Osa-miR439 Negatively Regulates Rice Immunity Against Magnaporthe oryzae
2021-03-20LuJunhuaYangXuemeiChenJinfengLiTingtingHuZijinXieYingLiJinluZhaoJiqun1PuMei1FengHui1FanJing1HuangYanyan1ZhangJiwei1WangWenming1LiYan1
Lu Junhua, Yang Xuemei, Chen Jinfeng, Li Tingting, Hu Zijin, Xie Ying, Li Jinlu, Zhao Jiqun1, , Pu Mei1, , Feng Hui1, , Fan Jing1, , Huang Yanyan1, , Zhang Jiwei1, ,Wang Wenming1, , Li Yan1,
Research Paper
Osa-miR439 Negatively Regulates Rice Immunity Against
Lu Junhua2, #, Yang Xuemei2, #, Chen Jinfeng2, Li Tingting2, Hu Zijin2, Xie Ying2, Li Jinlu2, Zhao Jiqun1, 2, Pu Mei1, 2, Feng Hui1, 2, Fan Jing1, 2, Huang Yanyan1, 2, Zhang Jiwei1, 2,Wang Wenming1, 2, Li Yan1, 2
(State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu 611130, China; Rice Research Institute and Key Laboratory for Major Crop Diseases, Sichuan Agricultural University, Chengdu 611130, China; These authors contributed equally to this work)
Osa-miR439 is a rice-specific microRNA family. Here we showed that Osa-miR439 acted as a negative regulator in rice immunity against blast fungus. Osa-miR439 differentially responded tobetween susceptible and resistant rice accessions. The accumulation of Osa-miR439 was constitutively more in the susceptible accession than in the resistant one. Transgenic lines overexpressing Osa-miR439a (OX439a) showed higher susceptibility associating with lower induction of defense-related genes and less hydrogen peroxide (H2O2) accumulation at the infection sites than the control plants. In contrast, transgenic lines expressing a target mimic of Osa-miR439 (MIM439) displayed compromised susceptibility associating with increased H2O2accumulation. Furthermore, we found that the expression of three predicted target genes was decreased in OX439a but increased in MIM439 in comparison to control plants, and this expression was differential in susceptible and resistant accessions uponinfection, indicating that Osa-miR439a may regulate rice blast resistance via these genes. Our results unveiled the role of Osa-miR439a in rice blast resistance and provided the potentiality to improve the blast resistance via miRNA.
Osa-miR439; rice blast; resistance; target gene
Plants possess an immunity system consisting of two perception layers, i.e. pathogen/microbe-associated molecular pattern (PAMP/MAMP)-triggered immunity (PTI/MTI) and effector-triggered immunity (ETI), to fight against pathogen invasion (Jones and Dangl, 2006). The recognition between PAMPs/MAMPs and plant pattern recognition receptors (PRRs) activates PTI/MTI, which effectively protects plants from the invasion of potential pathogens (Jones and Dangl, 2006; Boller and He, 2009). However, adapted pathogens can block PTI/MTI by secreting virulent effectors. In turn, host plants have evolved resistance proteins to trigger ETI by recognizing the cognate effectors to counterattack their virulence (Jones and Dangl, 2006). ETI is often associated with strong defense responses, including the hypersensitive response (HR) to protect plants against pathogens (Cui et al, 2015).
Rice is one of the most critical food crops. Rice blast disease caused by fungusis the most severe disease threating rice production worldwide (Baldrich and San Segundo, 2016). Both PTI and ETI are involved in rice immunity against.(Liu et al, 2013; Liu and Wang, 2016). In rice, several PPRs engaging in PTI have been identified. For example, OsCEBiP, OsLYP4 and OsLYP6 act as the receptor of chitin derived by fungi, and OsFLS2 recognizes bacterial flagellin (Chen and Ronald, 2011; Liu et al, 2013). Besides, ETI is well-studied in rice and found to mount strong defense responses against. ETI is usually mediated by nucleotide- binding site/leucine-rich repeat (NBS-LRR) proteins (Liu et al, 2013). To date, 102 rice blast resistance (R) genes have been identified, out of which more than 30 genes have been functionally characterized, and many of these R genes have been used in rice breeding (Xie et al, 2019).
Except for R genes, miRNAs act as regulators in rice immunity against pathogen invasion (Li et al, 2019). miRNAs are a kind of 20–24-nt noncoding RNAs that play vital roles in plant development and defense responses via negatively regulating target gene expression (Padmanabhan et al, 2009; Katiyar- Agarwal and Jin, 2010; Baldrich et al, 2014). Increasing evidence reveals that miRNAs modulate rice immunity againstvia regulating the expression of target genes (Campo et al, 2013; Li et al, 2014; Baldrich and San Segundo, 2016). For example, miR7695 promotes immunity againstby negatively regulating(Campo et al, 2013). miR398b positively contributes to resistance by repressing the expression of Cu/Zn-Superoxide Dismutases (CSDs) (Li et al, 2019). The transgenic lines overexpressing miR160a show enhanced resistance associating with decreased expression of three Auxin Response Factors (ARFs) (Li et al, 2014). The transgeniclines overexpressing miR166h-166k polycistron display enhanced resistance and activated ethylene signaling accompanying with suppressed expression of ethylene-insensitive 2 (EIN2) family genes (Salvador-Guirao et al, 2018). In contrast, several miRNAs act as negative regulators in rice blast resistance. miR164a negatively regulates rice resistance againstvia targeting NAC/ATAF/CUC (NAC) genes (Wang et al, 2018). miR167d compromises rice immunity againstby suppressing target genes(Zhao et al, 2019). miR169a facilitates blast fungus invasion and compromises the induction of defense-related genes and accumulation of hydrogen peroxide (Li et al, 2017). miR319 targets(), a transcription factor acting as a positive regulator of the rice blast resistance, to facilitateinvasion (Zhang et al, 2018). Over- expression of miR444b.2 leads to higher susceptibility toaccompanying with suppressed expression of MADS family genes (Xiao et al, 2017). Intriguingly, miR1873 fine-tunes rice immunity againstand yield traits via(Zhou et al, 2019). miR396 regulates both immunity and growth. Overexpression of miR396 isoforms leads to higher susceptibility toby suppressing multiple, includingand, which positively regulate growth and yield (Chandran et al, 2019). Therefore, the miR1873-module and miR396-module can be potential targets to demolish fitness costs in rice blast resistance breeding. However, whether other miRNAs are involved in rice-interaction remains unknown.
Osa-miR439 is a rice-specific miRNA family containing 10 loci generating the identical mature 21-nt sequences (Sunkar et al, 2005). Alignment analysis indicates that Osa-miR439 derives from the insertion of the rice transposable element MuDR4-OS, which is an inverted repeat followed by multiple repeats (Yu et al, 2010). Small RNA-seq data revealed that the amounts of Osa-miR439 are changed uponinvasion, indicating a role in responses to this pathogen (Li et al, 2014). We made transgenic lines overexpressing Osa- miR439a (OX439a) and its target mimic (MIM439) to detect the roles of Osa-miR439a in rice blast resistance. Our results revealed that Osa-miR439a negativelyregulated rice immunity against, whereasblocking its suppression on target genes via expressing a target mimic improved blast resistance. Thus, this study identified a miRNA that had potentiality in the improvement of blast resistance.
RESULTS
Osa-miR439a was responsive to blast fungus and chitin
We clarified the responses of Osa-miR439a toinfection by analyzing the accumulation of Osa-miR439a in highly susceptible Lijiangxintuanheigu (LTH) and monogenic resistance accession International Rice Blast Line Pyricularia-Kanto51-m-Tsuyuake (IRBLkm- Ts). Initially, we spray-inoculated LTH and IRBLkm- Ts seedlings at three-leaf-stage withstrain Guy11. LTH displayed acute susceptible phenotype with many large disease lesions at 5 d post-inoculation (dpi), whereas IRBLkm-Ts showed resistant phenotype with high resistance lesions (Fig. 1-A). We next examined the accumulation pattern of Osa-miR439ain the inoculated leaves of both accessions. Intriguingly, withoutinfection, the accumulation of Osa-miR439a was significantly higher in LTH than in IRBLkm-Ts. Upon.infection, the amounts of Osa-miR439awere significantly increased at 12 and 24 h post-inoculation (hpi) in LTH but decreased at 48 hpi (Fig. 1-B). In contrast, the accumulation of Osa- miR439awas increased dramatically at 12 hpi but kept unchanged at 24 and 48 hpi in IRBLkm-Ts (Fig. 1-B). These results indicated that Osa-miR439a was responsive to.and possibly negatively regulated rice blast resistance because its amounts were higher in the susceptible accession than in the resistance accession regardless of.infection. Moreover, we detected Osa-miR439a abundance with or without chitin treatment to detect its involvement in PTI. Osa-miR439a was decreased at 1 and 6 hpi but increased at 3 hpi in LTH (Fig. 1-C), indicating Osa- miR439a was involved in the regulation of rice PTI.
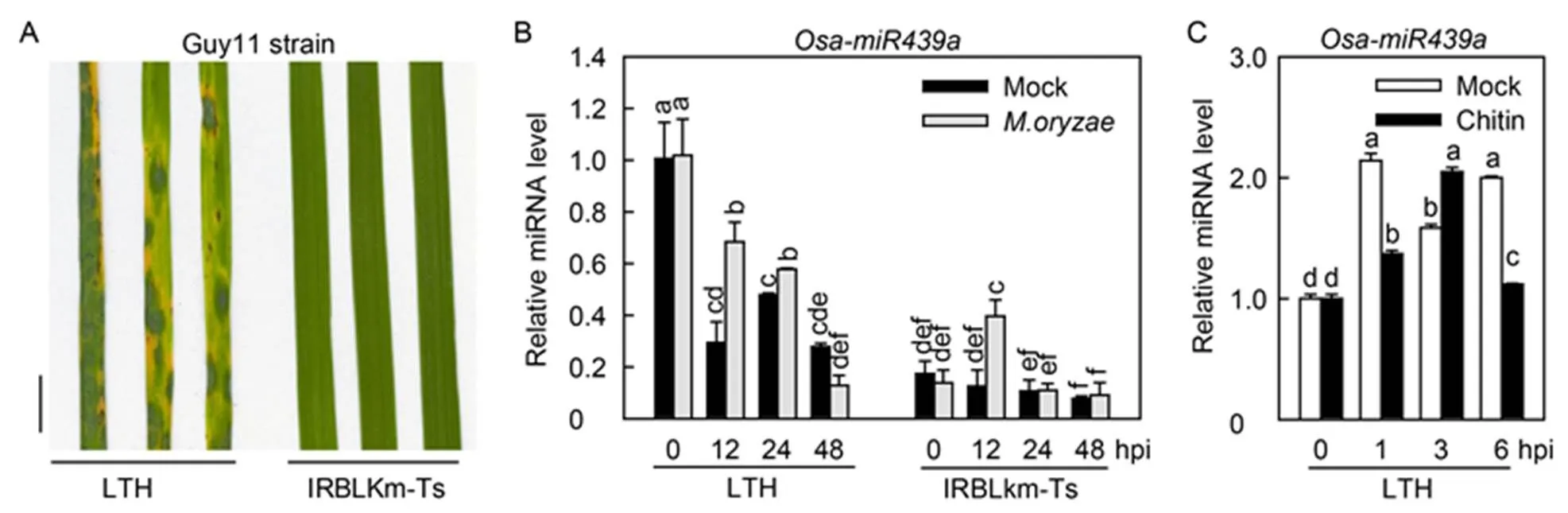
Fig. 1. Osa-miR439a is differentially responsive toin susceptible and resistant rice accessions.
A, Blast diseasephenotype of representative leaf sections from Lijiangxintuanheigu (LTH) and International Rice Blast Line Pyricularia-Kanto51- m-Tsuyuake (IRBLkm-Ts) upon spray-inoculation ofstrain Guy11 (1 × 105spore/mL) at 5 d post- inoculation (dpi). Scale bar, 1 cm.
B, Accumulation of Osa-miR439a in LTH and IRBLkm-Ts uponmock infection at indicated time points.
C, Accumulation of Osa-miR439a in LTH with or without chitin treatment at indicated time points.
hpi, Hours post-inoculation. In B and C, Error bars indicate SD (= 3). Different letters above the bars indicate significant differences (< 0.01) as determined by one-way analysis of variance followed byTukey’s Honestly Significant Difference analysis.
Overexpression of Osa-miR439a resulted in enhanced susceptibility to M. oryzae
We constructed transgenic rice lines OX439a, in which Osa-miR439a was overexpressed by 35S promoter within Nipponbare (NPB) background to identify the role of Osa-miR439a in rice immunity against. We selected two lines that showed up-regulated accumulation of Osa-miR439a for blast disease assay (Fig. 2-A). Three-leaf-stage seedlings were spray- inoculated with.virulent strain 97-27-2, and the disease phenotype was recorded at 5 dpi. OX439a lines (OX439a#1 and OX439a#2) displayed enhanced susceptibility associating with more and larger necrotic disease lesions than wild type control (Fig. 2-B). Consistently, OX439a lines supported significantly more fungal growth than WT (Fig. 2-C). Meanwhile, we also conducted disease assay via punch-inoculation with.strains eGFP-tagged Zhong8-10-14 (GZ8) and TJ13. Both strains formed larger disease lesions and proliferated more fungal biomass in OX439a than WT (Fig. S1). These results indicated that Osa-miR439a negatively regulated rice immunity against.
To understand how OX439a lines supported more fungal growth, we observed the infection process of eGFP-tagged strain GZ8 on sheath cells. Consistent with the disease phenotypes, the infection progress was faster in OX439a than in WT. In the WT plants, about 60% of spores formed invasive hyphae at 24 hpi, and approximately 40% invasive hyphae expanded into neighbor cells at 36 hpi. In contrast, in OX439a, over 80% inoculated spores formed invasive hyphae in sheath cells at 24 hpi, and approximately 60% of invasive hyphae expanded into neighbor cells at 36 hpi (Fig. 2-D and -E). These observations indicated that Osa-miR439a facilitated the invasion and proliferation of.
Overexpression of Osa-miR439a compromised defense responses
To clarify why overexpression of Osa-miR439a led to enhanced susceptibility, we examined defense responses, including the induction of defense-related genesand, as well as production of H2O2.is a PTI-related gene, the expression of which can be induced by chitin andat early time points (Park et al, 2012; Li et al, 2014), whereasis involved in phytoalexin biosynthesis (Miyamoto et al, 2018). The expression ofwas increased in the WT plants upon 97-27-2 infection, but decreased in OX439a lines at 48 hpi;was increased at all the three time points in WT, whereas increased at 12 hpi but decreased at 24 and 48 hpi in OX439a, suggesting that overexpression of Osa-miR439a compromised the induction of defense-related genes (Fig. 3-A and -B). H2O2plays a key role in rice resistance against(Shimono et al, 2012). Consistent with the suppressed induction of defense- related genes, the infectionofinduced H2O2accumulation in the infected leaves of WT plants but triggered less H2O2in OX439a lines (Fig. 3-C). These results indicated that overexpression of Osa-miR439a compromised-induced defense responses.

Fig. 2. Overexpressing Osa-miR439a resulted in enhanced susceptibility to.
A, Accumulation of Osa-miR439a in wild type (WT) control (Nipponbare) and the transgenic lines harboring(OX439a#1 and OX439a#2).
B, Blast disease phenotypes on leaves of the indicated lines at 6 d post-inoculation ofstrain 97-27-2. Scale bar, 1 cm.
C, Quantification analysis of the blast disease in B. Relative fungal biomass was determined by detecting the relative levels ofDNA against riceDNA.
D, Representative laser scanning confocal microscopy images showing the growth of thestrain GZ8 at 24 and 36 h post- inoculation (hpi) on sheath cells of the indicated lines. AP, Appressorium; IH, Invasive hyphal. Scale bars, 20 µm.
E, Quantification analysis of the progress of fungal infection at 24 and 36 hpi. Over 200 conidia in each line were analyzed.
In A and C, error bars indicate SD (= 3), and the letters above the bars indicate significant differences (< 0.01) as determined by one- way analysis of variance followed byTukey’s Honestly Significant Difference analysis.
Blocking Osa-miR439a enhanced rice resistance against M. oryzae
A target mimic of miRNA contains reversed complementary sequences of miRNA with insertion of three nucleotide acids between 10 and 11 nucleotide acids and acts as a sponge to capture miRNAs to prevent the silencing of targets (Wu et al, 2013). To confirm the role of Osa-miR439a in rice immunity against blast fungus, we made transgenic lines MIM439, in which a target mimic of Osa-miR439a was overexpressed under the NPB background. The accumulation of Osa-miR439a was significantly reduced in MIM439 in comparison with WT (Fig. 4-A). We next performed a disease assay by punch-inoculation ofstrain 97-27-2. The lesions in MIM439 were partially limited compared with those in WT (Fig. 4-B), indicating blocking Osa-miR439a led to enhanced resistance against. Consistently, the fungal biomass in the inoculated sites of MIM439 was significantly less than that in WT (Fig. 4-C). Similar results were obtained when inoculated with.strain GZ8 (Fig. S2-A and -B). Besides, 3,3′- diaminobenzidine (DAB)-staining showed that MIM439 produced more H2O2than WT following spray-inoculation ofspores (Fig. 4-D). These results indicated that blocking Osa-miR439a by expressing a target mimic enhanced rice immunity against
Predicted target genes of Osa-miR439a were differentially responsive to M. oryzae
We tried to identify Osa-miR439a target genes and examined whether these genes were responsive to. Previously, four genes were predicted as the target genes of Osa-miR439a, including_,_,_and_(Wu et al, 2009). However,_is no longer available in the database, and_does not express in leaves. Therefore, we excluded them in further analysis. Also, we conducted prediction again at a website (http:// plantgrn.noble.org/psRNATarget/) and selected three more predicted genes for further study, including_,_and_. The binding sites of the predicted target genes located in the coding region (Fig. 5-A). We then examined the expression of these genes in both OX439a and MIM439. Intriguingly, three candidates, namely_,_and_, showed significantly lower mRNA abundance in OX439a but statistically higher amounts in MIM439 than those in WT (Fig. 5-B and -C), indicating that Osa-miR439a probably targeted these three genes. Conversely, another two candidates,_and_, showed no converse alteration in OX439a and MIM439 (Fig. 5-B and -C), suggesting that Osa-miR439a may not regulate these genes at the post-transcriptional level.

Fig. 3. Overexpression of Osa-miR439a results in compromised disease-related defense responses.
A and B, Expression of the defense-related genes (and) in wild type (WT) and overexpression transgenic lines (OX439a) upon the infection ofstrain 97-27-2. RNA was extracted at the indicated time points for qRT-PCR assay. All the mRNA levels were normalized to that in the WT at 0 h post-inoculation (hpi). Error bars indicate SD (= 3). Different lowercase letters above the bars indicate significant differences (< 0.01) as determined by one-way analysis of variance followed byTukey’s Honestly Significant Difference analysis.
C, 3,3′-diaminobenzidine staining showing H2O2accumulation at 2 d post-inoculation ofstrain 97-27-2. The photos at the top portion were taken with a stereo-microscope (Scale bars, 500 µm), while the photos at down portion were taken with a Zeiss-microscope (Zeiss imager A2) (Scale bars, 20 µm). AP, Appressorium.

Fig. 4. Overexpression of a target mimic of Osa- miR439a leads to compromised susceptibility to.
A, Accumulation of Osa-miR439a in wild type (WT, Nipponbare) and transgenic lines expressing the target mimic of Osa-miR439a (MIM439).
B, Blast disease phenotypes on leaves of the indicated lines at 7 d post-inoculation (dpi) ofstrain 97-27-2 (1 × 103spore/mL). Scale bar, 1 cm.
C, Quantification analysis of the blast disease in B. Relative fungal biomass was determined by detecting the expression level ofgene against riceDNA level.
D, 3,3′-diaminobenzidine staining showing H2O2accumulation at 2 dpi ofstrain 97-27-2. The photos at the top portion were taken with a stereo-microscope (Scale bars, 500 µm). The photos at down portion were taken with a Zeiss- microscope (Zeiss imager A2) (Scale bars, 20 µm). AP, Appressorium.
In A and C, error bars indicate SD (= 3), and different lowercase letters above the bars indicate significant differences (< 0.01) as determined by one-way analysis of variance followed byTukey’s Honestly Significant Difference analysis.
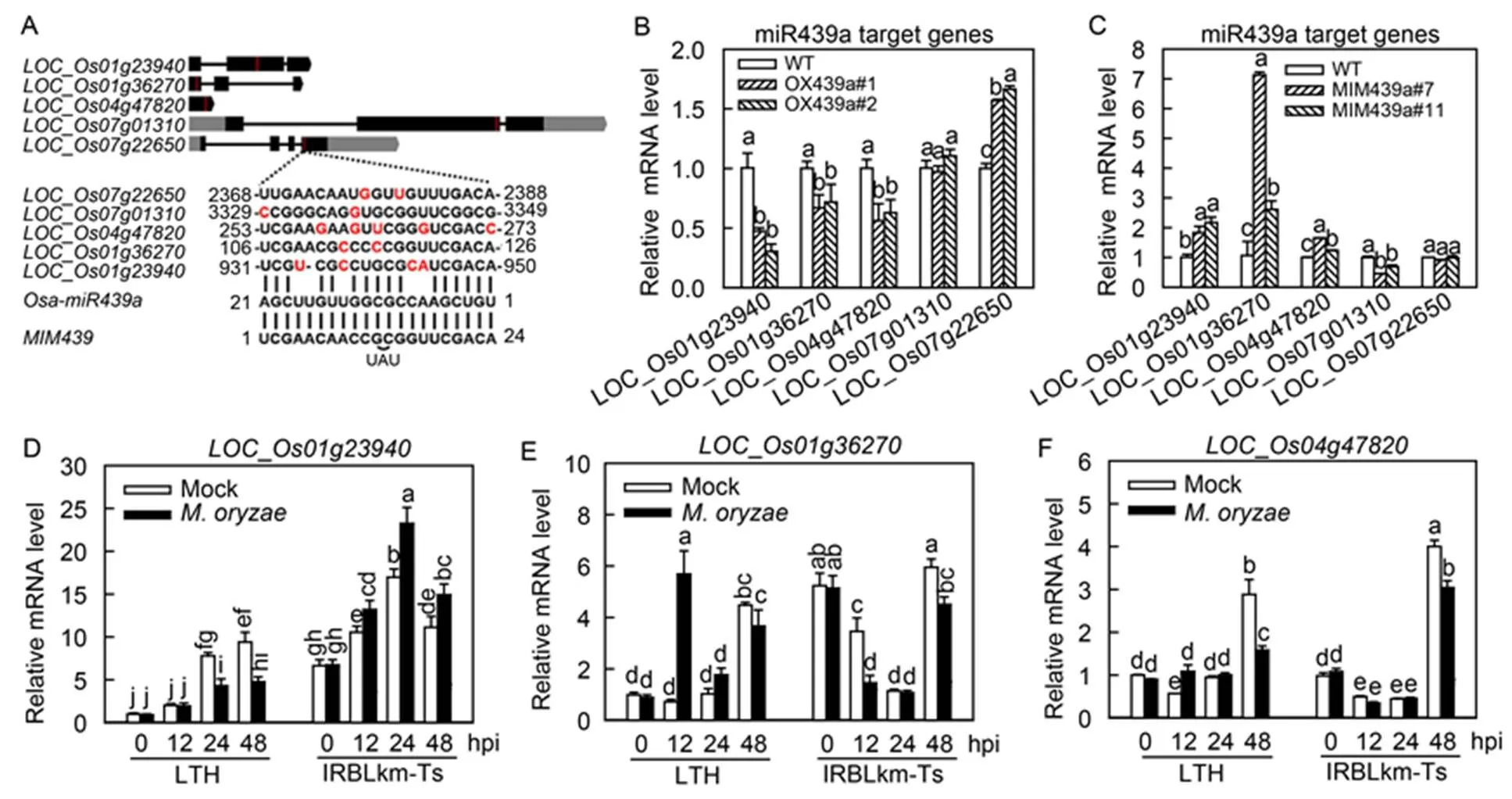
Fig. 5. Expression of Osa-miR439a predicted target genes was differently responsive toin susceptible and resistant rice accessions.
A, Graphs show the gene structure of the indicated Osa-miR439a target genes and the alignment of the target sequences with Osa-miR439a and MIM439 (target mimic of Osa-miR439a). Red lines indicate the target sites in the exon of target genes. Mismatched nucleotides are highlighted in red colors. UAU are insert bases.
B and C, Relative mRNA levels of the target genes in the wild type (WT) and the indicated transgenic lines.
D–F, Expression patterns of the indicated target genes in Lijiangxintuanheigu (LTH) and International Rice Blast Line Pyricularia-Kanto51-m- Tsuyuake (IRBLkm-Ts) uponinfection. RNA was extracted at the indicated time points for qRT-PCR analysis. mRNA level was normalized to that in untreated samples (0 h post-inoculation, 0 hpi).
In B–F, Different lowercase letters above the bars indicate significant differences (< 0.01) as determined by one-way analysis of variance followed byTukey’s Honestly Significant Difference (HSD) analysis. Error bars indicate SD (= 3).
Next, we hypothesized that if Osa-miR439a targeted_,_and_to regulate immunity, these genes should be responsive to. To test this hypothesis, we performed a time-course study to examine their expression in LTH and IRBLkm-Ts uponinfection. All the three genes were differentially responsive toin LTH and IRBLkm-Ts. The mRNA amountsof_kept stable at 12 hpi and was down-regulated significantly at 24 and 48 hpi in LTH, whereas up-regulated at all time points in IRBLkm-Ts (Fig. 5-D)._was increased at 12 hpi but kept stable at 24 and 48 hpi in LTH, whereas decreased at 12 and 48 hpi but kept stable at 24 hpi in IRBLkm-Ts (Fig. 5-E). The expressionof_was increased at 12 hpi but decreased at 48 hpi in LTH, whereas kept stable at 12 and 24 hpi but decreased at 48 hpi in IRBLkm-Ts (Fig. 5-F). These results indicated that all the three genes were involved in rice immunity against, and_most likely contributed to the resistant phenotype of IRBLkm-Ts because its induction was decreased in LTH but enhanced in IRBLkm-Ts. However, the expression patterns of all the three genes were not reversely associated with the expression pattern of Osa-miR439a, indicating Osa- miR439a and some other factors cooperated in regulation of the expression of these genes duringinfection.
To further detect whether Osa-miR439a directly targeted these responsive genes, we made constructs expressing yellow fluorescence protein (YFP) fused with the cDNA or target sites of these target genes at its 5′-terminus. We got YFP-fused wild type cDNA () or with a mutated target site() that abolished recognition by Osa-miR439a. The YFP intensity expressed either from wild type or mutated target sites was unchanged with or without co-expression of Osa-miR439a in(Fig. S3-A), and also stable when co-expressed with Osa-miR439aand MIM439 (Fig. S3-B), indicating that miR439a did not repress the expression of_. Thus,_was possibly not the target of Osa-miR439a. Unfortunately, we failed in the construction of the fused reporter genes of the other two predicted target genes. Whether_and_are real target genes of Osa-miR439a need further study.
DISCUSSION
A subset of miRNAs is responsive to.or its elicitors, and many of them act as candidates functioning in rice immunity against(Campo et al, 2013; Li et al, 2014). Among these miRNAs, 11 miRNAs have been functionally characterized to fine-tune rice blast resistance, including four miRNAs playing positiveroles, miR7695 (Campo et al, 2013), miR160a (Li et al, 2014), miR398b (Li et al, 2019) and miR166k-166h (Salvador-Guirao et al, 2018), and seven miRNAs playing negative roles, miR169 (Li et al, 2017), miR444b.2 (Xiao et al, 2017), miR319b (Zhang et al, 2018), miR164a (Wang et al, 2018), miR167d (Zhao et al, 2019), miR396 (Chandran et al, 2019) and miR1873 (Zhou et al, 2019). In this study, we characterized Osa-miR439a as a novel negative regulator in rice blast resistance.
We obtained evidence showing that Osa-miR439a played negative role in rice immunity against.. First, the accumulation of Osa-miR439a was constitutively higher and more significantly up-regulated byinfection in the susceptible accession than in the resistant one (Fig. 1), which was consistent with the previous high-throughput small RNA-seq data (Li et al, 2014). Second, overexpression of Osa-miR439a resulted in enhanced susceptibility, increased hypha proliferation at the early invasion stage, reduced induction of defense-related genes, and less production of H2O2upon.inoculation (Figs. 2 and 3, and Fig. S1). In contrast, transgenic plants expressing a target mimic of Osa-miR439a displayed enhanced resistance associating with increased H2O2accumulation (Fig. 4 and Fig. S2). At the current stage, however, we cannot explain how Osa-miR439a achieved its regulatory roles in rice immunity against..
It seemed quite challenging to identify the target genes of Osa-miR439a. Osa-miR439 is a rice-specific miRNA family. The expression of Osa-miR439a is abundant in seedlings and lower in leaves than in roots and inflorescence (Sunkar et al, 2005). Osa-miR439 family contains 10 members that derive from transposable elements (Yu et al, 2010). The 10 loci generate identical mature sequences, and hence suppress the same target genes. Theoretically, the expression of a target gene should be reversely altered between in OX439a and in MIM439. Accordingly, we found that the mRNA abundances of_,_and_were significantly decreased in OX439a and increased in MIM439 (Fig. S3), indicating that Osa-miR439a targeted these genes. We demonstrated that the protein level ofwas not suppressed by Osa-miR439a via transient expression assay, indicating Osa-miR439a did not directly target_. However, we failed to demonstrate that the other two predicted candidates were targeted directly by Osa-miR439a. We further speculated that the target genes should be responsive to.if they contributed to Osa- miR439a-mediated regulatory roles. Thus, we examined their expression patterns in LTH and IRBLkm-Ts upon.infection. We found that they were all differentially responsive to.(Fig. 5-D and -E). However, only_displayed patterns that imply a positive role in rice immunity against.. Therefore, future work should focus on confirmation of the authentic target genes of Osa-miR439a and the characterization of their contribution to Osa-miR439a-mediated regulation on rice immunity against..
This study functionally characterized that Osa- miR439a played negative roles in rice immunity against. Overexpression of Osa-miR439a led to enhanced susceptibility, whereas overexpression of MIM439 resulted in enhanced resistance. These results provided valuable information about the roles of Osa-miR439a in rice immunity, and also offered valuable candidate targets in breeding programs aiming to improve rice blast resistance.
METHODS
Plant materials and growth conditions
subsp.accession NPB, susceptible accession LTH and resistant accession IRBLkm-Ts were used. IRBLkm-Ts carries the R locusand exhibits high resistance toisolates carrying(Tsumematsu et al, 2000). NPB was the background material for transgenicanalysis. We planted all rice plants in a growth room maintained at 26 ºC and 70% relative humidity with 12 h light, followed by 12 h dark.
Plasmid construction and genetic transformation
We amplified the genomic sequence offrom NPB genomic DNA using the specific primers Osa-miR439a-I-F and Osa-miR439a-I-R (Table S1) to construct the transgenic line overexpressing. The amplified fragments contained 186 bp upstream and 162 bp downstream sequences. We cloned the fragments into the binary vector 35S- pCAMBIA1300 viaI andI sites. To make the transgenic lines overexpressing a target mimic of, we inserted the artificial target mimic sequence ofinto() gene which contains a mismatched loop at thecleavage site (Franco-Zorrilla et al, 2007) to substitute thetarget site as described previously (Zhao et al, 2019). The target mimic ofwas formed via annealing with primers MIM439a-F and MIM439a-R (Table S1). Then, the mutated fragments were cloned intoHI/II sites of the binary vector 35S-pCAMBIA1300, resulting in a clone oftarget mimic. We transformed both constructs into NPB viastrain GV3101. Positive transgenic lines were screened through hygromycin B resistance analysis.
Pathogen infection and microscopy analysis
Fourstrains, Guy11, 92-27-2, TJ13 and GZ8, were used. We incubated these strains on oatmeal and tomato agar media at 28 ºC with 12 h light followed by 12 h dark treatment. Ten days later, the hyphae were scratched, and the plates were further incubated at 28ºC with 24 h light treatment for 3 d to promote sporulation. Then, the spores were collected for spray- inoculation and punch-inoculation. For spray-inoculation, we sprayed 3 × 105spore/mL onto three-leaf-stage seedlings and examined the disease phenotypes on leaves at 6 dpi. For punch- inoculation, we drop-inoculated 5 µL of spore suspension (1 × 105spore/mL) at wound sites on leaves of three-leaf-stage seedlings following Kong et al (2012). We examined the lesion formation at 4–6 dpi. The relative fungal biomass was calculated using the DNA concentration ofagainst the DNA levels of riceby quantitative PCR (Park et al, 2012).
We observed the fungalinfection processusingstrain GZ8. Spore suspension (2 × 105spore/mL) was inoculated on 5-cm-long leaf sheaths, as described previously (Kankanala et al, 2007). We excised the inoculated epidermal layer from the leaf sheaths at 24 and 36 hpi to observe fungal invasion process using a laser scanning confocal microscopy (Nikon A1, Japan). The quantitative analysis of the infestation stage was conducted as described previously (Li et al, 2014). The DAB stain method was used to detect H2O2levels in the cells of the infected leaves by a stereomicroscope (Zeiss V20, Germany) and a microscope (Zeiss imager A2, Germany) following Li et al (2014).
Quantitative real-time polymerase chain reaction (qRT-PCR)
The leaves of OX439a and MIM439 were collected at three- leaf-stage to detect miRNA amounts and the mRNA levels of target genes. To examine the expression of defense-related genes, we spray-inoculated three-leaf-stage seedlings with indicatedstrains. For chitin treatment, we cut the leaves of three-leaf-stage plants into 1-cm-long strips and incubated these strips in water with or without 1 mg/mL chitin for 0, 1, 3 and 6 h. Then, the treated strips were collected for RNA extraction. The total RNA was extracted from collected samples using TRIzol reagent (Thermo Fisher, America). The RNA quality and quantity were determined by a NanoDrop 2000 UVevis spectrometer (Thermo Fisher, America). We carried out reverse transcription to cDNA using the SuperScript First-Strand Synthesis Kit (Thermo Fisher, America). To analyze the expression of miRNA, we performed stem-loop pulse qRT-PCR following Varkonyi-Gasic et al (2007) and selected U6 snRNA as an internal reference to normalize miRNA amounts (Turner et al, 2013).
Agrobacterium-mediated transient expression assay in N. benthamiana
Target gene detection and accumulation were assayed according to Li et al (2017). To express LOC_Os04g47820-fused YFP, we fused YFP with the full CDS sequence ofat its 5′-terminal () or with mutated target site () that cannot be recognized by Osa-miR439. The sequences ofandwere amplified from NPB cDNA using indicated primers, respectively (Table S1). The isolated fragments were then fused to the N-terminus of YFP, and the fused fragments were inserted intoII sites of binary vector 35S- pCAMBIA1300.strain GV3101 was used forinfection assay in. In brief, we incubatedthestrains harboring the respectiveexpression constructs (,,and) at 28 ºC overnight in Luria-Bertani (LB)media (50 mg/mL of, 50 mg/mL ofand 50 mg/mL of). Then, we collected the bacteria at 4 000 r/min for 5 min and resuspended the bacteria in anMMA buffer [10 mmol/L of 2-morpholino- ethanesulfonic acid (MES), 10 mmol/L of MgCl2, 100 µmol/L of acetosyringone (AS)]. We infiltrated the collected bacteriainto leaves offortransient expression assay, and examined the accumulation of the fused proteins at 48 hpi via image acquisition using a confocal laser scanning microscope (Nikon A1, Japan).
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31471761, 31430072 and 31672090). We thank Dr. Lei Cailin (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the monogenic resistant line IRBLkm-Ts.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/ rice-science; http://www.ricescience.org.
Fig. S1. Overexpression of Osa-miR439a results in enhanced susceptibility to
Fig. S2. Overexpression of a target mimicry of Osa-miR439a leads to compromised susceptibility to.
Fig. S3. Osa-miR439a has no effects on the expression of its target geneat the translational level.
Table S1. Primers used in this study.
Table S1. Primers used in the study.

PrimersSequence(5′–3′)Objective miR439a-F-Sac1AATGAGCTCTCTGCGAGCACTGTAGCATCACMake OX439a miR439a-R-Spe1ACGACTAGTTCCACCAGAGAGCGATGAGAAGMake OX439a MIM439a-BamH1-FGATCCTCGAACAACCGTAT CGGTTCGACAAMake MIM439a MIM439a-BglII-RGATCTTGTCGAACCGATACGGTTGTTCGAGMake MIM439a miR439a RT-FAGTGCCCGTGTCGAACCGCqRT-PCR miR439a RT-RCAGTGCAGGGTCCGAGGTATqRT-PCR miR439a-3'stem loop GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCGAACAACCqRT-PCR miR439LOC_Os01g23940-FCGTCCACAACCACTCTACGGqRT-PCR miR439LOC_Os01g23940-RTGTAGCCGAAGTCAGGAAGCqRT-PCR miR439LOC_Os01g36270-FAACTACCGGAATGTCCCTGCTqRT-PCR miR439LOC_Os01g36270-RTGTCCAGATGTCTATGACCGGATqRT-PCR miR439LOC_Os04g47820-FTTCCCCGTGGAGTCGAAGAAqRT-PCR miR439LOC_Os04g47820-RACTTGCAGTATCGGTCGGCqRT-PCR miR439LOC_Os07g01310-FCTTCTTCCTTTTCCTCCGTCTAqRT-PCR miR439LOC_Os07g01310-RACACGGCTAATCATCGGATAATqRT-PCR miR439LOC_Os07g22650-FTACGGTTGTTTGATAGGGGCqRT-PCR miR439LOC_Os07g22650-RATTGTACCCCTTTGCCTGCTTqRT-PCR OsNAC4-F for RTTCCTGCCACCATTCTGAGATG3qRT-PCR OsNAC4-R for RTTTGCAGAATCATGCTTGCCAG3qRT-PCR Os04g10010(OsMAS1)-F for RTAAATGATTTGGGACCAGTCGqRT-PCR Os04g10010(OsMAS1)-R for RTGATGGAATGTCCTCGCAAACqRT-PCR U6 RT-FCGATAAAATTGGAACGATACAGAqRT-PCR U6 RT-RATTTGGACCATTTCTCGATTTGTqRT-PCR ubi-F for RTGCCCAAGAAGAAGATCAAGAACqRT-PCR ubi-R for RTAGATAACAACGGAAGCATAAAAGTCqRT-PCR MoPot2_F ACGACCCGTCTTTACTTATTTGG M. oryzea quantility identification gene MoPot2_R AAGTAGCGTTGGTTTTGTTGGAT M. oryzea quantility identification gene

Baldrich P, Kakar K, Sire C, Moreno A B, Berger A, Garcia-Chapa M, Lopez-Moya J J, Riechmann J L, San Segundo B. 2014. Small RNA profiling reveals regulation ofmiR168 and heterochromatic siRNA415 in response to fungal elicitors., 15(1): 1083.
Baldrich P, San Segundo B. 2016. MicroRNAs in rice innate immunity., 9: 6.
Boller T, He S Y. 2009. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens., 324: 742–744.
Campo S, Peris-Peris C, Sire C, Moreno A B, Donaire L, Zytnicki M, Notredame C, Llave C, Segundo B S. 2013. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the() gene involved in pathogen resistance., 199(1): 212–227.
Chandran V, Wang H, Gao F, Cao X L, Chen Y P, Li G B, Zhu Y, Yang X M, Zhang L L, Zhao Z X, Zhao J H, Wang Y G, Li S C, Fan J, Li Y, Zhao J Q, Li S Q, Wang W M. 2019. miR396- OsGRFs module balances growth and rice blast disease- resistance.,9: 1999.
Chen X W, Ronald P C. 2011. Innate immunity in rice., 16(8): 451–459.
Cui H T, Tsuda K, Parker J E. 2015. Effector-triggered immunity: From pathogen perception to robust defense.,66: 487–511.
Franco-Zorrilla J M, Valli A, Todesco M, Mateos I, Puga M I, Rubio-Somoza I, Leyva A, Weigel D, Garcia J A, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity., 39(8): 1033–1037.
Jones J D G, Dangl J L. 2006. The plant immune system.,444: 323–329.
Kankanala P, Czymmek K, Valent B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus., 19(2): 706–724.
Katiyar-Agarwal S, Jin H L. 2010. Role of small RNAs in host- microbe interactions., 48: 225–246.
Kong L A, Yang J, Li G T, Qi L L, Zhang Y J, Wang C F, Zhao W S, Xu J R, Peng Y L. 2012. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus., 8(2): e1002526.
Li Y, Cao X L, Zhu Y, Yang X M, Zhang K N, Xiao Z Y, Wang H, Zhao J H, Zhang L L, Li G B, Zheng Y P, Fan J, Wang J, Chen X Q, Wu X J, Zhao J Q, Dong O X, Chen X W, Chern M, Wang W M. 2019. Osa-miR398b boosts H2O2production and rice blast disease-resistance via multiple superoxide dismutases., 222(3): 1507–1522.
Li Y, Lu Y G, Shi Y, Wu L, Xu Y J, Huang F, Guo X Y, Zhang Y, Fan J, Zhao J Q, Zhang H Y, Xu P Z, Zhou J M, Wu X J, Wang P R, Wang W M. 2014. Multiple rice microRNAs are involved in immunity against the blast fungus., 164(2): 1077–1092.
Li Y, Zhao S L, Li J L, Hu X H, Wang H, Cao X L, Xu Y J, Zhao Z X, Xiao Z Y, Yang N, Fan J, Huang F, Wang W M. 2017. Osa-miR169 negatively regulates rice immunity against the blast fungus., 8: 2.
Liu W D, Liu J L, Ning Y S, Ding B, Wang X L, Wang Z L, Wang G L. 2013. Recent progress in understanding PAMP- and effector- triggered immunity against the rice blast fungus.,6(3): 605–620.
Liu W D, Wang G L. 2016. Plant innate immunity in rice: A defense against pathogen infection., 3(3): 295–308.
Miyamoto K, Fujita M, Shenton M R, Akashi S, Sugawara C, Sakai A, Horie K, Hasegawa M, Kawaide H, Mitsuhashi W, Nojiri H, Yamane H, Kurata N, Okada K, Toyomasu T. 2018. Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice., 87: 293–304.
Padmanabhan C, Zhang X M, Jin H L. 2009. Host small RNAs are big contributors to plant innate immunity., 12(4): 465–472.
Park C H, Chen S B, Shirsekar G, Zhou B, Khang C H, Songkumarn P, Afzal A J, Ning Y S, Wang R Y, Bellizzi M, Valent B, Wang G L. 2012. Theeffector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice., 24(11): 4748–4762.
Salvador-Guirao R, Hsing Y I, San Segundo B. 2018. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2., 9: 337.
Shimono M, Koga H, Akagi A, Hayashi N, Goto S, Sawada M, Kurihara T, Matsushita A, Sugano S, Jiang C J, Kaku H, Inoue H, Takatsuji H. 2012. Rice WRKY45 plays important roles in fungal and bacterial disease resistance., 13(1): 83–94.
Sunkar R, Girke T, Jain P K, Zhu J K. 2005. Cloning and characterization of microRNAs from rice., 17: 1397–1411.
Tsumematsu H, Yanoria M J T, Ebron L A, Hayashi N, Ando I, Kato H, Imbe T, Khush G S. 2000. Development of monogenic lines of rice for blast resistance., 50(3): 229–234.
Turner M, Adhikari S, Subramanian S. 2013. Optimizing stem-loop qPCR assays through multiplexed cDNA synthesis of U6 and miRNAs.,8(8): e24918.
Varkonyi-Gasic E, Wu R M, Wood M, Walton E F, Hellens R P. 2007. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs., 3: 12.
Wang Z Y, Xia Y Q, Lin S Y, Wang Y R, Guo B H, Song X N, Ding S C, Zheng L Y, Feng R Y, Chen S L, Bao Y L, Sheng C, Zhang X, Wu J G, Niu D D, Jin H L, Zhao H W. 2018. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus.,95(4): 584–597.
Wu H J, Wang Z M, Wang M, Wang X J. 2013. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants., 161(4): 1875–1884.
WuL, Zhang Q Q, Zhou H Y, Ni F R, Wu X Y, Qi Y J. 2009. Rice microRNA effector complexes and targets., 21(11): 3421–3435.
Xiao Z Y, Wang Q X, Zhao S L, Wang H, Li J L, Fan J, Li Y, Wang W M. 2017. MiR444b.2 regulates resistance toand tillering in rice., 47(2): 511–522. (in Chinese with English abstract)
Xie Z, Yan B X, Shou J Y, Tang J, Wang X, Zhai K R, Liu J Y, Li Q, Luo M Z, Deng Y W, He Z H. 2019. A nucleotide-binding site- leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice., 374: 20180308.
Yu S W, Li J J, Luo L J. 2010. Complexity and specificity of precursor microRNAs driven by transposable elements in rice., 28: 502–511.
Yu Y, Jia T R, Chen X M. 2017. The ‘how’ and ‘where’ of plant microRNAs., 216(4): 1002–1017.
Zhang X, Bao Y L, Shan D Q, Wang Z H, Song X N, Wang J Y, Wang J S, He L Q, Wu L, Zhang Z G, Niu D D, Jin H L, Zhao H W. 2018.induces the expression of a microRNA to suppress the immune response in rice., 177(1): 352–368.
Zhao Z X, Feng Q, Cao X L, Zhu Y, Wang H, Chandran V, Fan J, Zhao J Q, Pu M, Li Y, Wang W M. 2019. Osa-miR167d facilitates infection ofin rice.,62(5): 702–715.
Zhou S X, Zhu Y, Wang L F, Zheng Y P, Chen J F, Li T T, Yang X M, Wang H, Li X P, Ma X C, Zhao J Q, Pu M, Feng H, Li Y, Fan J, Zhang J W, Huang Y Y, Wang W M. 2019. Osa-miR1873 fine-tunes rice immunity againstand yield traits.,62(8): 1213–1226.
3 October 2019;
13 May 2020
Li Yan (jiazaihy@163.com)
Copyright © 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2021.01.005
(Managing Editor: Fang Hongmin)
杂志排行
Rice Science的其它文章
- Genetic Interaction of Hd1 with Ghd7, DTH8 and Hd2 Largely Determines Eco-Geographical Adaption of Rice Varieties in Southern China
- Drought Tolerance in Rice: Focus on Recent Mechanisms and Approaches
- Genome Editing Strategies Towards Enhancement of Rice Disease Resistance
- RAVL1 Activates IDD3 to Negatively Regulate Rice Resistance to Sheath Blight Disease
- Exogenous Peroxidase Mitigates Cadmium Toxicity, Enhances Rhizobial Population and Lowers Root Knot Formation in Rice Seedlings
- Effects of Early- and Late-Sowing on Starch Accumulation and Associated Enzyme Activities During Grain Filling Stage in Rice
