BLV-miRNA跨界调控人类靶基因预测及生物信息学分析
2021-03-09王雍李思妍何思锐张迪连帅王建发武瑞
王雍,李思妍,何思锐,张迪,连帅,王建发,武瑞
BLV-miRNA跨界调控人类靶基因预测及生物信息学分析
王雍,李思妍,何思锐,张迪,连帅,王建发,武瑞
黑龙江八一农垦大学动物科技学院/黑龙江省牛病防制重点实验室,黑龙江大庆 163319
【】评估牛白血病病毒(BLV)来源的miRNAs跨界调控人源基因的风险。对BLV-miRNA可能带来的食品安全问题及对人体健康可能造成何种影响进行前瞻性研究,为未来实际生产中地方流行性白血病防控措施执行的必要性研究奠定基础,对BLV与人类疾病间关联性的研究提供理论指导。首先使用mirbase网站对BLV miRNA的成熟序列进行查询,通过miRanda软件对BLV编码的10种miRNA(BLV-miR-B1-3P,5P、BLV-miR-B2-3P,5P、BLV-miR-B3-3P,5P、BLV-miR-B4-3P,5P、BLV-miR-B5-3P,5P)进行靶基因预测,并选取每个BLV-miRNA评分前10的候选靶基因(去除重复基因后共88个)进行功能分析,对受到多个BLV miRNA共同调控的候选靶基因使用RNAhybrid软件进行二次预测验证,并对其功能进行分析。BLV编码的10种miRNA经预测后分别获得1 630—16 383个靶基因不等。对评分前十的共计88个候选靶基因进行功能分析后发现,其中18个基因无相关功能报道;36个候选靶基因与肿瘤性疾病的发生发展存在相关性。2个候选靶基因可以对细胞周期起调控作用;16个候选靶基因参与细胞信号转导的调控;14个候选靶基因在细胞结构/骨架蛋白的形成中发挥作用;细胞的增殖与凋亡的功能表现成拮抗关系,往往促进增殖的基因同时也可以抑制细胞凋亡,共有13个基因对细胞的增殖和凋亡起调节作用,有趣的是,这13的候选靶基因对细胞增殖凋亡功能的调节是双向性的,但不能明确BLV miRNA对细胞的调节到底是更趋向于增殖还是凋亡,因此仍需要后续研究深入探讨;2个候选靶基因对细胞分化起调节作用;16个候选靶基因对细胞的迁移/侵袭功能起调节作用,再次提示BLV miRNA与肿瘤性疾病可能存在更重要的关联性。7个候选靶基因可能在乳腺细胞的分化、迁移、侵袭过程中发挥重要作用,提示BLV与人乳腺癌相关性的研究中,可以从BLV miRNA的角度深入探讨;BLV-B4-3P的2个候选靶基因Ⅰ型胶原α1链基因(COL1A1)、断裂点簇集区(BCR)对人急性淋巴细胞白血病(ALL)具有调节作用。此外,可以被多个BLV miRNA共同靶向的候选靶基因均属于黏蛋白家族(MUC5B、MUC12和 MUC16),且均可以在结肠中表达,对结肠黏膜的形成产生影响。外源性BLV miRNA可能跨界调控细胞周期、信号转导、结构/骨架、增殖、凋亡、分化、迁移/侵袭相关等细胞功能相关基因,破坏细胞结构;BLV miRNA与人乳腺癌的相关性可能表现在人乳腺癌细胞的分化、迁移、和侵袭过程中;而BLV-miR-B4-3p本身与白血病相关miR 29a共享同一种子区域,可能对人急性淋巴细胞白血病的发生发展造成影响;外源性BLV miRNA具有靶向抑制黏蛋白基因(MUC5B、MUC12、MUC16)表达,通过破坏肠黏膜形成这一途径,跨界调控人源基因的风险。
生物信息学;牛白血病病毒;跨界调控;人源基因;miRNA
0 引言
【研究意义】牛白血病病毒(BLV)是牛群中常见的传染性病毒之一,在世界各地均有分布,阳性牛群会呈持续感染状态,通常为亚临床型,只有约5%的BLV阳性牛会表现出淋巴瘤的症状[1]。最新的调查结果显示,BLV在我国牛群中的发病率约为49%[2]。牛群中出现BLV感染后,会出现产奶量下降[3]、平均寿命降低[4]、免疫失败[5]等诸多问题。基于BLV临床症状不明显的特点,在目前的实际生产中并不能引起饲养者的重视,而BLV编码的miRNA具有靶向调控人源基因的风险,可能会对乳制品食用者的健康产生威胁,因此对BLV miRNA的研究对乳制品安全风险的预防具有重要意义。【前人研究进展】乳汁中发现的BLV可能引发全球性的公共卫生问题,1974年,McClure等用自然感染BLV奶牛乳汁(未巴氏消毒)饲喂6只初生黑猩猩,连续饲喂至第30周后全部黑猩猩出现了嗜睡、厌食、白细胞增多、贫血和进行性肺炎症状,第35周和第46周时有两只黑猩猩分别死亡,病死黑猩猩被确诊患有红细胞白血病(erythroleukemia)[6]。1976年,Graves等发现BLV可以体外感染牛、羊、猫、蝙蝠、类人猿和人类细胞(人胚肺二倍体细胞)。2015年以来,美国、澳大利亚和阿根廷乳腺癌患者乳腺组织样品中相继被检出BLV核酸片段[7]。2019年4月,Buehring等在美国加州当地招募了95名妇女志愿者,对其血液中BLV核酸和BLV IgA、IgM、IgG进行了检测,发现BLV核酸阳性数为33,IgG和IgA抗体阳性数均为30,IgM抗体阳性数为55[8]。因此,虽然尚无充分证据表明BLV是导致人乳腺癌的元凶,但BLV引发的公共卫生问题愈发受到关注。BLV作为逆转录病毒,但已经有研究者通过高通量测序技术证实BLV可编码miRNA,且在BLV阳性牛的肉和乳汁中发现了BLV miRNAs的存在[9-10]。机体内BLV主要依靠miRNA调控牛淋巴细胞功能,当BLV感染后,其核酸序列整合到B淋巴细胞基因组中,导致B细胞在宿主体内异常增殖[11]。但是由于B细胞淋巴瘤中缺乏5′LTR驱动的RNA聚合酶II(PolII),BLV在原代B细胞淋巴瘤中出现转录沉默,无法检测到病毒mRNA和蛋白质[12]。基于此,研究者在对BLV致病机制的研究过程中发现,白血病牛B淋巴细胞中存在大量高度保守的由RNA聚合酶III(PolIII)转录的BLV编码的miRNA[9],也打破了RNA病毒不编码miRNA以避免其基因组或mRNA的非生产性切割的传统认知。miRNA相比mRNA具有极高的稳定性,乳品中BLV-miRNA具有危害人类健康的潜在风险,BLV可编码5类共计10种miRNA,其中miR-B4-3p、miR-B2-5p、miR-B5-5p、miR-B1-3p、miR-B5-3p与牛白血病的发生相关[13],这些miRNA主要靶向颗粒酶A(GZMA)、棕榈酰蛋白硫酯酶1(PPT1)、膜联蛋白A1(ANXA1)、促分裂原活化蛋白激酶激酶1(MAP2K1)、磷酸肌醇3激酶(PIK3CG)、FBJ鼠科骨肉瘤病毒原癌基因(FOS)等癌相关基因,清除上述miRNA后BLV不能诱导白血病发生[14]。BLV与人类T淋巴细胞白血病病毒1型(HTLV-1)存在共同的致瘤机制和结构高度相似的致瘤miRNA[15]。miRNA可以跨物种、跨代际稳定存在,并在不同物种间、代际间跨界调控基因表达,且miRNA跨界调控的普适性规律已得到广泛认可[16]。BLV修饰后的牛树突状细胞外泌体携带有致瘤miRNA,并且牛奶外泌体所携miRNA可跨界调控人类免疫细胞功能[17]。【本研究切入点】前人对BLV可能对人类健康产生风险的研究主要集中于BLV与人乳腺癌之间的联系,本文以miRNA跨界调控的普适性作为切入点, BLV作为一种可以编码miRNA的逆转录病毒,其所编码的miRNA可以在牛奶的外泌体中发现[10],具有跨界调控人源基因的食品安全风险。【拟解决的关键问题】通过miranda软件,预测了BLV miRNAs可能调控的人源靶基因,并对BLV miRNAs的候选靶基因功能进行查询并分析,对进一步认识BLV miRNAs跨界调控人源基因风险的研究具有重要的科学意义和实际价值。
1 材料与方法
1.1 信息采集
通过mirbase(www.mirbase.org)查询BLV-miR- B1-3P,5P、BLV-miR-B2-3P,5P、BLV-miR-B3-3P,5P、BLV-miR-B4-3P,5P、BLV-miR-B5-3P,5P成熟序列(表1)。

表1 BLV miRNAs成熟序列
1.2 靶基因预测
miRanDa软件主要通过miRNA和mRNA间的序列互补匹配程度和形成复合结构的自由能两个因素来判断miRNA与候选靶基因的结合位点,根据序列匹配的打分值和对应的自由能预测结合位点的可能性,当分值大于150时,可以被认定为该miRNA 的候选靶基因,由于不依赖结合位点的保守性,可以更加广泛的对miRNA的结合位点进行评估,因此我们在哈尔滨医科大学李春权老师帮助下,于2019年7月在哈尔滨医科大学大庆分校区应用miRanda(V1.9)软件对BLV-miR-B1-3P,5P、BLV-miR-B2-3P,5P、BLV- miR-B3-3P,5P、BLV-miR-B4-3P,5P、BLV-miR-B5-3P,5P序列进行靶基因预测分析。
1.3 靶基因功能查询
通过NCBI、Bing检索工具检索每个BLV-miRNA所预测的靶基因评分前十位的靶基因功能。通过RNAhybrid[18]对多个BLV-miRNA共同调控的靶基因进行二次预测验证,当最小自由能(△G)≤-15 kcal·mol-1时结合的可能性较高。
2 结果
2.1 BLV miRNAs靶向人源基因预测
通过miRanda软件对BLV miRNAs进行靶基因预测,BLV miRNAs分别获得了1 630—16 383个靶基因不等(图1)。
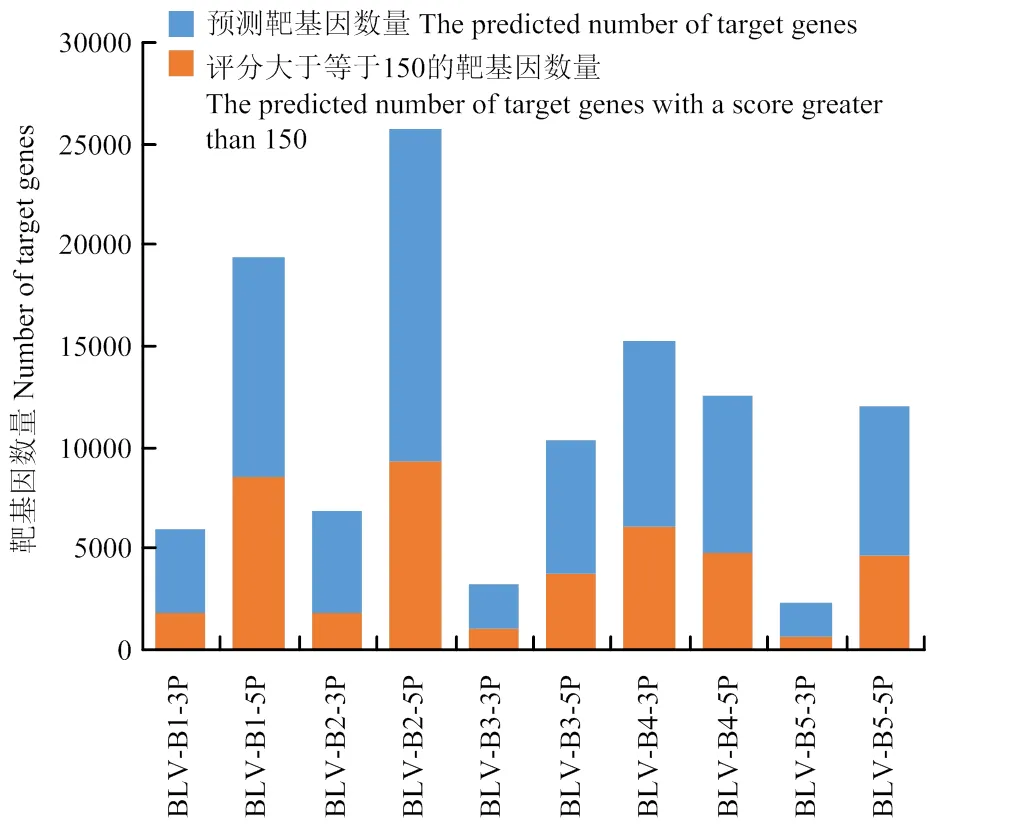
图1 BLV miRNAs靶基因预测
2.2 BLV miRNAs靶向评分前十位基因功能分析
对BLV-miRNAs的靶基因进行了分别预测,并对每个BLV-miRNA评分排名前十的靶基因(去除重复基因后共88个)功能进行了整理分析,由于18个候选靶基因没有相关功能报道,最终本研究对70个靶基因的功能进行了分析,其中有36个基因的功能与肿瘤的发生发展存在相关性。这些候选靶基因所表达的蛋白在细胞的周期、信号转导、结构/骨架、增殖、凋亡、分化、迁移/侵袭功能中发挥了重要作用(表2—7)。
2.3 BLV miRNAs靶向调控人源白血病的候选靶基因
BLV属于反转录病毒科,丁型反转录病毒属,在分类学上与人类T淋巴细胞白血病病毒I型最为接近。而BLV miRNAs进入人体后仍然存在调控人源白血病细胞的可能性,BLV-miR-B4-3p作为患病牛体内表达量最高的BLV miRNA,在所有评分排名前十的候选靶基因中有两个靶基因可以在人急性淋巴细胞白血病(ALL)的发生发展中发挥作用,且均为BLV-miR-B4-3p的候选靶基因(表8)。
2.4 BLV miRNAs靶向调控人源乳腺癌的候选靶基因
发现BLV miRNAs靶向调控的评分前十的候选靶基因中,有5个miRNA调控的7个候选靶基因在乳腺细胞的分化、迁移、侵袭生命活动中发挥了重要作用(表9)。
2.5 多个BLV miRNA共同靶向基因的最小自由能和二级结构
在评分前10的候选靶基因中,可以同时被多个BLV miRNA共同靶向的候选靶基因聚类于MUC5B、MUC12、MUC16基因(同属于黏蛋白家族)。我们通过RNAhybrid对这3个基因进行了二次预测验证的结果显示,这些BLV miRNA与靶基因结合的自由能均小于-15 kcal/mol(图2—4)。

红色核酸序列:候选靶基因序列;绿色核酸序列:miRNA序列;mfe:最小自由能
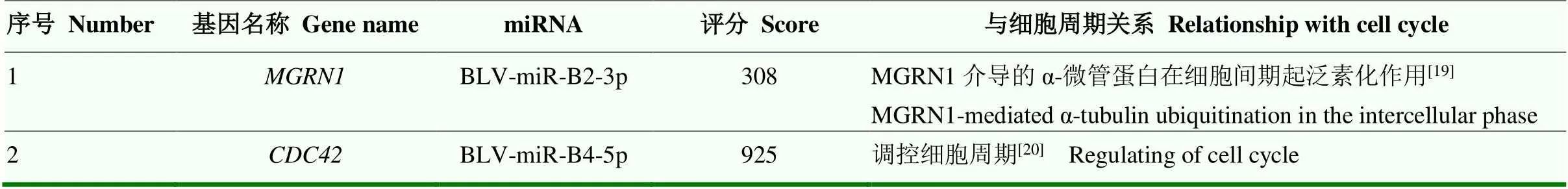
表2 参与调控细胞周期的候选靶基因
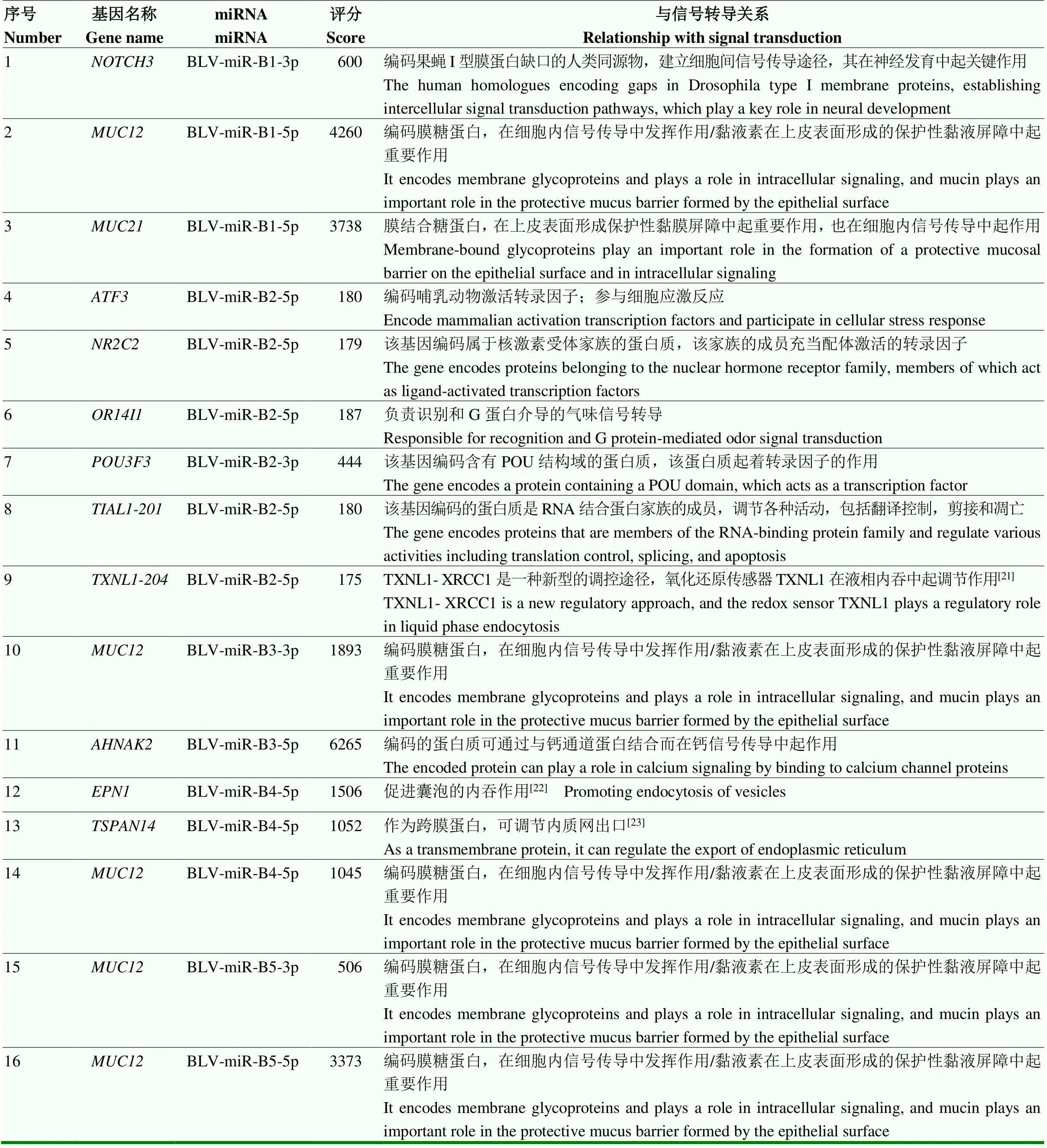
表3 参与细胞信号转导的候选靶基因

表4 参与合成细胞结构/骨架蛋白的候选靶基因

表5 参与细胞增殖/凋亡的候选靶基因

红色核酸序列:候选靶基因序列;绿色核酸序列:miRNA序列;mfe:最小自由能

表6 参与细胞分化的候选靶基因

表7 参与细胞迁移/侵袭的候选靶基因

表8 参与调控人急性淋巴细胞白血病的候选靶基因
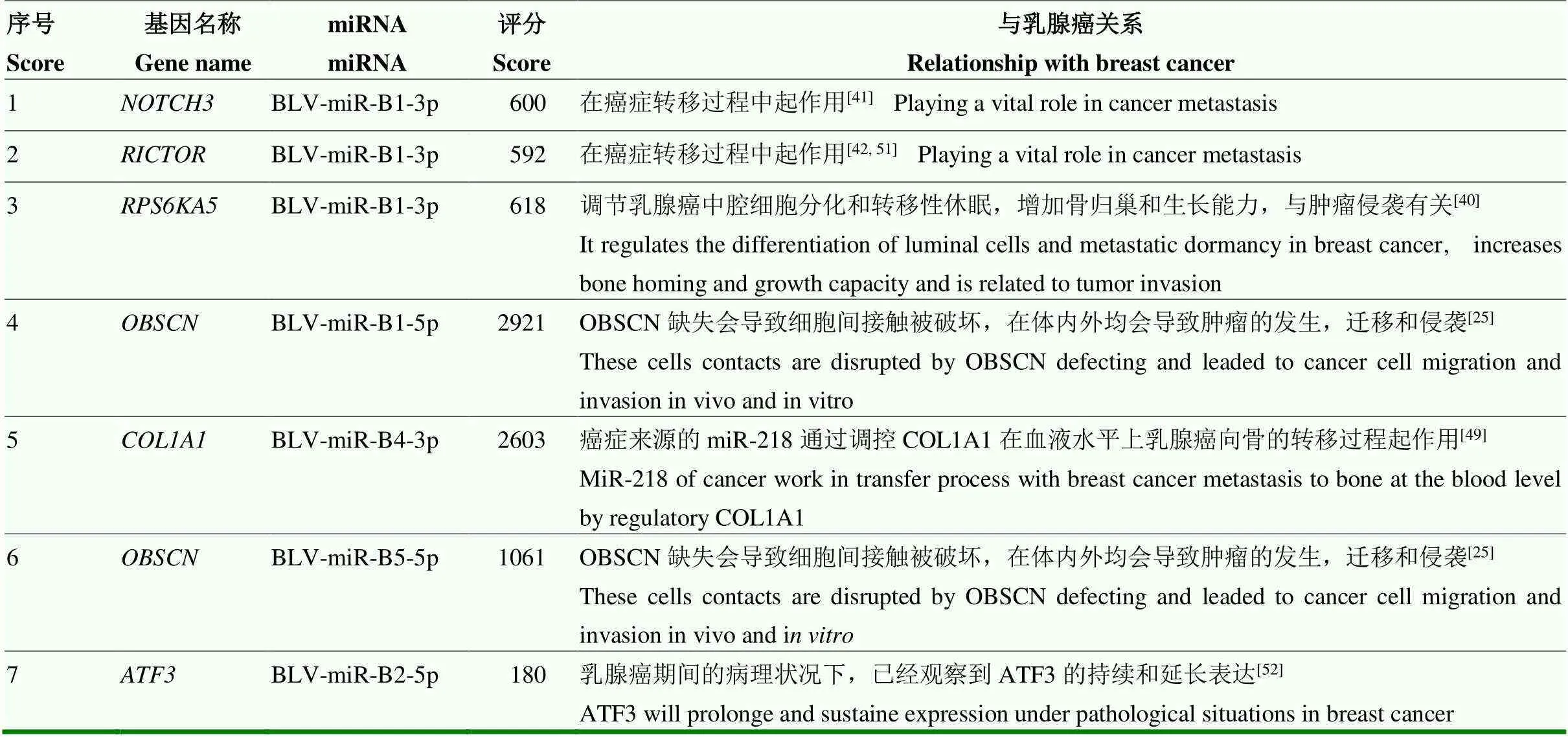
表9 参与调控人乳腺癌的候选靶基因

红色核酸序列:候选靶基因序列;绿色核酸序列:miRNA序列;mfe:最小自由能
3 讨论
20世纪90年代,URSIN等提出了一项前瞻性研究,研究了牛奶消费与癌症之间存在可能联系,评估了包括白血病、淋巴癌等癌症类型,虽然此项调查没有确定牛奶消费与癌症总发病率之间的关系,但是牛奶消费量(每天两杯)和淋巴器官癌症,特别是淋巴癌之间存在密切联系[53],虽然它们之间是通过何种方式进行互作还需进一步研究,但这与我们所探究的BLV miRNAs通过食源途径跨界调控人源基因的风险可以相互印证。
笔者查询分析的70个候选靶基因中,36个候选靶基因的功能与肿瘤的发生发展存在相关性。这些候选靶基因所表达的蛋白在细胞的周期、信号转导、结构/骨架、增殖、凋亡、分化、迁移/侵袭功能中发挥重要作用。值得注意的是,调控细胞增殖与凋亡的候选靶基因均具有双向性,即促进与抑制细胞增殖和凋亡功能的基因共同存在,但对凋亡和增殖功能起抑制作用的基因相对较多,被miRNA靶向后可能增强细胞的代谢速度。有16个候选靶基因对细胞的迁移/侵袭功能产生影响,迁移/侵袭作为癌细胞最主要的功能表达形式,同样证实了BLV miRNAs与癌症存在相关性。
BLV-miR-B4-3p作为BLV编码的10种miRNA中,是患病牛体内表达量最高的一个[10],与白血病相关miR 29a共享同一种子区域[9],人急性淋巴细胞白血病的融合转录本同样作为BLV-miR-B4-3p的候选靶基因,提示BLV miRNA跨界进入人体后可能对人白血病的发生发展产生影响,虽然BLV与人类T淋巴细胞白血病病毒I型和Ⅱ(HTLV-1,2)同源性较高,但目前未见两种疾病间相关性的研究,这也可能是未来BLV的研究方向之一。但一直以来,BLV最受大家关注的公共卫生问题就是其与人乳腺癌之间的关系,虽然目前尚无证据表明BLV就是人源乳腺癌的重要致病因素之一,但近年来美国、澳大利亚、阿根廷等国家已经相继在乳腺癌患者的组织样品中检测到BLV基因片段[7],BLV miRNAs作为BLV在机体内致病功能的主要发动者[9, 54],我们的研究显示BLV miRNA在乳腺癌细胞的分化、迁移、侵袭过程中,均可以发挥重要作用,这一结论对BLV与人源乳腺癌之间相关性的研究有重要意义与实际价值。
BLV miRNAs的候选靶基因中,可以同时被多个BLV miRNA共同靶向的候选靶基因聚类于黏蛋白家族的MUC5B、MUC12、MUC16。其中MUC12可以编码一种完整的膜糖蛋白,可以在结肠表面的凝胶状分泌物中作为关键组成部分;虽然MUC5B是呼吸道黏蛋白的主要贡献者,MUC16是一种众所周知的卵巢癌标志物,但MUC5B在结肠中同样存在大量表达,MUC16也可以作为结肠癌的独立预后标志物[55]。鉴于miRNA所存在的跨界调控机制[16],而结肠作为机体吸收水分的重要肠段,我们推测BLV miRNAs极有可能通过靶向调控结肠组织细胞,破坏黏蛋白功能,进而跨界进入人体,并通过这一途径靶向调控人源基因。基于此,我们认为牛白血病病毒来源的miRNAs具有跨界调控人源基因的风险。
4 结论
外源性牛白血病病毒 miRNA可能跨界调控细胞周期、信号转导、结构/骨架、增殖、凋亡、分化、迁移/侵袭相关等细胞功能相关基因,破坏细胞结构。
牛白血病病毒miRNA与人乳腺癌的相关性可能表现在人乳腺癌细胞的分化、迁移和侵袭过程中;而BLV-miR-B4-3p本身与白血病相关miR 29a共享同一种子区域,可能对人急性淋巴细胞白血病的发生发展造成影响。
外源性牛白血病病毒 miRNA具有靶向抑制黏蛋白基因(MUC5B、MUC12、MUC16)表达,通过破坏肠黏膜形成这一途径,跨界调控人源基因的风险。
[1] GILLET N, FLORINS A, BURTEAU C, NIGRO A, VANDERMEERS F, BALON H, BOUZAR A, DEFOICHE J, BURNY A, REICHERT M, KETTMANN R, WILLEMS L. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology, 2007, 4: 18.
[2] 杨奕. 牛白血病病毒分子流行病学调查及其致病性的研究[D]. 扬州:扬州大学, 2018.
YANG Y. Molecular epidemiological investigation and pathogenicity of bovine leukemia virus[D]. Yangzhou: Yangzhou University, 2018. (in Chinese)
[3] OTT S, JOHNSON R, WELLS S. Association between Bovine- Leukosis virus seroprevalence and herd-level productivity on US dairy farms. Preventive Veterinary Medicine, 2003, 61(4): 249-262.
[4] ERSKINE R, BARLETT P C, BYREM T M, RENDER C L, FEBVAY C, HOUSEMAN J T. Association between bovine leukemia virus, production, and population age in Michigan dairy herds. Journal of Dairy Science, 2012, 95: 727-734.
[5] FRIE M, SPORER K, WALLACE J, MAES R, SORDILLO L, BARTLETT P, COUSSENS P. Reduced humoral immunity and atypical cell-mediated immunity in response to vaccination in cows naturally infected with bovine leukemia virus. Veterinary Immunology and Immunopathology, 2016, 182: 125-135.
[6] MCCLURE H M, KEELING M E, CUSTER R P, MARSHAK R R, ABT D A, FERRER J F. Erythroleukemia in two infant chimpanzees fed milk from cows naturally infected with the bovine C-type virus. Cancer Research, 1974, 34(10): 2745-2757.
[7] MARTINEZ C L, PAMELA L, NIETO F M, DOLCINI G L, CERIANI C. Can bovine leukemia virus be related to human breast cancer? A review of the evidence. Journal of Mammary Gland Biology and Neoplasia, 2018, 23(3): 101-107.
[8] BUEHRING G C, DELANEY A, SHEN H, et al. Bovine leukemia virus discovered in human blood. BMC Infectious Diseases, 2019, 19(1): 297.
[9] KINCAID R P, BURKE J M, SULLIVAN C S, CHU D L C, RAZAVIAN N, SCHWARTZ D A, DEMKOVICH Z R, BATES M N. RNA virus microRNA that mimics a B-cell oncomiR. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(8): 3077-3082.
[10] NICOLAS R, MÉLANIE M, KEITH D, HARUKO T, FLORIAN C, YVETTE C, CÉLINE V, FRANCK M, ERIC W, ARSÈNE B, MICHEL G, ANNE V. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(6): 2306-2311.
[11] MOULES V, POMIER C, SIBON D, GABET A S, REICHERT M, KERKHOFS P, WILLEMS L, MORTREUX F, WATTEL E. Fate of premalignant clones during the asymptomatic phase preceding lymphoid malignancy. Cancer Research, 2005, 65(4): 1234-1243.
[12] MERIMI M, KLENER P, SZYNAL M, CLEUTER Y, KERKHOFS P, BURNY A, MARTIAT P, VAN DEN BROEKE A. Suppression of viral gene expression in bovine leukemia virus-associated B-cell malignancy: interplay of epigenetic modifications leading to chromatin with a repressive histone code. Journal of Virology, 2007, 81(11): 5929-5939.
[13] SAFARI R, HAMAIDIA M, DE BROGNIEZ A, GILLET N, WILLEMS L. Cis-drivers and trans-drivers of bovine leukemia virus oncogenesis. Current Opinion in Virology, 2017, 26: 15-19.
[14] GILLET N A, HAMAIDIA M, DE BROGNIEZ A, GUTIÉRREZ G, RENOTTE N, REICHERT M, TRONO K, WILLEMS L. Bovine leukemia virus small noncoding rnas are functional elements that regulate replication and contribute to oncogenesis. PLoS Pathogens, 2016, 12(4): e1005588.
[15] ROSEWICK N, DURKIN K, ARTESI M, MARÇAIS A, HAHAUT V, GRIEBEL P, ARSIC N, AVETTAND-FENOEL V, BURNY A, CHARLIER C, HERMINE O, GEORGES M, VAN DEN BROEKE A. Cis-perturbation of cancer drivers by the HTLV-1/BLV proviruses is an early determinant of leukemogenesis. Nature Communications, 2017, 8(1): 15264.
[16] ZHANG L, HOU D, LI D, ZHU L Y, ZHANG Y J, LI J, BIAN Z, LIANG X Y, CAI X, YIN Y, WANG C, ZHANG T F, ZHU D H, ZHANG D M, XU J, CHEN Q, BA Y, LIU J, ZHANG C Y. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Research, 2011, 22: 107-126.
[17] IZUMI H, TSUDA M, SATO Y, KOSAKA N, OCHIYA T, IWAMOTO H, NAMBA K, TAKEDA Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. Journal of Dairy Science, 2015, 98(5): 2920-2933.
[18] REHMSMEIER M, STEFFEN P, HOCHSMANN M, GIEGERICH R. Fast and effective prediction of microRNA/target duplexes. RNA -A Publication of The RNA Society, 2004, 10(10): 1507-1517.
[19] MUKHERJEE R, MAJUMDER P, CHAKRABARTI O. MGRN1- mediated ubiquitination of alpha-tubulin regulates microtubule dynamics and intracellular transport. Traffic, 2017, 18(12): 791-807.
[20] XIAO X H, LV L C, DUAN J, WU Y M, HE S J, HU Z Z, XIONG L X. Regulating Cdc42 and its signaling pathways in cancer: Small molecules and microrna as new treatment candidates. Molecules, 2018, 23(4): 787.
[21] FeLBERBAUM-CORTI M, MOREL E, CAVALLI V, VILBOIS F, GRUENBERG J. The redox sensor TXNL1 plays a regulatory role in fluid phase endocytosis. PLoS ONE, 2007, 2(11): e1144.
[22] LIU Z, ZHENG Y. A requirement for epsin in mitotic membrane and spindle organization. Journal of Cell Biology, 2009, 186(4): 473-480.
[23] DORNIER E, COUMAILLEAU F, OTTAVI J F, et al. TspanC8 tetraspanins regulate ADAM10 / Kuzbanian trafficking and promote Notch activation in flies and mammals. Journal of Cell Biology, 2012, 199(3): 481-496.
[24] QUINTERO O A, DIVITO M M, ADIKES R C, KORTAN M B, CASE L B, LIER A J, PANARETOS N S, SLATER S Q, RENGARAJAN M, FELIU M, CHENEY R E. Human Myo19 is a novel myosin that associates with mitochondria. Current Biology, 2009, 19(23): 2008-2013.
[25] SHRIVER M, STROKA K M, VITOLO M I, MARTIN S, HUSO D L, KONSTANTOPOULOS K, KONTROGIANNI-KONSTANTOPOULOS A. Loss of giant obscurins from breast epithelium promotes epithelial- to-mesenchymal transition, tumorigenicity and metastasis. Oncogene, 2015, 34(32): 4248-4259.
[26] JANG S I, KALININ A, TAKAHASHI K, MAREKOV L N, STEINERT P M. Characterization of human epiplakin: RNAi-mediated epiplakin depletion leads to the disruption of keratin and vimentin IF networks. Journal of Cell Science, 2005, 118(Pt 4): 781-793.
[27] JUNG J, KIM J, ROH S H, JUN I, SAMPSON R D, GEE H Y, CHOI J Y, LEE M G. The HSP70 co-chaperone DNAJC14 targets misfolded pendrin for unconventional protein secretion. Nature Communications, 2016, 7: 11386.
[28] AU F K, JIA Y, JIANG K, GRIGORIEV I, HAU B K, SHEN Y, DU S, AKHMANOVA A, QI R Z. GAS2L1 Is a centriole-associated protein required for centrosome dynamics and disjunction. Developmental Cell, 2017, 40(1): 81-94.
[29] BRANCOLINI C, BOTTEGA S, SCHNEIDER C. Gas2, a growth arrest-specific protein, is a component of the microfilament network system. Journal of Cell Biology, 1992, 117(6): 1251-1261.
[30] FU X, FAN X, HU J, ZOU H, CHEN Z, LIU Q, NI B, TAN X, SU Q, WANG J, WANG L, WANG J. Overexpression of MSK1 is associated with tumor aggressiveness and poor prognosis in colorectal cancer. Digestive and Liver Disease, 2017, 49(6): 683-691.
[31] ZHANG M, HUANG N, YANG X, LUO J, YAN S, XIAO F, CHEN W, GAO X, ZHAO K, ZHOU H, LI Z, MING L, XIE B, ZHANG N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene, 2018, 37(13): 1805-1814.
[32] LIU F, LIU X, XU Z, YUAN P, ZHOU Q, JIN J, YAN X, XU Z, CAO Q, YU J, CHENG Y, WAN R, HONG K. Molecular mechanisms of Ellisvan Creveld gene variations in ventricular septal defect. Molecular Medicine Reports, 2018, 17(1): 1527-1536.
[33] WANG Z, LUO H, FANG Z, FAN Y, LIU X, ZHANG Y, RUI S, CHEN Y, HONG L, GAO J, ZHANG M. MiR-204 acts as a potential therapeutic target in acute myeloid leukemia by increasing BIRC6-mediated apoptosis. BMB Reports, 2018, 51(9): 444-449.
[34] MCCLURE H M, KEELING M E, CUSTER R P, MARSHAK R R, ABT D A, FERRER J F. Erythroleukemia in two infant chimpanzees fed milk from cows naturally infected with the bovine C-type virus. Cancer Research, 1974, 34(10): 2745-2757.
[35] BEDAL K B, GRASSEL S, SPANIER G, REICHERT T E, BAUER R J. The NC11 domain of human collagen XVI induces vasculogenic mimicry in oral squamous cell carcinoma cells. Carcinogenesis, 2015, 36(11): 1429-1439.
[36] XIONG W, DENG Z, TANG Y, DENG Z, LI M. Downregulation of KMT2D suppresses proliferation and induces apoptosis of gastric cancer. Biochemical and Biophysical Research Communications, 2018, 504(1): 129-136.
[37] FB U B, CAU L, TAFAZZOLI A, MECHIN M C, WOLF S, ROMANO M T, VALENTIN F, WIEGMANN H, HUCHENQ A, KANDIL R, et al. Mutations in three genes encoding proteins involved in hair shaft formation cause uncombable hair syndrome. American Journal of Human Genetics, 2016, 99(6): 1292-1304.
[38] AO R, GUAN L, WANG Y, WANG J N. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. Journal of Cellular Biochemistry, 2018, 119(6): 4420-4434.
[39] ROHN J L, PATEL J V, NEUMANN B, BULKESCHER J, MCHEDLISHVILI N, MCMULLAN R C, QUINTERO O A, ELLENBERG J, BAUM B. Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Current Biology, 2014, 24(21): 2598-2605.
[40] GAWRZAK S, RINALDI L, GREGORIO S, ARENAS E J, SALVADOR F, UROSEVIC J, FIGUERAS-PUIG C, ROJO F, DEL BARCO BARRANTES I, CEJALVO J M, et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nature Cell Biology, 2018, 20(2): 211-221.
[41] LEONTOVICH A A, JALALIRAD M, SALISBURY J L, MILLS L, HADDOX C, SCHROEDER M, TUMA A, GUICCIARDI M E, ZAMMATARO L, GAMBINO M W, et al. NOTCH3 expression is linked to breast cancer seeding and distant metastasis. Breast Cancer Reserach, 2018, 20(1): 105.
[42] EL SHAMIEH S, SALEH F, MOUSSA S, KATTAN J, FARHAT F. RICTOR gene amplification is correlated with metastasis and therapeutic resistance in triple-negative breast cancer. Pharmacogenomics, 2018, 19(9): 757-760.
[43] BROCKSCHMIDT A, TROST D, PETERZIEL H, ZIMMERMANN K, EHRLER M, GRASSMANN H, PFENNING P N, WAHA A, WOHLLEBER D, BROCKSCHMIDT F F, et al. KIAA1797/FOCAD encodes a novel focal adhesion protein with tumour suppressor function in gliomas. Brain, 2012, 135(Pt 4): 1027-1041.
[44] NGUYEN T T, PARK W S, PARK B O, KIM C Y, OH Y, KIM J M, CHOI H, KYUNG T, KIM C H, LEE G, et al. PLEKHG3 enhances polarized cell migration by activating actin filaments at the cell front. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(36): 10091-10096.
[45] RATZINGER S, EBLE J A, PASOLDT A, OPOLKA A, ROGLER G, GRIFKA J, GRASSEL S. Collagen XVI induces formation of focal contacts on intestinal myofibroblasts isolated from the normal and inflamed intestinal tract. Matrix Biology, 2010, 29(3): 177-193.
[46] DERYUGINA E I, ZAJAC E, ZILBERBERG L, MURAMATSU T, JOSHI G, DABOVIC B, RIFKIN D, QUIGLEY J P. LTBP3 promotes early metastatic events during cancer cell dissemination. Oncogene, 2018, 37(14): 1815-1829.
[47] ZHU C, YANG Q, XU J, ZHAO W, ZHANG Z, XU D, ZHANG Y, ZHAO E, ZHAO G. Somatic mutation of DNAH genes implicated higher chemotherapy response rate in gastric adenocarcinoma patients. Journal of Translational Medicine, 2019, 17(1): 109.
[48] MIYOSHI N, ISHII H, MIMORI K, TANAKA F, NAGAI K, UEMURA M, SEKIMOTO M, DOKI Y, MORI M. ATP11A is a novel predictive marker for metachronous metastasis of colorectal cancer. Oncology Reports, 2010, 23(2): 505-510.
[49] ZHANG Z, FANG C, WANG Y, ZHANG J, YU J, ZHANG Y, WANG X, ZHONG J. COL1A1: A potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. International Journal of Oncology, 2018, 53(5): 1869-1880.
[50] CHOPRA A, SONI S, VERMA D, KUMAR D, DWIVEDI R, VISHWANATHAN A, VISHWAKAMA G, BAKHSHI S, SETH R, GOGIA A, KUMAR L, KUMAR R. Prevalence of common fusion transcripts in acute lymphoblastic leukemia: A report of 304 cases. Asia-Pacific Journal of Clinical Oncology, 2015, 11(4): 293-298.
[51] SCHMIDT K M, DIETRICH P, HACKL C, GUENZLE J, BRONSERT P, WAGNER C, FICHTNER-FEIGL S, SCHLITT H J, GEISSLER E K, HELLERBRAND C, LANG S A. Inhibition of mTORC2/RICTOR impairs melanoma hepatic metastasis. Neoplasia, 2018, 20(12): 1198-1208.
[52] ROHINI M, HARITHA MENON A, SELVAMURUGAN N. Role of activating transcription factor 3 and its interacting proteins under physiological and pathological conditions. International Journal of Biological Macromolecules, 2018, 120(Pt A): 310-317.
[53] URSIN G, BJELKE E, HEUCH I, VOLLSET S E. Milk consumption and cancer incidence: a Norwegian prospective study. British Journal of Cancer, 1990, 61(3): 454-459.
[54] SANTANAM U, ZANESI N, EFANOV A, COSTINEAN S, PALAMARCHUK A, HAGAN J P, VOLINIA S, ALDER H, RASSENTI L, KIPPS T, CROCE C M, PEKARSKY Y. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(27): 12210-12215.
[55] BJORKMAN K, MUSTONEN H, KAPRIO T, HAGLUND C, BOCKELMAN C. Mucin 16 and kallikrein 13 as potential prognostic factors in colon cancer: Results of an oncological 92-multiplex immunoassay. Tumour Biology, 2019, 41(7): 1010428319860728.
Prediction and Bioinformatics Analysis of BLV-miRNA Transboundary Regulation of Human Target Genes
WANG Yong, LI SiYan, HE SiRui, ZHANG Di, LIAN Shuai, WANG JianFa, WU Rui
College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University/Heilongjiang Provincial Key Laboratory of Prevention and Control of Bovine Diseases, Daqing 163319, Heilongjiang
【】To assess risk of regulation of human-derived genes by miRNAs derived from bovine leukemia virus (BLV), the prospective research on the possible food safety problems and the possible impact on human health caused by BLV-miRNA were carried out, which would lay the foundation for the necessary research on the implementation of Enzootic Bovine Leukosis (EBL) prevention and control measures in actual production in the future, and provide theoretical guidance for the study of the relationship between BLV and human diseases.【】In this study, the mature sequence of BLV miRNAs was first queried using mirbase website, and the miRanda software was used to predict target genes. The predictive 10 miRNAs (BLV-miR-B1-3P,5P, BLV-miR-B2-3P, 5P, BLV-miR-B3-3P,5P, BLV-miR-B4-3P,5P, and BLV-miR-B5-3P,5P) were encoded by BLV. The top 10 candidate target genes of each BLV-miRNA score were selected for functional analysis, including a total of 88 duplicated genes. The candidate target genes co-regulated by multiple BLV miRNAs were verified by secondary prediction using RNAhybrid software, and their functions were analyzed. 【】The ten miRNAs encoded by BLV were predicted to obtain 1 630-16 383 target genes, respectively. After functional analysis of eighty-eight candidate target genes in the top ten, it was found that eighteen of them had no relevant functional reports. Thirty-six candidate target genes were related to the occurrence and development of neoplastic diseases. Two candidate target genes could regulate cell cycle. Sixteen candidate target genes were involved in the regulation of cell signal transduction. Fourteen candidate target genes played a role in the formation of structure/cytoskeleton proteins. The function of cell proliferation and apoptosis showed an antagonistic relationship and the genes that often promoting proliferation could also suppress apoptosis. A total of thirteen genes played a regulatory role in cell proliferation and apoptosis. Interestingly, the regulation of the thirteen candidate target genes on cell proliferation and apoptosis was bidirectional. However, it was not clear whether the regulation of BLV miRNA towards cells was more prone to proliferation or apoptosis, so further studies were still needed to discuss in depth. Two candidate target genes could regulate cell differentiation. The sixteen candidate target genes played a role in regulating cell migration/invasion function, again suggesting that BLV miRNA might have a more important correlation with neoplastic diseases. The seven candidate target genes might play an important role in the differentiation, migration and invasion of breast cells, suggesting that the study on the correlation between BLV and human breast cancer could be further discussed from the perspective of BLV miRNA. Two candidate target genotypes of BLV-B4-3P, Collagen 1 chain gene (COL1A1), had a regulatory effect on human acute lymphoblastic leukemia (ALL). In addition, candidate target genes that could be co-targeted by multiple BLV miRNAs belong to the mucin family (MUC5B, MUC12 and MUC16), and be expressed in the colon, influencing the formation of colon mucosa.【】Exogenous BLV miRNA might transboundary regulate cell cycle signal transduction structure/cytoskeleton proliferation apoptosis differentiation migration/invasion related cell function related genes and destroy cell structure. The correlation between BLV miRNA and human breast cancer might be shown in the process of differentiation, migration and invasion of human breast cancer cells. BLV-miR-B4-3p shared a seed sequence with miR 29a, which might affect the occurrence and development of human acute lymphoblastic leukemia. Exogenous BLV miRNA had the target of inhibiting the expression of mucin genes, such as MUC5B, MUC12, and MUC16, through the destruction of intestinal mucosa formation to achieve transboundary regulation of human gene risk.
bioinformatics; bovine leukemia virus; transboundary regulation; human-derived genes; miRNA

10.3864/j.issn.0578-1752.2021.03.019
2020-02-23;
2020-07-29
国家自然科学基金(2041340046)、黑龙江省自然科学基金(YQ2019C014)、研究生创新科研项目(YJSCX2019-Y39)
王雍,Tel:13251599676;E-mail:bywy0209@126.com。通信作者武瑞,Tel:13836961026;E-mail:fuhewu@126.com
(责任编辑 林鉴非)
