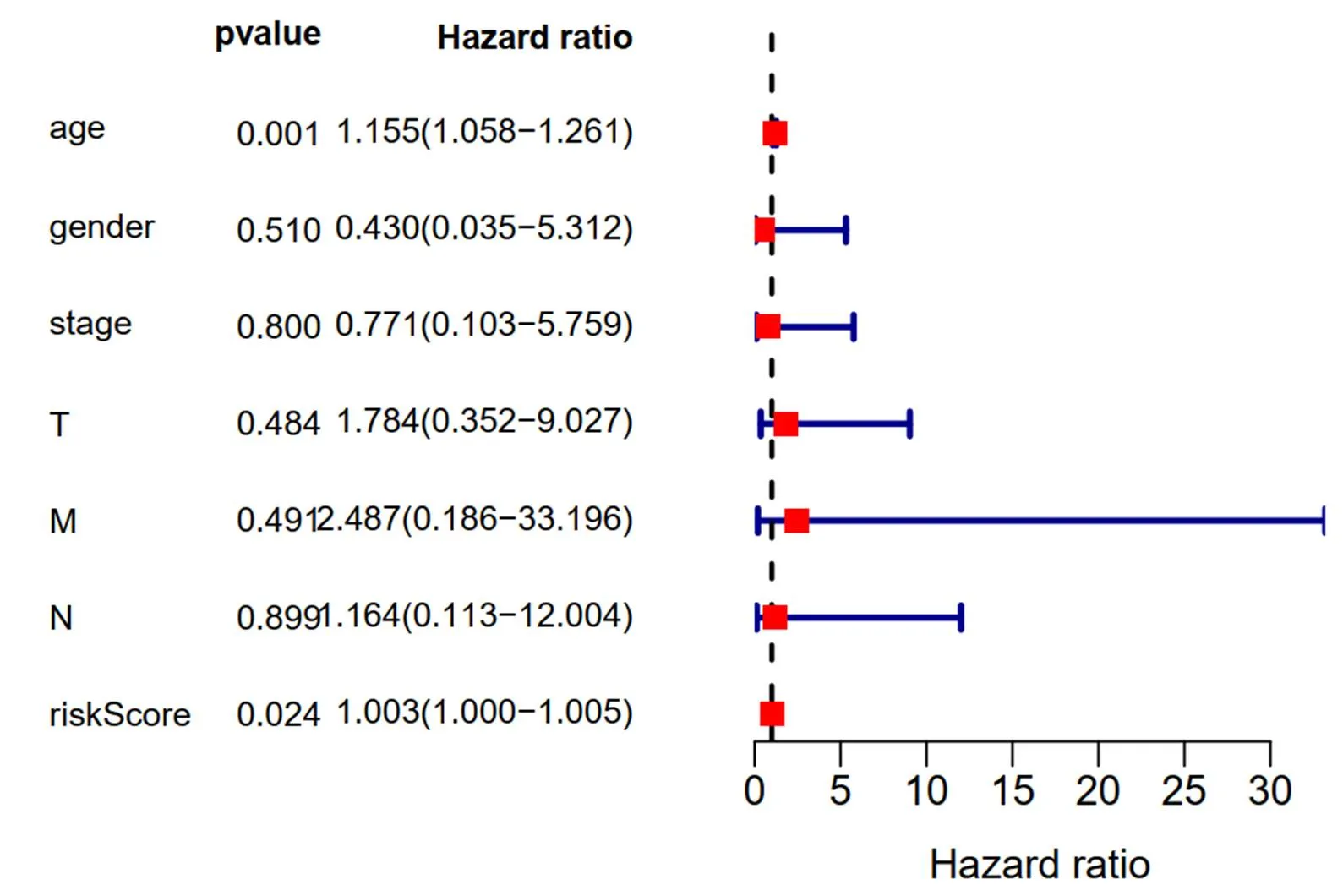
Figure 12 Multivariate analysis results of long non-coding RNAs related prognostic risk model

Table3 Gene set enrichment analysis results based on the signature of 10 autophagy long non-coding RNAs.

Table3 Gene set enrichment analysis results based on the signature of 10 autophagy long non-coding RNAs. (continued)

Figure 13 Gene set enrichment analysis results of The Cancer Genome Atlas- thyroid carcinoma data
Discussion
Recently, the incidence rate of THCA has been increasing gradually due to the increasing popularity of diagnostic equipment. Incidence rate of the disease was increasing worldwide. Although some well differentiated THCA subtypes have a good prognosis, a considerable number of patients still have metastasis [10−12]. In addition, TNM cannot fully predict the risk of THCA. Therefore, autophagy has attracted great attention, and autophagy regulators have great potential in THCA [13−15]. The exploration of autophagy mechanism provides a new perspective for the study of THCA. Although there have been many reports on THCA gene expression profiles and prognosis models, such as autophagy by studying signal genes, the prognosis models of autophagy-related genes and lncRNAs have not been reported. In order to obtain THCA relevant genes from the angle of autophagy, we screened ARGs and identified 7 key prognostic ARGs and 10 key prognostic lncRNAs, which may provide potential therapeutic targets based on autophagy. We further studied the relationship between mRNA and lncRNA. This comprehensive study contributed to promising ideas of THCA biology and potential therapeutic biomarkers and approaches [16−18].
In order to get knowledge of the influence on tumor gene composition of clinical consequences, TCGA has been established to discover gene characteristics. We then concluded the expression profiles of ARGs and lncRNAs to find biomarkers THCA patients. We screened 26 ARGs differentially expressed in THCA and non-tumor tissues, among which 15 were up-regulated and 11 were down regulated. Then we used GO and KEGG analysis on these targeted genes. And what's interesting to us is that feature analysis indicated that the most important KEGG pathway was related to autophagy, apoptosis and p53 signaling pathway. On this basis, we speculated that autophagy might be a tumor suppressor in THCA.
By univariate survival analysis, 22 ARGs were found to be related to OS in TCGA database. Multivariate survival analysis determined seven key prognostic parameters (,,,,,and) to develop risk scores, which could be independent prognostic indicators of THCA patients. With univariate as well as multivariate analyses, ARGs with significant prognostic value were selected to perform the risk-score model. According to the risk score median evaluation, THCA patients were divided into two groups. The risk score and clinical parameters were combined to analyze the prognostic characteristics, results showed that the expressions of,andwere closely related to clinical features. 5-aminoimidazol-4-formamide ribonucleotide formyltransferase/inosine monophosphate cyclohydrolase (atic) is a bifunctional protease that catalyzes the last two steps of de novo purine biosynthesis pathway [19]. Li M and colleges found that that, as a oncogene, promotes proliferation, migration and survival by targeting AMPK-mTOR-S6 K1 signaling [18]. Zhu J et al [20] focused on lung adenocarcinoma and lung squamous cell carcinoma. Their results showed thathad better AUC than other clinical parameters, showed as a promising marker focused on predict the survival rate of patients. The explanation ofmay not only be limited to THCA related autophagy, but also play a vital role in lung cancer.
In addition, Liu X and others believe thatis an effective and previously unrecognized chemical radiosensitization target. More generally, the role of purine level in regulating the efficiency of DNA damage response may not be fully understood, and DNA damage response can be used as radiosensitization strategy [21]. It is suggested thatis an important factor in the treatment of cancer, especially chemotherapy and radiotherapy. It has been reported thatcould induce autophagy in a variety of malignant tumors [22, 23]. It is well known that THCA is closely related to diabetes. Banerjee J et al found thatis significantly associated with aging in patients with T2DM and controls. The correlation strength of MCP-1 and CKDN2A with aging in T2DM patients was stronger than that in the control group, stating thatmight be an important link between T2DM and THCA [24].mutation is also common in pancreatic ductal adenocarcinoma. Wu C et al. explainedparticipated in the development of pancreatic ductal adenocarcinoma, and also regarded as the basis of gene targeting with a correlation coefficient ofabove 0.9 on the STRING website. The role of these genes andin pancreatic ductal adenocarcinoma may provide a new direction, which will update and supplement the current treatment options, because the progress of cancer is achieved through cell-cell interaction [25]. Gang Hu et al. reported autophagy induced byin papillary thyroid carcinoma [26, 27], and our study also had the same results.
was verified specifically bind to, which regulate p53, further studies revealed thatis stable inat the post-translational level [28]. Our KEGG enrichment analysis also showed that p53 signaling pathway was significant, suggesting thatmay plays an important role. However, there is no report of autophagy induced byin THCA.
All the lncRNAs we found regulate autophagy directly or indirectly, and many of them regulate miRNAs. Innovatively, we performed lncRNA and mRNA co-expression analysis to evaluate lncRNAs function. Therefore, we concluded 10 autophagy-related lncRNAs, and further identified the potential possibility of therapeutic targets. Moreover, ten autophagy-associated lncRNA were identified, among which 4 lncRNAs were favorable factors (TONSL-AS1, AC008063.1, AC092279.1and LINC00900) and 6 lncRNAs were confirmed as unfavorable prognostic factors for thyroid cancer (FAM201A, LINC02454, AL162231.2, AC004918.3, AC011297.1, TONSL-AS1, AC008063.1, AC092279.1and CRNDE) , which could split into high-risk and low-risk groups. LncRNAs involved in THCA process regulation has been widely studied [29−31]. Recently, Peng x et al reported that lncRNA TNRC6C-AS1 could promote STK4 methylation to inhibit thyroid carcinoma cell autophagy as well as apoptosis through Hippo signalling pathway [32]. Wang Yong and colleagues confirmed that BRAF activated lncRNAs contributes to cell proliferation as well as activates autophagy in papillary THCA [33]. Gou Lisha also verified that knockout of lncRNA MALAT1 may inhibit the progression of anaplastic THCA by regulating miR-200a-3p/FOXA1 axis [34]. Above studies showed that autophagy related lncRNAs have essential effect in regulating THCA progression, and of great significance, out consequence of 10 lncRNAs also show favorable predictive ability with great relevance to clinicians based on to predict individual patient outcomes.
In conclusion, by constructing the network of co-expression of ARGs and lncRNAs, we identified the characteristics of 7 ARGs and 10 autophagy-relevant lncRNAs, which may provide prognostic value for THCA patients. Although the role of autophagy in various environments of THCA needs to be further elucidated, autophagy regulators also might be of great potential for doctors and researchers. And the prognostic risk model was constructed to provide decision-making basis for individualized therapeutic regimen design.
1. Acevo-Rodríguez PS, Maldonado G, Castro-Obregón S, Hernández G. Autophagy Regulation by the Translation Machinery and Its Implications in Cancer.. 2020;10:322.
2. Deng S, Shanmugam MK, Kumar AP, Yap CT, Sethi G, Bishayee A. Targeting autophagy using natural compounds for cancer prevention and therapy.. 2019;125(8):1228−1246.
3. Chen HT, Liu H, Mao MJ, et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy.2019;18(1):101.
4. Hönscheid P, Datta K, Muders MH. Autophagy: detection, regulation and its role in cancer and therapy response.. 2014;90(8):628−635.
5. Wentao Wei, Qinjiang Liu, Wei Ya. Factors influencing the presence of circulating differentiated thyroid cancer cells in the thyroidectomy perioperative period., 2015, 1(005):208−211.
6. Wang JY, Yao WX, Wang Y, Fan YL, Wu JB. Network analysis reveals crosstalk between autophagy genes and disease genes.2017;7:44391.
7. Schwertheim S, Theurer S, Jastrow H, et al. New insights into intranuclear inclusions in thyroid carcinoma: Association with autophagy and with BRAFV600E mutation.2019;14(12):e0226199.
8. Tan H, Li Z, Li N, et al. Thyroid imaging reporting and data system combined with Bethesda classification in qualitative thyroid nodule diagnosis.2019;98(50):e18320.
9. Zhao Y, Zhong L, Yi H. A review on the mechanism of iodide metabolic dysfunction in differentiated thyroid cancer.2019;479:71−77.
10. Wei W, Hardin H, Luo QY. Targeting autophagy in thyroid cancers.2019;26(4):R181−R194.
11. Lim AM, Solomon BJ. Immunotherapy for Anaplastic Thyroid Carcinoma.2020;38(23):2603−2604.
12. Lan X, Bao H, Ge X, et al. Genomic landscape of metastatic papillary thyroid carcinoma and novel biomarkers for predicting distant metastasis.2020;111(6):2163−2173.
13. Morani F, Titone R, Pagano L, et al. Autophagy and thyroid carcinogenesis: genetic and epigenetic links.2013;21(1):R13−R29.
14. Li LC, Liu GD, Zhang XJ, Li YB. Autophagy, a novel target for chemotherapeutic intervention of thyroid cancer.2014;73(3):439−449.
15. Connerty P, Lock RB, de Bock CE. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer.. 2020;10:285.
16. Barangi S, Hayes AW, Reiter R, Karimi G. The therapeutic role of long non-coding RNAs in human diseases: A focus on the recent insights into autophagy.2019;142:22−29.
17. Yang L, Wang H, Shen Q, Feng L, Jin H. Long non-coding RNAs involved in autophagy regulation.. 2017;8(10):e3073.
18. Yao H, Han B, Zhang Y, Shen L, Huang R. Non-coding RNAs and Autophagy.2019;1206:199−220.
19. Lv L, Cao L, Hu G, Shen Q, Wu J. Methylation-Driven Genes Identified as Novel Prognostic Indicators for Thyroid Carcinoma.2020;11:294.
20. Li M, Jin C, Xu M, Zhou L, Li D, Yin Y. Bifunctional enzymepromotes propagation of hepatocellular carcinoma by regulating AMPK-mTOR-S6 K1 signaling.2017;15(1):52.
21. Zhu J, Wang M, Hu D. Development of an autophagy-related gene prognostic signature in lung adenocarcinoma and lung squamous cell carcinoma.2020;8:e8288.
22. Liu X, Paila UD, Teraoka SN, et al. Identification ofas a Novel Target for Chemoradiosensitization.2018;100(1):162−173.
23. Bernard M, Yang B, Migneault F, et al. Autophagy drives fibroblast senescence through MTORC2 regulation.2020;16(11):2004−2016.
24. Jeong EH, Lee TG, Ko YJ, et al. Anti-tumor effect of CDK inhibitors on-defective squamous cell lung cancer cells.2018;41(6):663−675.
25. Banerjee J, Dhas Y, Mishra N. Middle-Aged Indians with Type 2 Diabetes Are at Higher Risk of Biological Ageing with Special Reference to Serum.2020;2020:7569259.
26. Wu C, Yang P, Liu B, Tang Y. Is there a-centric network in pancreatic ductal adenocarcinoma?.2020;13:2551−2562.
27. Hu G, Feng HF, Zhan H. Identification of an Autophagy-Related Signature Predicting Overall Survival for Papillary Thyroid Carcinoma.2020;18(1):1559325819899265.
28. Qi M, Zhang J, Zeng W, Chen X.stabilizesand contributes to cancer cell proliferation in a p53-dependent manner.2014;1839(1):62−69.
29. Zhao Y, Zhao L, Li J, Zhong L. Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2.. 2019;234(11):20980−20991.
30. Qin Y, Sun W, Zhang H, et al. LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer.2018;59(3):555−564.
31. Yang LX, Wu J, Guo ML, Zhang Y, Ma SG. Suppression of long non-coding RNA TNRC6C-AS1 protects against thyroid carcinoma through DNA demethylation of STK4 via the Hippo signalling pathway.2019;52(3):e12564.
32. Peng X, Ji C, Tan L, et al. Long non-coding RNA TNRC6C-AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway.2020;24(1):304−316.
33. Wang Y, Guo Q, Zhao Y, et al. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma.2014;8(5):1947−1952.
34. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1.2019;20(11):1355−1365.
10.12032/TMRCR20210516008
1Xiao-Jiang Li. Department of Oncology, First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, No.88, Changling Road, Liqizhuang Street, Xiqing District, Tianjin 300381, China. Email: li_xj2020@126.com.2Deng Hao. Tianjin Key Laboratory of Translational Research of TCM Prescription and Syndrome, First Teaching Hospital of Tianjin University of Traditional Chinese Medicine, No.88, Changling Road, Liqizhuang Street, Xiqing District, Tianjin 300381, China. Email: lujiang-denghao@163.com.
:THCA, thyroid carcinoma; ARGs, autophagy-related genes; lncRNAs, long non-coding RNAs; TCGA, The Cancer Genome Atlas; GSEA, gene set enrichment analysis; OS, overall survival; GO, gene ontology; KEGG, Kyoto Encyclopedia of genes and genomes; HR, hazard ratio; AUC, areas under the curve;
:Tianjin Education Commission Research Project (Grant 2019KJ055). Extension Project of First Teaching Hospital of Tianjin University of Traditional Chinese Medicine (Grant 201911).
:Guo Shanqi, the main author of study, conceived and designed the analysis and wrote the manuscript. Li Xiaojiang designed the study, Deng Hao revised the manuscript. Jia Yingjie analyzed the data and conducted the results. All authors read and approved the final manuscript.
: The authors declare that they have no conflict of interest.
: Guo SQ, Jia YJ, Hao D, Li XJ. Evaluation of autophagy-related genes and lncRNAs signature for prognositic prediction in thyroid carcinoma via bioinformatics analysis.. 2021;4(2):8.
:Ying Chen.
: 20 October 2020,
15 May 2020,
: 16 May 2021
© 2021 By Authors. Published by TMR Publishing Group Limited. This is an open access article under the CC-BY license (http://creativecommons.org/licenses/BY/4.0/)
杂志排行
Clinical Research Communications的其它文章
- Role of herbal medicines to treat the symptoms of COVID-19 disease
- Cardiac exercise rehabilitation for heart failure: a review of the literature
- Screening of acupuncture treatment for simple obesity
- Clinical observation of Shengxuebao Mixture in treating anemia after concurrent chemoradiotherapy for cervical cancer
