黄条生长性状全基因组关联分析*
2021-03-05崔爱君徐永江柳学周
崔爱君 徐永江 王 滨 姜 燕 柳学周
崔爱君1,2徐永江1,2①王 滨1姜 燕1柳学周1
(1. 中国水产科学研究院黄海水产研究所 青岛海洋科学与技术试点国家实验室海洋渔业科学与食物产出过程功能实验室 青岛 266071;2. 上海海洋大学水产与生命学院 上海 201306)


全基因组关联分析(Genome-wide association study, GWAS)是应用全基因组范围内的大量分子标记(一般为SNP),将标记基因型结合性状表型进行联合分析,统计每个标记与目标性状之间的关联性大小(一般用值表示),鉴定出与目标性状密切相关且具有特定功能和育种潜力的基因位点或分子标记,主要用于物种经济性状相关SNP分子标记以及功能基因的鉴定,从而达到缩短育种周期和提高育种效率的目的,目前已在畜禽等脊椎动物育种中广泛应用(Tavares, 2020; Müller, 2019; Cui, 2016; Zhang, 2019)。近年来,随着基因组高通量测序技术的发展及测序成本的降低,GWAS开始应用于水产养殖动物的育种研究,如在大黄鱼()、鲶鱼()、凡纳滨对虾()、龙胆石斑鱼()、虾夷扇贝()等物种的生长性状关联SNP位点、候选基因的挖掘和鉴定(Zhou, 2019; Li, 2017; Yu, 2019; Wu, 2019; Ning, 2019)方面应用并取得了一定进展。但是,与陆生脊椎动物相比,GWAS在水产动物育种中的应用尚处于起步阶段。
1 材料与方法
1.1 实验材料
1.2 基因组DNA提取与质量检测
使用QIAGEN公司生产的动物基因组DNA提取试剂盒(DP121221),参照试剂盒使用说明,提取鳍条基因组DNA。用NanoDrop 2000分光光度计(Thermo, 美国)测定基因组DNA浓度,通过1%琼脂糖凝胶电泳检测DNA的完整性,通过260 nm/280 nm的比值来判断DNA的质量。将质检合格的DNA浓度稀释至100 ng/µl,于–20℃条件保存备用。
1.3 文库构建与测序
将≥200 ng的各样品基因组DNA采用IIB型限制性内切酶XI进行酶切,酶切产物分别加入5组不同的接头,使用T4 DNA Ligase连接,然后PCR扩增连接产物,最后根据5组接头信息,将5个标签按顺序串联,连接产物添加barcode序列,混库,使用Illumina Hiseq测序平台对混合好的文库进行Paired-end测序。
1.4 数据分析

1.4.2 测序数据分析与SNP分型 Illumina HiSeq测序平台得到的原始图像数据文件经碱基识别转化为Raw Reads,过滤删除含有接头序列的Reads,得到Clean Reads,过滤删除含有N碱基比例大于8%的Reads,过滤删除低质量Reads(质量值低于Q30的碱基超过15%);利用Pear (Zhang, 2014)软件(V0.9.6)将成对的Clean Reads拼接,提取出各样品对应的Reads,过滤删除不含酶切识别位点的Reads后,得到各样品的Enzyme Reads;利用电子酶切从参考基因组中提取含有酶切识别位点的标签,作为参考序列,利用SOAP软件将各样品的Enzyme Reads比对到参考序列上,主要参数为-r0–M4–v2 (-r0指唯一比对;–M4指最优比对;–v2指比对允许2个错配),对比对到相同标签的reads聚类,得到unique标签深度,选择样品深度>3×且深度<500、标签长度为27 bp的标签,利用SOAP软件(V 2.21) (Li, 2008)将测序数据比对到参考序列,利用最大似然法(ML)进行位点的分型(Fu, 2013),过程中使用的RAD分型软件包(RAD typing),包含10余个软件组分,覆盖了从数据预处理至最终分型结果输出的全过程。
1.4.3 全基因组关联分析 使用EMMA eXpedited (EMMAX)高效混合模型(Kang, 2010),通过方差分量方法进行SNP分子标记和表型性状的全基因组关联分析,所用模型:
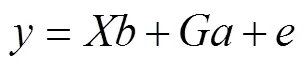
式中,为表型值;为固定效应关联矩阵,为固定效应向量,为通过SNP标记计算得到的关系矩阵,为随机加性遗传方差的参数,为剩余效应的向量。
每个SNP位点能得到1个关联值。对GWAS给出的值划定2条显著性水平线,其中1条经Bonferroni校正=0.05/来确定全基因组显著性阈值(Bonferroni, 1936),为SNP标记的个数,2个性状经Bonferroni校正后显著关联阈值–lg=5.726;另一条使用R软件包中的p.adjust()函数计算得到经FDR校正后的阈值,体质量性状潜在显著关联阈值–lg= 4.091,全长性状潜在显著关联阈值–lg=4.413,挑选Scaffold长度的前30使用R软件包的qqman绘制曼哈顿图,绘制QQ图对关联分析进行评价,判断关联分析结果是否可靠。
1.5 候选基因鉴定及功能分析


式中,为所有基因中具有KEGG注释的基因数目,为中差异表达基因中具有的KEGG注释的基因数目,为所有基因中注释为某特定KEGG的基因数目,为注释某特定KEGG的差异表达基因的数目。计算的结果会返回一个富集显著性的值,小的值表示基因在该Pathway中出现富集,当≤0.05表示显著富集。
2 结果
2.1 表型性状描述性统计


Tab.1 Descriptive statistics of growth traits of yellowtail kingfish
2.2 SNP分型
对2b-RAD简化基因组测序数据按照以下指标进一步过滤。剔除所有样品中低于80%个体可以分型的位点;剔除MAF低于0.05的位点,剔除等位基因大于2的位点。最终,测序获得26665个SNP位点进行GWAS分析。
2.3 全基因组关联分析

图1 黄条体质量性状GWAS关联分析的QQ检验

图2 黄条全长性状GWAS关联分析的QQ检验
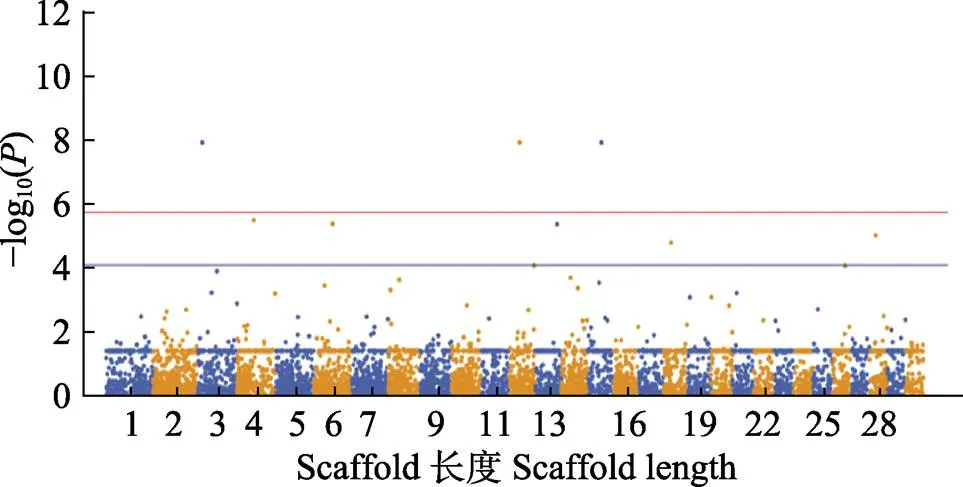
图3 黄条体质量性状GWAS分析的曼哈顿图
红色实线代表全基因显著关联阈值:–log10=5.726, 蓝色实线代表潜在显著关联阈值: –log10=4.091
The red solid line indicates the genome wide significant threshold: –log10=5.726. The blue solid line indicates the threshold for the significance of “suggestive association”: –log10=4.091

图4 黄条全长性状GWAS分析的曼哈顿图
红色实线代表全基因显著关联阈值–log10=5.726, 蓝色实线代表潜在显著关联阈值–log10=4.413
The red solid line indicates the genome wide significant threshold: –log10=5.726. The blue solid line indicates the threshold for the significance of “suggestive association”: –log10=4.413
2.4 候选基因生物信息学分析
3 讨论

表4 体质量性状和全长性状KEGG注释结果

Tab.4 KEGG annotation results of body weight and total length trait

Bonferroni CE. Teoria statistical delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commericiali di Firenze, 1936, 8: 3–62
Chen SL, Xu WT, Liu Y. Fish genomic research: Decade review and prospect. Journal of Fisheries of China, 2019, 43(1): 1– 14 [陈松林, 徐文腾, 刘洋. 鱼类基因组研究十年回顾与展望. 水产学报, 2019, 43(1): 1–14]
Chen ZD, Wang WH. Genome-wide association study on feet weight in chicken (). Journal of Agricultural Biotechnology, 2016, 24(10): 1569–1577 [陈则东, 王文浩. 鸡脚重性状的全基因组关联分析. 农业生物技术学报, 2016, 24(10): 1569–1577]
Cingolani P, Platts A, Wang LL,. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome ofstrain w1118; iso-2; iso-3. Fly, 2012, 6(2): 80–92
Cui ZH, Luo JD, Qi CY,Genome-wide association study (GWAS) reveals the genetic architecture of four husk traits in maize. BMC Genomics, 2016, 17(1): 946
Fu X, Dou J, Mao J,RAD typing: An integrated package for accuratecodominant and dominant RAD genotyping in mapping populations. PLoS One, 2013, 8(11): e79960
Johannessen M, Moller S, Hansen T,. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cellular and Molecular Life Sciences, 2006, 63(3): 268–284
Kang HM, Sul JH, Service SK,. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics, 2010, 42(4): 348–354
Li N, Zhou T, Geng X,. Identification of novel genes significantly affecting growth in catfish through GWAS analysis. Molecular Genetics and Genomics, 2017, 293(3): 1–13
Li R, Li Y, Kristiansen K,. SOAP: Short oligonucleotide alignment program. Bioinformatics, 2008, 24(5): 713–714


Matthias C, Edwige T, Bernadette B,. FHL2 interacts with both ADAM-17 and the cytoskeleton and regulates ADAM- 17 localization and activity. Journal of Cellular Physiology, 2006, 208: 363–372
Müller BSF, de Almeida Filho JE, Lima BM,. Independent and joint-GWAS for growth traits inby assembling genome-wide data for 3373 individuals across four breeding populations. New Phytologist, 2019, 221(2): 818–833
Naha BC, Prasad A, Sailo L,, Concept of genome wide association studies and its progress in livestock. International Journal of Science and Nature, 2016, 7(1): 39–42
Nguyen NH, Premachandra HKA, Kilian A,. Genomic prediction using DArT-Seq technology for yellowtail kingfish. BMC Genomics, 2018a, 19(1): 107
Nguyen NH, Rastas PMA, Premachandra HKA,. First high- density linkage map and single nucleotide polymorphisms significantly associated with traits of economic importance in yellowtail kingfish. Frontiers in Genetics, 2018b, 9: 127
Ning XH, Li X, Wang J,. Genome-wide association study reveals E2F3 as a candidate gene for scallop growth. Aquaculture, 2019, 73(4): 734216
Ohara E, Nishimura T, Nagakura Y,Genetic linkage maps of two yellowtails (and). Aquaculture, 2005, 244: 41–48
Pi X, Ren R, Kelley R,Sequential roles for myosin-X in BMP6 dependent filopodial extension, migration, and activation of BMP receptors. Journal of Cell Biology, 2008, 179(7): 1569–1582
Premachandra HKA, De la Cruz FL, Takeuchi Y,. Genomic DNA variation confirmedcomprises three different populations in the Pacific, but with recent divergence. Scientific Reports, 2017, 7(1): 9386
Raise A, Stefanie W, Ralf J,. Hunting for the function of orphan GPCRs-beyond the search for the endogenous ligand. British Journal of Pharmacology, 2015, 172(13): 3218–3228
Sepulveda FA, Gonzalez M. Spatio-temporal patterns of genetic variations in populations of yellowtail kingfishfrom the southeastern Pacific Ocean and potential implications for its fishery management. Journal of Fish Biology,2017, 90(1): 249–264

Sicuro B, Luzzana U. The state ofspp. other than yellowtail () farming in the world. Reviews in Fisheries Science and Aquaculture, 2016, 24(4): 314–325
Swart BL, Merwe BVD, Kerwath SE,Phylogeography of the pelagic fishat different scales: Confirmationof inter-ocean population structure and evaluation of southern African genetic diversity. South African Journal of Marine Science, 2016, 38(4): 513–524
Symonds JE, Walker SP, Pether S,. Developing yellowtail kingfish () and hāpuku () for New Zealand aquaculture. New Zealand Journal of Marine and Freshwater Research, 2014, 48(3): 371–384
Tao L, He XY, Di R,. Research progress on genome-wide association study for growth-related traits in livestock and poultry. Chinese Journal of Animal Science, 2019, 55(11): 34–41 [陶林, 贺小云, 荻冉, 等. 畜禽生长发育相关性状的全基因组关联分析研究进展. 中国畜牧杂志, 2019, 55(11): 34–41]
Tavares V, Pinto R, Assis J,. Venous thromboembolism GWAS reported genetic makeup and the hallmarks of cancer: Linkage to ovarian tumour behavior. Biochimica et Biophysica Acta - Reviews on Cancer, 2020, 1873(1): 188331
Wang B, Xu Y, Liu X, et al. Molecular characterization and expression profiles of insulin-like growth factors in yellowtail kingfish () during embryonic development. Fish Physiology and Biochemistry, 2019, 45(1): 375-390
Whatmore P, Nguyen NH, Miller A,. Genetic parameters for economically important traits in yellowtail kingfish. Aquaculture, 2013, 400(25): 77–84
Woolner S, O'Brien LL, Wiese C, etMyosin-10 and actin filaments are essential for mitotic spindle function. Journal of Cell Biology, 2008, 182(1): 77–88
Wu LN, Yang Y, Li, BJ,. First genome-wide association analysis for growth traits in the largest coral reef-dwelling bony fishes, the giant grouper (. Marine Biotechnology, 2019, 21(5): 707–717
Yu Y, Wang QC, Zhang Q, Genome scan for genomic regions and genes associated with growth trait in pacific white shrimpMarine Biotechnology, 2019, 21(3): 374–383
Zhang J, Kobert K, Flouri T,. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics, 2014, 30(5): 614–620
Zhang YF, Zhang JJ, Gong HF,. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Science, 2019, 150: 47–55
Zhou, Z. Han, K. Wu, Y,Genome-wide association study of growth and body-shape-related traits in large yellow croaker () using ddRAD sequencing. Marine Biotechnology, 2019, 21(5): 655–670
Genome-Wide Association Analysis of Growth Traits in Yellowtail Kingfish ()
CUI Aijun1,2, XU Yongjiang1,2①, WANG Bin1, JIANG Yan1, LIU Xuezhou1
(1.Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266071; 2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306)
The genetic resources available for the commercially important pelagic yellowtail kingfish () are relative sparse. 2b-RAD simplified genome sequencing technology was applied to screen single nucleotide polymorphisms (SNPs) in yellowtail kingfish, and a total of 26,665 SNPs were obtained. A genome-wide association study was carried out to detect body weight- and total length-associated SNPs in 119 individuals from the yellowtail kingfish population in the Yellow Sea. The results showed that 17 SNPs associated with body weight and with potential genome-wide significance were found. Genes in the candidate regions with 1 Mb windows were screened, and 17 candidate genes were obtained. A total of 12 SNPs associated with total length and with potential genome-wide significance were identified, and 12 candidate genes were found. For these candidate genes, KEGG pathway analysis showed that they are mainly involved in the metabolic regulation pathway of growth and development in other vertebrates, which may be important candidate SNP loci and functional genes closely related to the growth traits of yellowtail kingfish. The present results could provide genetic information for the sustainable utilization of germplasm resources and genetic breeding of yellowtail kingfish in the in the future.
; Growth trait; Genome-wide association study (GWAS); 2b-RAD; Simplified genome
XU Yongjiang, E-mail: xuyj@ysfri.ac.cn
S917; Q78
A
2095-9869(2021)02-0071-08
10.19663/j.issn2095-9869.20200205002
http://www.yykxjz.cn/

Cui AJ, Xu YJ, Wang B, Jiang Y, Liu XZ. Genome-wide association analysis of growth traits in yellowtail kingfish (). Progress in Fishery Sciences, 2021, 42(2): 71–78
*山东省支持青岛海洋科学与技术试点国家实验室重大科技专项(2018SDKJ0303-1)、中国水产科学研究院基本科研业务费(2019GH15)、山东省重点研发计划项目(2018GHY115044)、国家重点研发计划项目(2019YFD0900901; 2018YFD0901204)和国家海水鱼产业技术体系(CARS-47)共同资助 [This work was supported by Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0303-1), Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2019GH15), Key Research and Development Program of Shandong Province (2018GHY115044), National Key Research and Development Program of China (2019YFD0900901; 2018YFD0901204) and China Agriculture Research System (CARS-47)]. 崔爱君,E-mail: aijun0218@126.com
徐永江,研究员,E-mail: xuyj@ysfri.ac.cn
2020-02-05,
2020-03-02
(编辑 冯小花)
