RAVL1 Activates IDD3 to Negatively Regulate Rice Resistance to Sheath Blight Disease
2021-03-20SunQianYangShuoGuoXiaofanWangSitingJiaXintongLiShuangXuanYuanhu
Sun Qian, Yang Shuo, Guo Xiaofan, Wang Siting Jia Xintong Li Shuang, Xuan Yuanhu
Research Paper
RAVL1 Activatesto Negatively Regulate Rice Resistance to Sheath Blight Disease
Sun Qian1,#, Yang Shuo1,#, Guo Xiaofan2,#, Wang Siting1, Jia Xintong1, Li Shuang3, Xuan Yuanhu1
(Shaanxi Key Laboratory of Chinese Jujube / College of Life, Yan’an University,;)
Sheath blight disease (ShB) has a severe impact on the production of rice. ABI3/VP1-like 1 (RAVL1) negatively regulated the rice defense mechanism against ShB, however, this regulatory mechanism is not clearly understood. In this study, we identified that() was positively regulated by RAVL1. Further, chromatin immunoprecipitation (ChIP) assay, yeast one-hybrid assay and transient expression assay indicated a direct binding between RAVL1 and thepromoter region.was ubiquitously expressed in different tissues and at different stages, and its expression was significantly enhanced byinfection. IDD3 exhibited transcription activation activity in yeast and IDD3-GFP was found to be localized in the nucleus.mutants exhibited no significant differences in response to ShB, whileoverexpressors were more susceptible to ShB compared with wild type (WT) plants. Furthermore,repressors were less susceptible tothan WT plants. Interestingly, the expression of brassinosteroid-related genes (,and) was lower inrepressors and higher inoverexpressors compared with WT. However, the ChIP assay revealed that IDD3 did not directly bind to theandpromoters. Overexpression ofinmutantinhibited the activity of IDD3, reducing its susceptibility to ShB compared withoverexpressor and WT plants, indicating that IDD3 negatively regulated the rice defense mechanism against ShB by activating the BR signaling pathway. Thus, our analyses provided information to enhance the understanding of the rice defense mechanism against ShB.
sheath blight disease; AB13/VP1-like 1; indeterminate domain 3; rice; promoter
Sheath blight disease (ShB) is one of the three major diseases caused by the fungus,in rice () (Savary et al, 1995). It damages rice during the whole growth period and primarily infects the leaves, sheaths and panicles, eventually resulting in the withering and lodging of the whole plant (Savary et al, 1995). A severe form of ShB can reduce rice production by approximately 50% (Savary et al, 2000). Currently, the primary approach to control this disease involves the use of fungicides since there is a lack of resistant varieties against ShB (Savary et al, 2000; E et al, 2019). However, fungicides are not environment-friendly as they directly impact other microbes, and their use is also associated with an increased cost of cultivation. Thus, it is necessary to isolate resistant rice varieties and understand their defense mechanisms to develop resistance against ShB. Previous studies demonstrated that the overexpression of chitinase, β-1,3-glucanase or polygalacturonase- inhibiting protein (OsPGIP1) leads to enhanced rice resistance to(Shah et al, 2009; Mao et al, 2014; Wang et al, 2015). The overexpression of OsACS2, an ethylene synthesis enzyme, promotes the rice resistance to blast and sheath blight (Helliwell et al,2013). The overexpression of the() gene generatesand rice resistance to(Maeda et al, 2019). Also, salicylic acid-dependent immunity contributes to resistance againstin rice and(Kouzai et al, 2018). Additionally, mutation in() gene and overexpression of()/() can significantly protect rice against ShB (Gao et al, 2018). IDD13 interacts with LPA1 to enhance resistanceagainst ShB via the activation of(Sun et al, 2019).
The IDD contains two C2H2and two C2HC zinc finger motifs, and the IDD genes play diverse biological functions in plants. Previous studies have reported thatcontrols the flowering time in maize and rice (Colasanti et al, 1998; Park et al, 2008);/and/regulate the fate of root cells (Welch et al, 2007);/regulates seed maturation (Feurtado et al, 2011);modulates plant development (Seo et al, 2011);,andsynergistically regulate the lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport in(Cui et al, 2013); and/regulates shoot gravitropism and lamina joint angle (Wu et al, 2013; Liu et al, 2016). Additionally,, the regulator of, which encodes the IDD protein, activates/to regulate the tolerance against cold in rice (Dou et al, 2016).is known to regulate secondary cell wall formation in rice (Huang et al, 2018).repressor activates the immune response in(Volz et al, 2019). Previous studies have also identified the binding motifs of the transcription factor IDD in maize (, 5′-TTTGTCG/CTTTT-3′),(, 5′-TTTTGTCC-3′) and rice (, 5′-TTTGTCC/G-3′) (Kozaki et al, 2004; Seo et al, 2011; Xuan et al, 2013).
Generally, brassinosteroids (BRs) are recognized by the brassinosteroid insensitive 1 (BRI1) receptor, resulting in the dissociation of BRI1 kinase inhibitor 1 (BKI1) and association of the BRI1- associated receptor kinase 1 (BAK1). The activation of BRI1-BAK1 results in the dephosphoryla- tion and activation of two transcription factors, brassinazole-resistant 1 (BZR1) and BRI1- EMS-SUPPRESSOR 1 (BES1), which regulate the expression of target genes in response to BRs (Li and Chory, 1997; Li et al, 2002; Nam and Li, 2002; Kim and Wang, 2010; Yang et al, 2011). Theandgain-of-function mutants significantly enhance protein stability compared with BES1 and BZR1 ones, respectively, which rescues the dwarf and BR- insensitive phenotypesof theBR-receptor mutant (Wang et al, 2002; Yin et al, 2002). In rice, RAVL1 has been identified as an upstream regulator of BR homeostasis, since it binds to the E-box in the promoter sequences of BR-receptor and BR-biosynthesis genes (Je et al, 2010). Another study identified that RAVL1 modified rice defenses against ShB via the activation of BRs and ethylene signaling genes (Yuan et al, 2018). However, the regulatory role of RAVL1 in rice defense against ShB is unclear.
Thus, to analyze the regulatory mechanism of RAVL1 in modifying rice defense to sheath blight, we hypothesized that RAVL1 directly regulated, a downstream gene of RAVL1, and thus, tested the role ofin the modulation of rice resistance to ShB. Additionally, the connection between IDD3 and BR signaling was investigated. The results identified in this study will extend the understanding of the defense mechanism and RAVL1-mediated regulation of BR signaling in rice.
Results
RAVL1 directly activated IDD3
We previously identified that RAVL1 negatively regulates the rice defense mechanism against ShB (Yuan et al, 2018), and/protects rice against ShB (Sun et al, 2019). Additionally, the RNA-seq results showed that, another IDD member, is regulated by RAVL1 (Yuan et al, 2018). In this study, the qRT-PCR results showed that the expression ofwas downregulated inmutants (and), while it was upregulated in the RAVL1overexpressor plants (and) compared with the wild type (WT) (Fig. 1-A). The promoter sequence analysis identified that two E-box motifs, which were the putative RAVL1 binding sequences, appeared within 1.0 kb of thepromoter(Fig. 1-B). Results of the chromatin immuoprecipitation (ChIP) assay using 35S:GFP and 35S:RAVL1:GFP transgenic plant calli showed that the precipitation of RAVL1 enriched the P1 region but not the P2 region of thepromoter (Fig. 1-C).
Results of this binding assay were confirmed using a yeast one-hybrid assay, which indicated that RAVL1 can activate 1.0 kb() only if E-box promoters were not mutated at the P1 region of() (Fig. 1-D). These mutated promoters had the E-box element sequences CANNTG substituted with the sequence TTTTTT. Next, we performed a transactivation assay by the transient expression inprotoplasts to verify if these-elements were responsible for the transcriptional activation of thepromoter via RAVL1 pathway. Results of the transactivation assay suggested that RAVL1 transactivatedbut not(Fig. 1-E), indicating that RAVL1 might activateexpression via promoter binding.

Fig. 1. RAVL1 directly activated.
A, Expression ofwas examined andwas used as the reference gene to normalize gene expression level. The leaves from one-month-old seedlings of wild type (WT),mutants (and) and RAVL1 overexpressors (and) were used for RNA extraction using the TRIzol.
B, Diagrammatic representation of the 1.0 kbpromoter. P1 and P2 indicate the regions detected in the chromatin immunoprecipitation (ChIP)-PCR assay (C).
C, ChIP-PCR was performed to analyze the binding affinity of RAVL1 to P1 and P2 regions (B). The anti-GFP antibody was used for immune- precipitation.
D, Yeast one-hybrid assay was performed to analyze RAVL1 activation of the 1.0 kbpromoter. Yeast cells harboring either AD-RAVL1 or AD together withHis orHis were grown on synthetic dropout media lacking either Leu (-L) or Leu and His (-LH). mp, Mutated at the P1 region.
E, Transient expression assay was performed by co-transfection with p35S:RAVL1 and each of the vectors expressing the beta-glucuronidase gene () under the control of native () and E-box motif-mutated () promoters in protoplast cells. The luciferase gene driven by the 35S promoter was used as an internal control to normalize the expression of.
Data in A, C and E represent Mean ± SE (= 3). Different lowercase lowercase letters indicate significant differences at< 0.05.
Expression pattern of IDD3 and its transcriptional activity
We analyzed the expression patterns ofin leaves, shoot apecies, nodes, flowers and roots. ThemRNAs were detected in all tissues, with lower expression in the nodes and the flowers (Fig. 2-A). Next, we used transgenic plants with GUS driven by a 3.0 kb endogenouspromoterto investigate the expression pattern ofin plants. The expression of GUS in the leaf vasculatures, nodes, flowers, lateral root primordia and primary root vasculature was detected (Fig. 2-B). Then, qRT-PCR was performed usingAG1-IA inoculated leaf RNA to test whether the expression ofwas in response to ShB. The results showed that the expression ofwas enhanced at 24 and 72 h post-inoculation (hpi), while it was suppressed at 48 hpi (Fig. 2-C).
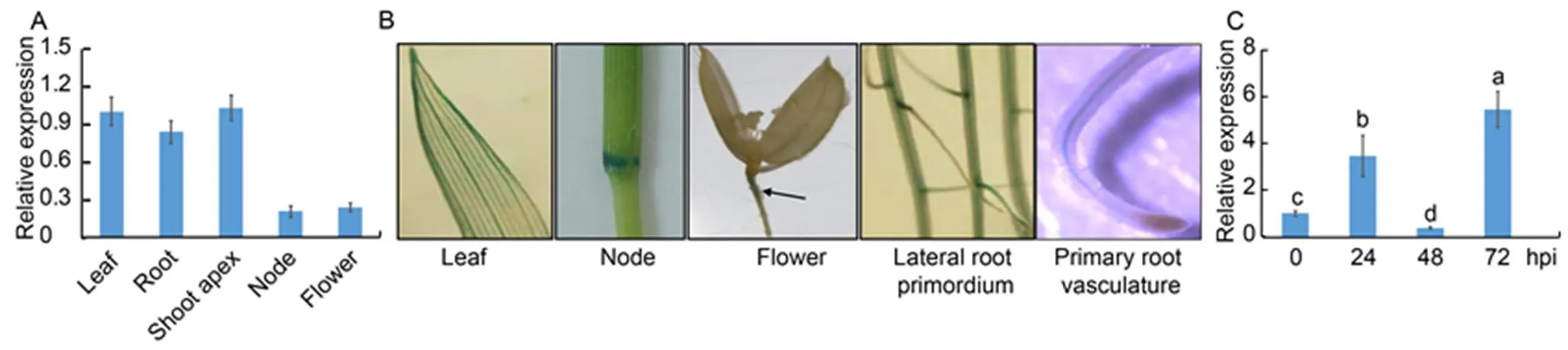
Fig. 2.expression pattern in rice.
A, Quantitative real-time PCR was performed on the mRNA extracted from the roots and leaves of one-week-old plants, shoot apecies of one- month-old plants, nodes and flowers of three-month-old plants.was used as the control.
B, Expression offrom one-week-old plant leaves or roots as well as nodes and flowers of three-month-old plants was analyzed using transgenic plants, in whichwas expressed from an endogenouspromoter. Black arrowhead indicates GUS staining site in the flower photo.
C,-infection mediated expression ofwas analyzed at 0, 24, 48 and 72 h post-inoculation (hpi). The RNA was extracted by using the TRIzol, andwas used as the reference gene to normalize gene expression level.
Data in A and C represent Mean ± SE (= 3). Different lowercase letters indicate significant differences at< 0.05.
We performed sequence alignment between IDD3 and ID1. The analyses showed that IDD3 and ID1 were highly conserved in the ID domain. However, they possessed different C-terminal regions (Fig. 3-A). Since IDDs are known to act as transcription factors, we examined the transcriptional activity of IDD3 using a yeast mono-hybrid system. In the transcriptional activation assay, vectors expressed the GAL4 DNA- binding domain (BD) were fused with the full-lengthcoding region, the N-terminal ID domain, or the C-terminal region in the yeast. The activation domain of anNAC genewas used as the positive control (Xie et al, 2000). It was observed that the N-terminal peptide, which contains the ID domain, showed no transcriptional activity.However, the C-terminal domain exhibited strong transcriptional activation (Fig. 3-B). Thus, a vector encoding GFP fused to the C-terminal end of IDD3 was expressed in transgenic plants under a non-specific promoter (35S) to detect the subcellular localization of IDD3. We detected a strong GFP signal in the nuclei, which indicated that IDD3 was a nuclear protein (Fig. 3-C).
IDD3 negatively regulated rice defense to ShB
We examinedmutants (and) andoverexpressors (and) to evaluate the function ofin the rice defense mechanism against ShB. The T-DNAs were inserted into the second intron of thelocus (Fig. 4-A). Results of qRT-PCR confirmed that themutants were the knock-out mutants and that the expression ofwas significantly highly expressed inoverexperesors compared to the WT plants (Fig. 4-B). The results ofinoculation showed thatmutants exhibited similar responses against ShB with WT.However,overexperesorsexhibited more susceptible symptoms toinfection compared with WT (Fig. 4-C). In WT, approximately 41.5% of the leaf area was covered with lesions, compared with 42.2% in, 40.9% in, 54.9% inand 55.7% inplants (Fig. 4-D).

Fig. 3. Transactivation and nuclear localization of IDD3.
A, Sequence alignment of rice IDD3 and ID1. Identical and similar amino acids are shown in black and gray boxes, respectively. Blue and red horizontal bars indicate the putative nuclear localization sequence and indeterminate domains, respectively. Asterisks in the red horizontal bar indicate the core amino acids of the two C2H2and two C2HC zinc fingers.
B, Transactivation activity of different regions of IDD3. DNA encoding the full-length IDD3 (495 aa), the N-terminal (212 aa), and the C-terminal (283 aa) regions of IDD3 was fused to the GAL4 DNA-binding domain and transformed into the yeast cells. AtNAC1 was used as the positive control.
C, Confocal microscopic images of GFP overlapped with propidium iodide (PI) staining in a lateral root of IDD3-GFP transgenic plants (top left). The lower left panel shows light microscopy of the same tissue (DIC). Bars, 20 µm.
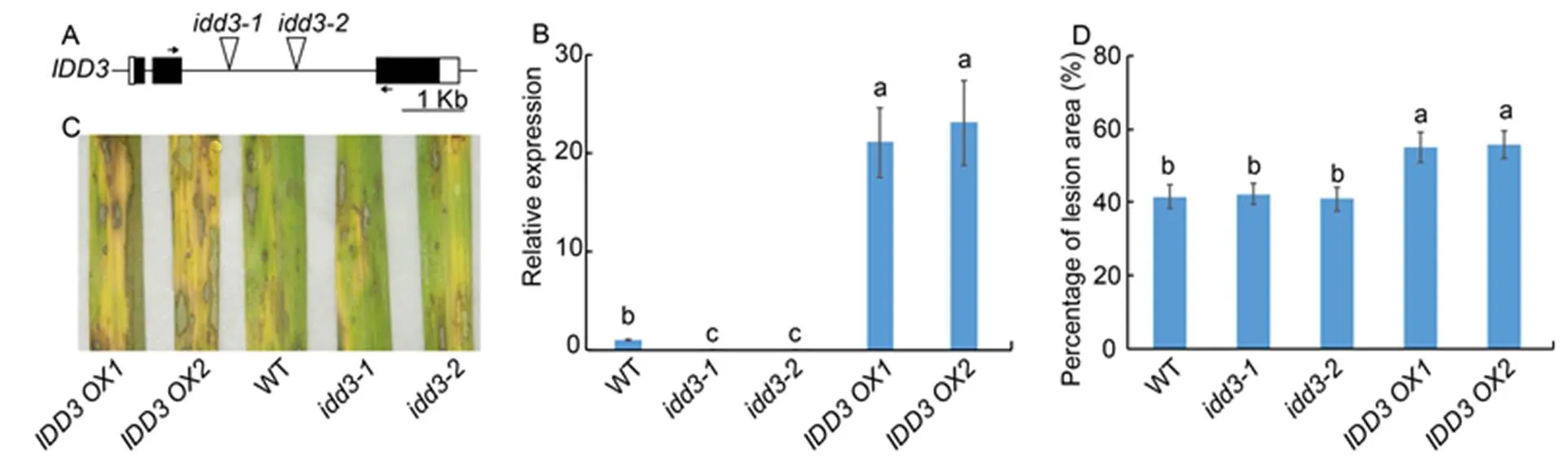
Fig. 4. Response ofmutants andoverexpressors against sheath blight.
A, Diagrammatic representation of the genomic structure along with the T-DNA insertion site. Black and white boxes indicate the exons and the untranslatedregions, respectively. The triangles in the second intron indicate the T-DNA insertion site in themutants (and). Short horizontal arrows indicate the location of primers that were used in quantitative real-time PCR (qRT-PCR).
B, qRT-PCR was performed to test the expression ofin wild type (WT),mutants andoverexpressors (and). The leaves from one-month-old plants were used for RNA extraction using the TRIzol.was used as the reference gene to normalize gene expression level.
C, Phenotype response of WT,mutants andoverexpressors.
D, Percentage of lesion area in WT,mutants andoverexpressors.
Data represent Mean ± SE (= 3 in B, and> 10 in D). Different lowercase letters indicate significant differences at< 0.05.
Themutants showed no significant difference in response to ShB compared to the WT plants, which indicated that IDD3 and the other IDD members might be functionally redundant. Since/repressor plants were similar toin exhibiting an enlarged lamina joint angle (Wu et al, 2013; Liu et al, 2016), therepressor lines were generated through the C-terminal fusion of SUPERMAN repressive domain X (SRDX) motif sequences to IDD3. In the IDD3 repressors (,,,and), we observed an upregulated expression ofcompared with the WT plants (Fig. 5-A). The results ofinoculation showed thatrepressors (and) were less susceptible to ShB compared with the WT plants (Fig. 5-B). In WT, 42.1% of the leaf area was covered with lesions, compared with 34.1% inrepressor, and 33.5% inrepressorplants (Fig. 5-C).

Fig. 5. Response ofrepressors to sheath blight.
A, Quantitative real-time PCR was performed to examine the expression ofin wild type (WT) andrepressors (,,,and). The leaves from one-month-old plants were used for RNA extraction using the TRIzol.was used as the reference gene to normalize gene expression level.
B, Phenotype response ofrepressors (and) toAG1-IA compared with WT.
C, Percentage of lesion area in therepressors (and) compared with WT.
Data represent Mean ± SE (= 3 in A, and> 10 in C). Different lowercase letters indicate significant differences at< 0.05.

Fig. 6. Regulatory role ofand brassinosteroid (BR) signaling in rice defense to sheath blight (ShB).
A, Quantitative real-time PCR was done to analyze,andexpression in wild type (WT),mutants (and) andoverexpressors (and). The leaves from one-month-old plants were used for RNA extraction using the TRIzol.was used as the reference gene to normalize gene expression level.
B, Schematic diagram of 1.0 kb ofandpromoters. D2P and D11P indicate the regions detected in the chromatin immunoprecipitation (ChIP)-PCR assay (C).
C, ChIP-PCR was done to analyze the binding affinity of IDD3 to D2P and D11P regions shown in (B). The anti-GFP antibody was used for immunoprecipitation.
D, Phenotype response of,,andtoAG1-IA compared with WT.
E, Percentage of the lesion area in,,andcompared with WT.
Data represent Mean ± SE (= 3 in A and C, and> 10 in E). Different lowercase letters indicate significant differences at< 0.05.
IDD3 positively regulated BR biosynthetic and signaling genes
RAVL1 is an upstream regulator of BR homeostasis (Je et al, 2010); therefore, we examined the role of IDD3 in the regulation of the expression of BR-related genes. We analyzed the expression of the BR receptor geneand biosynthetic genesandin therepressors and overexpressors. The results of the qRT-PCR analysis showed that the expression of,andwas downregulated in therepressors while upregulated in theover- expressors compared with the WT plants (Fig. 6-A). The promoter sequence analysis showed that the putative IDD protein binding motif (TTTGTCC/G) was located within 1.0 kb of theandpromoters (Fig. 6-B). Results of the ChIP assay using 35S:GFP and 35S:IDD3:GFP transgenic plant calli showed that IDD3 did not bind to the putative motifs located inandpromoter regions (Fig. 6-C).
Previously, we had identified that themutantandwere less susceptible to ShB compared with the WT plants (Yuan et al, 2018). Next, theoverexpressor was crossed with themutant to test whethervia activation of BR signaling genes to regulate rice defense against ShB. The results ofinoculation indicated that the susceptibility ofwas similar toplants, whileplants were more susceptible to ShB compared with the WT plants (Fig. 6-D). In WT, 40.7% of the leaf area was covered with lesions, compared with 23.1% in, 55.3% in, and 24.6% inplants (Fig. 6-E).
Discussion
ShB, caused by, is a major rice disease, which severely reduces grain yield. However, the host resistance mechanisms remain unclear. Our previous studies found that RAVL1 modifies the rice defense mechanism against ShB via the activation of BR and ethylene signaling pathways. The results indicated that BR and ethylene signaling pathways regulate rice defense against ShB negatively and positively, respectively (Yuan et al, 2018). However, the detailed mechanism of RAVL1 regulation in rice defense against ShB is not clearly understood.
Sun et al (2019) identified that LPA1/IDD14 protects rice against ShB via activation of. Interestingly, we identified another IDD member, is positivelyregulated by RAVL1, which is confirmed through themutants and overexpressors. RAVL1 is a transcriptional activator, which activates the downstream genes by binding to the E-box elements (Je et al, 2010). Further analysis identified that thepromoter region contains the E-box elements. The binding affinity of RAVL1 topromoter region was confirmed by ChIP assay, the yeast one-hybrid assay and the transient assay. The results indicated that RAVL1 bound to the P1 region containing a single E-box element, however, it did not bind to the P2 region also harboring one E-box element, indicating that RAVL1 directly activatedthrough promoter binding.
Next, we found that the expression ofwas enhanced at 24 and 72 hpi withAG1-IA, suppressed at 48 hpi, indicating a complex regulation mechanism betweenandtranscription. Thus, future research may involve in testing the protein levels. Inoculation withrevealed that twomutant lines showed no significant differences compared with the WT plants in response to ShB.is localized at the nuclei in IDD3-GFP transgenic plant roots, and our analyses showed that it has transcription activity in yeast cells, which confirmed thatmight function as a transcriptional activator. The fusion of SRDX with LPA1 functions as a repressor (Liu et al, 2016); thus,, a repressor of,was generated to address whether IDD3 was functionally redundant with other transcription factors for the regulation of downstream gene expression. The results ofinoculation showed that IDD3 repressors were less susceptible to ShB compared with the WT plants. Thus,overexpressors were generated, and their response to ShB was analyzed to further verify. The results indicated thatoverexpressors were more susceptible to ShB compared with the WT plants. These results suggested thatnegatively regulated rice defense against ShB.
is a transcriptional activator and a target of RAVL1, a key BR signaling transcription factor. Further analysis identified that,a BR receptor gene, andand,the BR biosynthesis genes,were repressed in therepressors, while they were highly expressed inoverexpressors, suggesting that IDD3 positively regulated the expression of BR-related gene. BR signaling negatively regulates rice defense against ShB (Yuan et al, 2018), indicating thatmight activate BR signaling to negatively regulate the rice defense mechanism against ShB. However, the putative IDD binding motifs appeared in theandpromoters, while the ChIP assay indicated thatdid not directly bind to the promoters ofand.mutantis less susceptible to ShB compared with the WT plants (Yuan et al, 2018). Next, a genetic combination betweenandwas generated to explore whetherregulation of the rice defense mechanism against ShB happened via BR signaling. The results ofinoculation indicated thatwas more susceptible, whileandwere less susceptible to ShB compared with the WT plants. Additionally,andplants exhibited similar defense against ShB, implying that IDD3 acted at the upstream ofand thatactivated BR signaling to regulate rice defense to ShB. Thus, our results proved thatnegatively regulated the rice defense mechanism against ShB by activating BR signaling, which is at the downstream of RAVL1. This study extended the knowledge of the signaling pathways adopted by rice to defend itself against ShB.
Methods
Plant growth and R. solani AG1-IA inoculation
Wild type (WT) (L. subsp., cultivar Dongjin),(PFG_3A-09378),(PFG_3A-14411),overexpressor (),,andoverexpressor () plants were used, and grown in a greenhouse at Shenyang Agricultural University, China, at 23 ºC–30 ºC. A 10-cm-long piece was cut from the second youngest leaf of the main tiller and placed on a moistened filter paper in a petri dish (diameter, 36 cm; height, 2.5 cm). Each replicate comprised of six leaves, and we used four replicates perline for a completely randomized design. Colonized potato dextrose agar (PDA) blocks (diameter, 7 mm) were excised and placed on the abaxial surface of each leaf. The leaves were incubated at 25 ºCfor 72 h in an incubator with continuous light, and the filter paper was kept moist using sterile water. After 72 h, the dimensions of the lesions (Length ×Width) within each leaf piece were measured using the Image J software (NIH, USA), and the percentage of the lesion area was calculated using the methods by Eizenga et al (2002) and Prasad and Eizenga (2008). Next, one-month- old wild-type plants were inoculated withAG1-IA, and their leaves were sampled after 0, 24, 48 and 72 hpi to analyze theAG1-IA infection-mediated expression of thegenes (Prasad and Eizenga, 2008).
RNA extraction and quantitative real-time PCR (qRT-PCR)
We used the TRIzol reagent (Takara, China) to extract total RNA from the one-month-old rice leaves, shoot apecies and roots, as well as the three-month-old rice node and whole panicle including flower, followed by the removal of the genomic DNA using RQ-RNase-free DNase (Promega, USA). Complementary DNA (cDNA) was synthesized using the GoScript Reverse Transcription Kit (Promega, USA) following the manufacturer’s instructions. Next, the qRT-PCR analysis was performed using a BIO-RAD CFX96 Real-time PCR system (Bio-Rad, USA) and SYBR-Green Master Mix (Takara, China). The gene expression levels were normalized to that of the level of. Table S1 provides the list of primers used for qRT-PCR.
Plasmid construction
Theopen reading frame (ORF) sequences were amplified and cloned into theII andI restriction enzyme sites of the pCAMBIA1302 binary vector to generate theoverexpressedtransgenic plants. Here, thecoding sequence was N-terminally fused to thecoding sequence, and theORF sequence was N-terminally fused to the (SUPERMAN repressive domain X) SRDX motif sequence to generate therepressor construct in the pGA1611 binary vector.
Transactivation assay
Transactivation assays were performed in the yeast strain PJ69-4A, which contains theandreporter genes. Using the pGBT9 vector (Clontech, USA), DNA encoding the GAL4 DNA-binding domain was fused to the followingDNA fragments: the complete ORF, a 5′-cDNA encoding the first 212 amino acids, or a 3′-cDNA encoding amino acids 213–495. The 3′-fragment, which encodes a peptide from amino acids 143–324, was used as the positive control. These constructs or the empty vector (pGBT9) were introduced individually into the yeast cells. Yeast transformants were grown on the synthetic dropout (SD)/Trp- and SD/His-plates (Rose et al, 1990). Table S1 provides the primers used for cloningcDNA fragments.
Chromatin-immunoprecipitation (ChIP) assay
Rice calli (8 g) were collected from the transgenic plants expressing 35S:GFP, 35S:RAVL1:GFP or 35S:IDD3:GFP for performing the ChIP assay. The ChIP assay and subsequent ChIP-PCR assayswere performed following a previously described protocol (Je et al,2010). Table S1 provides the primers used for the ChIP-PCR assay.
Transient expression assay
For the transient expression assay, the effector plasmid (35S:RAVL1) and the reporter plasmids (or mutated promoter,), along with an internal control plasmid (35S:LUC), were co-transformed into theprotoplast cells (Yamaguchi et al, 2010). The GUS activity analyses were performed following the method by Xuan et al (2013). The luciferase assay was evaluated using the Luciferase Assay Kit (Promega, USA). The experiments were repeated three times, and the polyethylene glycol (PEG)-mediated transformation and luciferase activity assays were performed following a previously described protocol (Yoo et al, 2007). Table S1 provides the list of the primers used for the transient expression assay.
Yeast one-hybrid analysis
A 1.0 kb fragment of thepromoter (normal or E-box mutated) was cloned into a pHISi vector. Next, theORF sequences were cloned into the pGAD424 vector. The constructedorempty vector was transformed into a yeast strain (YM4271), and the growth of the yeast cells was monitored on a synthetic dropout media (SD) lacking either Leu or Leu and His.
Vector construction
For the construction of a GFP fusion vector, cDNA, encoding the 1 608 bp ORF ofwas isolated through RT-PCR and was fused to GFPin the PCAMBIA1302 vector. For a GUS- fusion vector, the 3.0 kbpromoter region was fused to GUS in the pCAMBIA1381 vector. Table S1 provides the PCR primers for cloningcDNA and promoter.
Statistical analyses
Statistical analyses were performed using Prism 5 (GraphPad, USA). All data were expressed as Mean ± SE. One-way analysis of variance (ANOVA) was performed, followed by the Bonferroni’s multiple comparison tests.< 0.05 represented statistically significant results.
SUPPLEMENTAL DATA
The following material is available in the online version of this article at http://www.sciencedirect.com/science/journal/rice-secience; http://www.ricescience.org.
Table S1. Primer sequences used in this study.
Table S1. Primer sequences used in this study.

Primer SequencesSize of product IDD3 RT-FACCGGGATCAAGAAGCACTACTG254 bp IDD3 RT-RGATCAAACTGAGAGGCGCCATTG RAVL1-FTCCTCACCAACTCCACATTACGGT187 bp RAVL1-RCAGATCGAGATCCAACGAGGA D2-FATGTGATAACAGAGACGCTGCGGT213 bp D2-RTGGTGACCAAGTGGTGAAGGAAGA D11-FAGTGAAGAGGGAGCATGAAGGCAT193 bp D11-RATCTGCAGGGCTGAAATTGTTGGG BRI1-FCAGCTACTTGGCTATCTTGAAGCTCAGC167 bp BRI1-RCCATTCTTGTTGAAGGTGTACTCCGTGC UBQ-FCAAGATGATCTGCCGCAAATGC251 bp UBQ-RTTTAACCAGTCCATGAACCCG IDD3 PM-FGAATTCAGGTTTGCTGTCTCCCTTTC IDD3 PM-RGAGCTCTCTCGCTGCTTACTTTGTTG IDD3 cDNA-FGATTCATACAAGCTTATGGCGGCCGCCTCGTCCGCACCCTTC IDD3 cDNA-RAGATCTGCGGCGGCCATGTTTGCCGGGTCCAGTGAGCCGAC IDD3 N-FGAATTCATGGCGGCCGCCTCGTCCGC IDD3 N-RGTCGACGGGCGGCATGCGCGCGTTC IDD3 C-FCGTATACGCCGGCGCCGATGAATTC IDD3 C-RGTCGACTCAGTTCATGTTTGCCGGG IDD14 AD-FGAATTCATGGCACTGGTCAAGAGCCA IDD14 AD-RAGATCTGCATGCATGTACATATCAGCTA D2P-FGGTTGAAATAACGGGAAGCGT D2P-RGTTTAA AACAGGCCCTAAATCATC D11P-FGCGCATAAGCTTCATCAGATT C D11P-RCAGAGTAGCTAGCATCTAGGCTG IDD3P1-FTTCCTCTCTCCTTGATG IDD3P1-RCAAACGAGCAAAGAGAG IDD3P2-FGCATCATGGTCCCACTAGTC IDD3P2-RCTCTTTTATTCTAAATAGCTG
Acknowledgements
This study was supported by the Science and Technology Innovation Talents of Shenyang, China (Grant No. RC190489). We thank Professor Han Chang-deok from Gyeongsang National University, Korea, for providingmutant seeds.
Colasanti J, Yuan Z, Sundaresan V. 1998. Thegene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize., 93(4): 593–603.
Cui D Y, Zhao J B, Jing Y J, Fan M Z, Liu J, Wang Z C, Xin W, Hu Y X. 2013. TheIDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport., 9(9): e1003759.
Dou M Z, Cheng S, Zhao B T, Xuan Y H, Shao M L. 2016. The indeterminate domain protein ROC1 regulates chilling tolerance via activation of/in rice., 17(3): 233.
E Z G, Cheng B Y, Sun H W, Wang Y J, Zhu L F, Lin H, Wang L, Tong H H, Chen H Q. 2019. Analysis on Chinese improved rice varieties in recent four decades., 33(6): 523–531. (in Chinese with English abstract)
Eizenga G C, Lee F N, Rutger J N. 2002. Screeningspecies plants for rice sheath blight resistance., 86(7): 808– 812.
Feurtado J A, Huang D Q, Wicki-Stordeur L, Hemstock L E, Potentier M S, Tsang E W T, Cutler A J. 2011. TheC2H2zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation., 23(5): 1772–1794.
Gao Y, Zhang C, Han X, Wang Z Y, Ma L, Yuan D P, Wu J N, Zhu X F, Liu J M, Li D P, Hu Y B, Xuan Y H. 2018. Inhibition offunction in mesophyll cells improves resistance of rice to sheath blight disease., 19(9): 2149–2161.
Helliwell E E, Wang Q, Yang Y N. 2013. Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogensand., 11(1): 33–42.
Huang P, Yoshida H, Yano K, Kinoshita S, Kawai K, Koketsu E, Hattori M, Takehara S, Huang J, Hirano K, Ordonio R L, Matsuoka M, Ueguchi-Tanaka M. 2018. OsIDD2, a zinc finger and INDETERMINATE DOMAIN protein, regulates secondary cell wall formation., 60(2): 130–143.
Je B I, Piao H L, Park S J, Park S H, Kim C M, Xuan Y H, Park S H, Huang J, Do Choi Y, An G, Wong H L, Fujioka S, Kim M C, Shimamoto K, Han C D. 2010.maintains brass- inosteroid homeostasis via the coordinated activation ofand biosynthetic genes in rice., 22(6): 1777–1791.
Kim T W, Wang Z Y. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors., 61: 681–704.
Kouzai Y, Kimura M, Watanabe M, Kusunoki K, Osaka D, Suzuki T, Matsui H, Yamamoto M, Ichinose Y, Toyoda K, Matsuura T, Mori I C, Hirayama T, Minami E, Nishizawa Y, Inoue K, Onda Y, Mochida K, Noutoshi Y. 2018. Salicylic acid-dependent immunity contributes to resistance against, a necrotrophic fungal agent of sheath blight, in rice and., 217(2): 771–783.
Kozaki A, Hake S, Colasanti J. 2004. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties., 32(5): 1710–1720.
Li J, Wen J Q, Lease K A, Doke J T, Tax F E, Walker J C. 2002. BAK1, anLRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling., 110(2): 213–222.
Li J M, Chory J. 1997. A putative leucine-rich repeat receptor kinaseinvolved in brassinosteroid signal transduction., 90(5): 929–938.
Liu J M, Park S J, Huang J, Lee E J, Xuan Y H, Je B I, Kumar V, Priatama R A, Raj K V, Kim S H, Min M K, Cho J H, Kim T H, Chandran A K N, Jung K H, Takatsuto S, Fujioka S, Han C D. 2016.() determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice., 67(6): 1883–1895.
Maeda S, Dubouzet J G, Kondou Y, Jikumaru Y, Seo S, Oda K, Matsui M, Hirochika H, Mori M. 2019. The rice CYP78A geneconfers resistance toand affects seed size and growth inand rice., 9(1): 587.
Mao B Z, Liu X H, Hu D W, Li D B. 2014. Co-expression ofandconfers rice resistance to fungal sheath blightand blastand reveals impact on seed germination., 30(4): 1229–1238.
Nam K H, Li J M. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling., 110(2): 203–212.
Park S J, Kim S L, Lee S, Je B I, Piao H L, Park S H, Kim C M, Ryu C H, Park S H, Xuan Y H, Colasanti J, An G, Han C D. 2008.() is necessary for the expression of() regardless of photoperiod., 56(6): 1018–1029.
Prasad B, Eizenga G C. 2008. Rice sheath blight disease resistance identified inspp. accessions., 92(11): 1503–1509.
Rose M D, Winson F, Hieter P. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. New York, USA: Cold Spring Harbor Laboratory Press: 198.
Savary S, Castilla N P, Elazegui F A, McLaren C G, Ynalvez M A, Teng P S. 1995. Direct and indirect effects of nitrogen supply anddisease source structure on rice sheath blight spread., 85(9): 959–965.
Savary S, Willocquet L, Elazegui F A, Castilla N P, Teng P S. 2000. Rice pest constraints in tropical Asia: Quantification of yield losses due to rice pests in a range of production situations., 84(3): 357–369.
Seo P J, Ryu J, Kang S K, Park C M. 2011. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in., 65(3): 418–429.
Shah J M, Raghupathy V, Veluthambi K. 2009. Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from., 31(2): 239–244.
Sun Q, Li T Y, Li D D, Wang Z Y, Li S, Li D P, Han X, Liu J M, Xuan Y H. 2019. Overexpression ofincreases planting density and resistance to sheath blight disease via activation ofin rice., 17(5): 855–857.
Volz R, Kim S K, Mi J N, Mariappan K G, Siodmak A, Al-Babili S, Hirt H. 2019. A chimeric IDD4 repressor constitutively induces immunity invia the modulation of salicylic- and jasmonic acid homeostasis., 60(7): 1536–1555.
Wang R, Lu L X, Pan X B, Hu Z L, Ling F, Yan Y, Liu Y M, Lin Y J. 2015. Functional analysis ofin rice sheath blight resistance., 87: 181–191.
Wang Z Y, Nakano T, Gendron J, He J X, Chen M, Vafeados D, Yang Y L, Fujioka S, Yoshida S, Asami T, Chory J. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis., 2(4): 505–513.
Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. 2007.JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action., 21(17): 2196–2204.
Wu X R, Tang D, Li M, Wang K J, Cheng Z K. 2013. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice., 161(1): 317–329.
Xie Q, Frugis G, Colgan D, Chua N H. 2000.NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development., 14(23): 3024–3036.
Xuan Y H, Priatama R A, Huang J, Je B I, Liu J M, Park S J, Piao H L, Son D Y, Lee J J, Park S H, Jung K H, Kim T H, Han C D. 2013. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots., 197(3): 791–804.
Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. 2010. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in., 22(4): 1249–1263.
Yang C J, Zhang C, Lu Y N, Jin J Q, Wang X L. 2011. The mechanisms of brassinosteroids’ action: From signal transduction to plant development., 4(4): 588–600.
Yin Y H, Wang Z Y, Mora-Garcia S, Li J M, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation., 109(2): 181–191.
Yoo S D, Cho Y H, Sheen J. 2007.mesophyll protoplasts: A versatile cell system for transient gene expression analysis., 2(7): 1565–1572.
Yuan D P, Zhang C, Wang Z Y, Zhu X F, Xuan Y H. 2018.activates brassinosteroids and ethylene signaling to modulate response to sheath blight disease in rice., 108(9): 1104–1113.
7 February 2020;
6 July 2020
Xuan Yuanhu (xuanyuanhu115@syau.edu.cn)
Copyright © 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2021.01.004
(Managing Editor: Wang Caihong)
杂志排行
Rice Science的其它文章
- High-Quality de novo Genome Assembly of Huajingxian 74, a Receptor Parent of Single Segment Substitution Lines
- Growth and Photosynthesis Responses of a Super Dwarf Rice Genotype to Shade and Nitrogen Supply
- Lodging Resistance Related to Root Traits for Mechanized Wet-Seeding of Two Super Rice Cultivars
- Effects of Early- and Late-Sowing on Starch Accumulation and Associated Enzyme Activities During Grain Filling Stage in Rice
- Exogenous Peroxidase Mitigates Cadmium Toxicity, Enhances Rhizobial Population and Lowers Root Knot Formation in Rice Seedlings
- Osa-miR439 Negatively Regulates Rice Immunity Against Magnaporthe oryzae
