circRNA及其调控动物骨骼肌发育研究进展
2020-12-18郑婷甘麦邻沈林園牛丽莉郭宗义王金勇张顺华朱砺
郑婷,甘麦邻,沈林園,牛丽莉,郭宗义,王金勇,张顺华,朱砺
circRNA及其调控动物骨骼肌发育研究进展
郑婷1,甘麦邻1,沈林園1,牛丽莉1,郭宗义2,王金勇2,张顺华1,朱砺1
1. 四川农业大学动物科技学院,成都 611130 2. 重庆市畜牧科学院,荣昌 402460
环状RNA (circular RNA, circRNA)是一类反向剪接形成的闭合环状RNA分子,广泛存在于生物体内,近年来成为非编码RNA中的研究热点。骨骼肌在生物体中起到协调运动的作用,是维持体内正常代谢和内分泌的器官之一。随着测序技术和生物信息学分析技术的发展,circRNA在动物骨骼肌发育中的功能和调控机制逐渐被揭示。本文就当前circRNA的分子调控机制类型、circRNA经典研究思路及功能研究方法以及circRNA参与骨骼肌正常发育与骨骼肌疾病发生调控的研究进展进行了综述,以期为进一步深入研究circRNA参与骨骼肌生长发育调控的遗传机制以及circRNA相关研究方法提供参考。
环状RNA;骨骼肌发育;骨骼肌疾病;RNA测序
骨骼肌是动物体执行运动功能的重要器官,同时也是体内重要的内分泌和代谢器官,与动物有机体的正常生长发育密切相关[1,2]。骨骼肌中肌纤维是由大量单核成肌细胞融合成多核肌管,进一步分化成熟而来。肌纤维聚集形成肌束,大量肌束和结缔组织、毛细血管等共同构成了肌肉组织[3,4]。研究表明,MRFs (myogenic regulator factors)[5,6]、MEF2 (myocyte enhance factor 2)[7]、MSTN (myostatin)[8]和Pax基因家族[9]参与调控骨骼肌发育的基本过程。除了这些编码基因,越来越多的非编码RNA也参与调控骨骼肌生长发育过程,主要包括微小RNA (microRNA, miRNA)、长链非编码RNA (long non-coding RNA, lncRNA)和环状RNA (circular RNA, circRNA)[10~12]。
近年来,circRNA的相关研究日益增多,这种非编码RNA具有表达特异性,且在生物体内不易被降解,在生物体发育和肿瘤发生过程中的生物学功能被逐渐揭示。本文归纳了circRNA的分子调控机制,分析了circRNA经典研究方法,并结合circRNA在骨骼肌发育调控方面的最新研究,对circRNA在骨骼肌正常生长发育与相关疾病发生中的调控作用进行了总结,以期为进一步阐明circRNA参与骨骼肌发育的遗传调控机制提供参考,为探究circRNA生物学功能提供研究思路。
1 circRNA的产生及作用机制
1.1 circRNA产生方式及特征
circRNA通过反向剪接形成闭合环状RNA分子。目前发现的circRNA生物形成方式包括直接反向剪切、外显子跳跃、内含子的反义互补序列配对模式、依赖于RNA结合蛋白(RNA-binding proteins, RBPs)的环化模式以及类似于可变剪切的可变环化模式等。Gao等[13]对circRNA外显子结构和可变剪接事件进行了研究,发现circRNA普遍是通过可变剪接形成的,并且大部分circRNA由外显子剪接形成(ecircRNAs),主要存在于细胞质(图1A);由内含子剪接环化的circRNA (ciRNAs)主要存在于细胞核中(图1C);一小部分既含外显子也含内含子的circRNA (EIciRNAs),主要也存在于细胞核中(图1B)[14~16]。circRNA因其闭合环状的结构,在细胞中不易被RNA核酸外切酶降解,稳定性好。此外,大部分circRNA在不同物种间是进化保守的,circRNA在不同组织或发育阶段的表达模式具有较强的组织时空特异性[17]。另外,已经有大量研究发现circRNA与许多疾病的发生和发展有关[18,19],有潜力成为疾病诊断的生物标志物。
1.2 circRNA功能调控机制
目前普遍认可的circRNA分子调控机制包括:竞争性结合miRNA分子;调控基因的转录;与RBPs互作间接调控RNA或DNA;参与蛋白质翻译过程;作为蛋白质支架促进蛋白酶化学修饰(图1)[12,20~22]。
1.2.1 circRNA竞争性吸附miRNA
这类circRNA含有miRNA结合位点,能竞争性吸附miRNA进而调控miRNA下游靶mRNA表达(图1E)。在生物体中这类circRNA大多是由外显子剪接而成的ecircRNA。现已发现相当数量的竞争性内源circRNA,如Hansen等[23]在人()和小鼠()脑组织中发现一种高表达的,含有至少70个miR-7的结合位点,间接调控相关靶基因表达;由性别决定基因(sex- determining region Y)剪接形成的含有至少16个miR-138结合位点,作为miR-138分子海绵发挥调控作用[23]。
1.2.2 circRNA调控亲本基因转录
细胞核中的ciRNAs能够与RNA聚合酶II (RNA polymerase II, Pol II)相互作用,从而促进其自身编码基因的转录(图1D),如在人细胞中首次发现的ciRNA (),在亲本基因转录位点上大量积累,发挥顺式调控作用[15]。此外,EIciRNAs能够与Pol II和U1小核核糖核蛋白(U1 small nuclear ribonucleoproteins, snRNP)结合,促进其亲本基因的转录(图1D),如Li等[16]发现的一种EIciRNA ()。
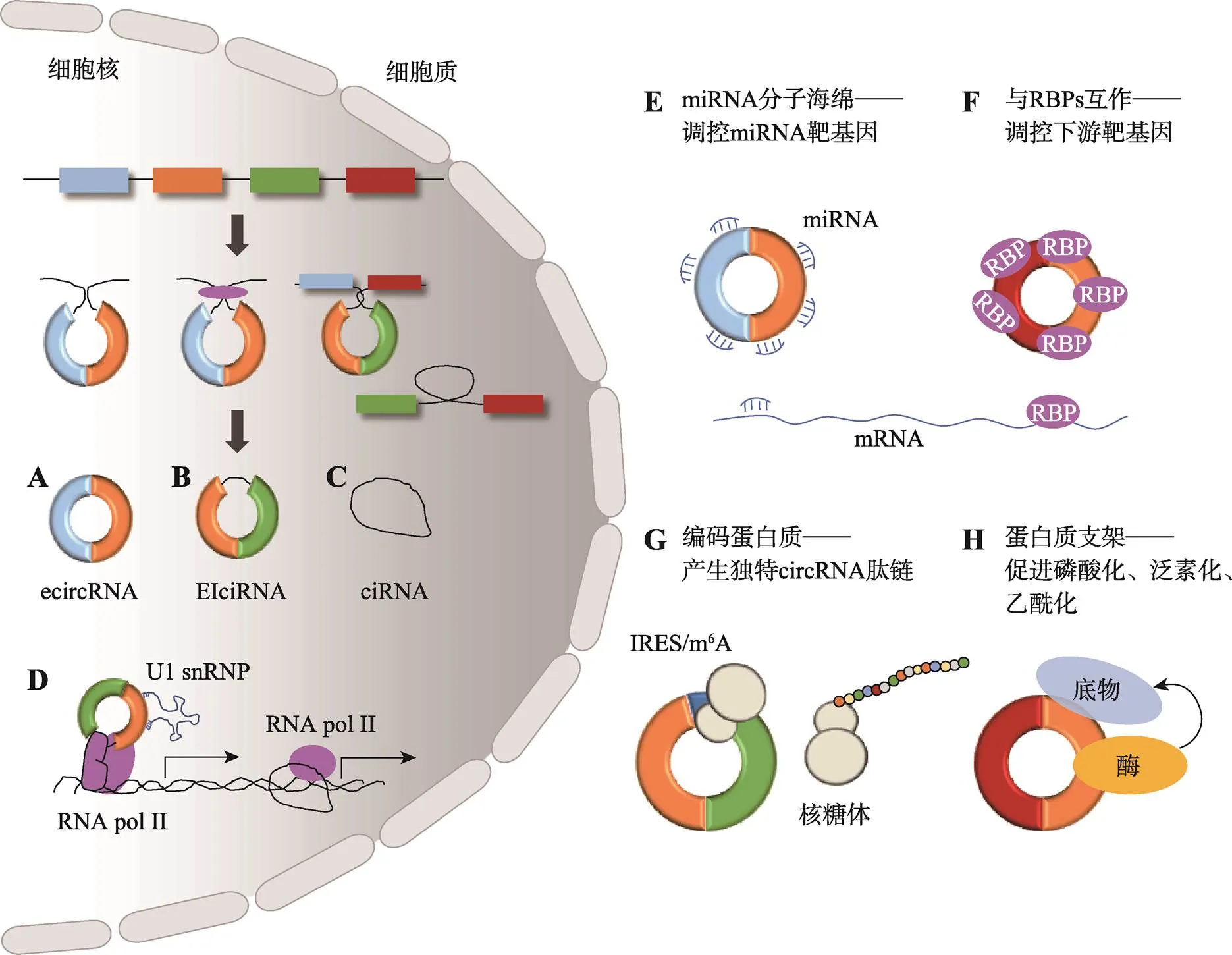
图1 circRNA产生的方式及生物学功能
A:外显子剪接形成的ecircRNA;B:既含外显子又含内含子的EIciRNA;C:内含子剪接形成的ciRNA;D:circRNA调控亲本基因转录;E:circRNA竞争性结合miRNA;F:circRNA与RBPs互作;G:circRNA自身翻译蛋白质;H:circRNA作为蛋白质支架,促进酶及其底物的共域化。参考文献[12,20~22]总结绘制。
1.2.3 circRNA与RBPs结合互作
circRNA可以直接与蛋白质相互作用形成RNA-蛋白质复合体(RNA-protein complex, RPC),进一步与RNA或DNA结合发挥调控作用(图1F)[24],如能与miRNA效应因子AGO蛋白相结合[25]。另外,Ashwal-Fluss等[26]发现在人和果蝇()盲肌基因(muscleblind)第二个外显子环化形成的,与MBL蛋白特异性结合,平衡体内和mRNA的水平进而发挥调控作用。
1.2.4 circRNA参与蛋白质翻译
有研究发现,具有内部核糖体进入位点(internal ribosome entry site,IRES)的circRNA可以招募核糖体启动翻译机制(图1G),如编码蛋白质FBXW7-185aa抑制了胶质瘤细胞的增殖[27];作为一种可以翻译蛋白质的circRNA,在肌生成过程中也发挥调控作用[28]。
1.2.5 circRNA作为蛋白质支架促进酶及其底物的共域化
与AKT1 (protein kinase B)和PDK1 (phosphoinositide dependent protein kinase 1)结合,促进AKT1的PDK1依赖性磷酸化,激活AKT1在小鼠模型中起到心脏保护作用[29];可以促进小鼠双微粒体基因(murine double minute 2)介导的p53突变体泛素化,促使其靶向蛋白酶体降解[30](图1H)。
2 circRNA研究策略
总结现有研究报道,circRNA的经典研究思路是基于生物学问题,通过转录组测序技术(RNA-seq)或微阵列芯片检测(Microarray)得到circRNA数据集,筛选出两种或多种差异表型样本间的差异circRNA,接着对差异circRNA进行鉴定和软件分析,补充相应的circRNA数据库信息,再结合生物信息学分析手段,预测circRNA生物学功能,完成后续功能验证。
2.1 circRNA的发现与鉴定手段
近年来,研究非编码RNA在特定细胞或组织中某一特定时刻的表达和功能,RNA-seq成为有力的研究工具,也是目前circRNA研究中最常见的测序手段[31]。针对circRNA研究,RNA-seq包括文库构建、上机测序、数据分析处理与功能预测等基本流程[32]。根据不同的研究目的与需求,研究者通常选择全转录组测序或circRNA测序来实现对circRNA的筛选。前者可同时探究编码和非编码RNA的表达模式,适用于circRNA生物学功能研究;后者则适用于circRNA的富集,有助于挖掘未知circRNA。两者主要差异在于测序文库的构建,circRNA测序文库除了要求去除绝大多数rRNA和poly(A)外,需用核糖核酸外切酶RNase R处理去除线性RNA的干扰。大多数研究报道显示去线性RNA后得到的circRNA丰度降低,因为某些circRNA对RNase R敏感而可能被消化[33,34]。目前已发现的circRNA数量差异很大,从几百到几千不等。这种可变性可能与RNA样本预处理的方式、测序的深度以及用于数据筛选的过滤器等因素有关。值得注意的是,circRNA的可变剪接对区分测序结果中的正反义链来源有要求,因此circRNA测序最好构建链特异性文库,能够更准确地检测转录表达水平[35]。
得到circRNA测序数据后,通常基于find_circ[36~38]、CIRCexplorer2[36,38]与CIRI36~38]等识别软件,对circRNA进行预测鉴定。Hansen等[39]建议结合使用MapSplice与circRNA finder这两个软件算法,更能提高预测的可靠性。由于软件算法使用的要求各不相同,假阳性率存在很大差异,故由算法检测到的circRNA需要进一步验证。为鉴定circRNA的存在,通常使用实时荧光定量PCR (quantitative real-time PCR, qRT-PCR)[40]、Northern印迹杂交(Northern blot)[27]、原位杂交(hybridization, ISH)[41]、RPAD(RNase R treatment, polyadenylation, and poly(A)+ RNA Depletion)[42]等技术检测circRNA。
Microarray芯片也是分析检测circRNA的有效工具,在许多疾病肿瘤的临床诊断和监测中广泛应用。Microarray的检测成本及分析难度都低于RNA-seq,它们主要区别在于:(1) Microarray检测circRNA必须有已知参考序列,而RNA-seq能检测未知circRNA;(2) Microarray检测的本质是核酸杂交,可定量circRNA表达,而RNA-seq则不能准确定量circRNA[43];(3) Microarray可高效检测反向剪接位点序列,相比RNA-seq可得到更大数量的circRNA[44]。Microarray为circRNA的发现提供了高灵敏度和特异性的平台,也提供了高效的circRNA标记系统。然而Microarray检测的缺点之一是对样品进行预处理需要高的总RNA输入,利用Microarray芯片也得不到全转录组测序所得对应的线性RNA数据[45]。在参考序列未知的情况下,很多研究通常先用RNA-seq测得全转录组序列,再通过Microarray对circRNA进行深入分析。
2.2 circRNA生物学功能的研究策略
如图1所示,circRNA的功能调控机制多样,包括竞争性吸附miRNA、与RNA结合蛋白互作、在翻译水平编码蛋白质等。研究人员通常利用实验技术调控circRNA表达水平,揭示circRNA与其他分子的相互作用以阐明circRNA的功能。
2.2.1 circRNA竞争性吸附miRNA
验证circRNA竞争性结合miRNA从而调控下游效应基因是目前circRNA功能研究中最为常见、且相对较为成熟的研究方向。通常情况下,利用RNA-seq数据找到可能具有生物学功能的差异表达circRNA,分别将预测的靶miRNA、靶mRNA和差异表达miRNA、mRNA取交集,构建circRNA- miRNA-mRNA网络调控图,再利用软件进行GO (gene ontology)功能注释和KEGG (Kyoto encyclopedia of genes and genomes)通路分析[46,47],预测相关circRNA的生物学功能和调控通路。结果表明,大量circRNA具有miRNA的结合位点,但它们的结合关系和相互作用还需要严谨的实验验证。通常利用双荧光素酶报告系统、RNA pull down等实验技术进行结合关系的验证<[48~51],通过circRNA功能获得缺失实验,验证其对靶miRNA和靶基因的功能调控作用[51,52]。
2.2.2 circRNA与RBPs互作
虽然与对应线性RNA相比,circRNA与RBPs的结合位点很少被发现有富集,但也有不少研究证实了两者之间的功能调控关系。目前circRNA与蛋白的相互作用主要通过RNA pull-down和RNA免疫沉淀法(RNA immunoprecipitation, RIP)分析[53,54]。根据circRNA反向剪接位点(back-splice junction region, BSJ)设计探针,获取目标circRNA,其相关蛋白可以利用Western Blot或质谱法进行分析鉴定[55,56]。另外,为了鉴定与感兴趣的蛋白相关的circRNA,也可以使用针对该蛋白的抗体进行RIP分析[57,58],并使用交联免疫沉淀法(crosslinking immunoprecipitation, CLIP)定位circRNA上确切的蛋白结合位点[54,57]。为避免线性RNA的干扰,在RNA pull-down和免疫沉淀之前,必须去除线性RNA。
2.2.3 circRNA编码蛋白质
目前已发现有的circRNAs能够被翻译成多肽,形成特殊的功能蛋白质。研究发现,编码蛋白的circRNA具有开放阅读框(open reading frame, ORF)、内部核糖体进入位点(internal ribosome entry site, IRES)或N6-甲基腺苷(N6-methyladenosine, m6A)基序[59~62]。目前ORFfinder[63]、IRESfinder[64]、m6Apred[65]等多种生物学工具能够辅助预测circRNA中潜在的ORF、IRES和m6A序列。预测circRNA编码潜能后,还需从多个方面对circRNA的编码功能进行验证。通过深度测序结合核糖体印记技术,可以筛选出与核糖体结合的circRNA[66];在circRNA预测的ORF终止密码子上游插入特异蛋白标签,通过检测标签蛋白免疫荧光探针来验证circRNA的翻译功能[27,61,67]。通过标签研究circRNA编码的肽段,既能识别可能与circRNA衍生肽段互作的RNA或蛋白,又能揭示circRNA肽段的亚细胞定位[27,28,68]。研究circRNA衍生肽的生物学功能,通过敲低或过表达目的circRNA或设计特定细胞刺激,检测circRNA蛋白产物水平,分析其对基因表达谱及表型的影响[68]。
3 circRNA参与骨骼肌发育调控
随着circRNA研究策略和方法的不断成熟,circRNA在动物体内发挥的多种生物学功能被逐渐揭示。在动物骨骼肌发育进程中,成肌细胞的增殖和分化、肌卫星细胞活化促进肌肉再生等一系列过程,都离不开肌生成相关编码基因家族的调控[8,9]。随着研究技术与手段的更迭发展,越来越多circRNA被发现与肌生成、肌纤维类型转化和骨骼肌疾病发生等过程密切相关。
3.1 circRNA参与骨骼肌正常生长发育的调控
动物骨骼肌从胚胎期到出生后期,经历了骨骼肌卫星细胞活化以及成肌细胞增殖、分化、凋亡等过程[2]。研究表明,人、模式动物和家养动物的骨骼肌中都存在circRNA,并且不同生长发育阶段的骨骼肌中circRNA呈时序表达特异性[69~71]。
3.1.1 不同circRNA参与骨骼肌不同生长发育进程
Wei等[72]从幼年到老年的恒河猴()骨骼肌样本中鉴定出12,007个circRNAs,发现并证实其中有5个circRNAs表达水平随年龄增长而下调。同样,猪()不同胚胎时期肌肉中circRNA也呈现动态表达,并随着胚胎骨骼肌的生长发育进程,部分circRNA表达水平显著下调[73]。这种动态表达模式提示不同circRNA在动物骨骼肌发育各阶段起到不同的调控作用。
从骨骼肌发育相关测序结果中发现,一个基因可转录剪接成多个circRNA亚型,表达量较高的亚型在骨骼肌发育调控中发挥作用,并且在不同的骨骼肌发育阶段,宿主基因会产生不同的circRNA亚型[69,70,73]。Ling等[69]首次揭示了山羊()骨骼肌中circRNA时序动态表达模式,发现多个基因都产生了不同circRNA亚型,其中SLX4相互作用蛋白基因()的两种circRNA亚型在山羊骨骼肌发育各阶段均表现出完全相反的表达趋势,表明不同circRNA亚型在骨骼肌发育过程中可能发挥相反作用,表明circRNA的多样性与其调控机制的复杂性。
3.1.2 circRNA参与成肌细胞增殖、分化、凋亡等过程调控
近年来,研究者们利用测序分析技术挖掘出许多参与调控生物体发育过程的miRNA、lncRNA和circRNA,也鉴定出相当数量的circRNA可能参与调控动物骨骼肌生长发育的生物学过程(图2)。研究发现,经功能验证的大部分circRNA作为miRNA的竞争性内源RNA (competing endogenous RNA, ceRNA),参与调控成肌细胞增殖、分化等过程(表1)。Ouyang等[74]鉴定了不同发育阶段肌肉中差异表达circRNAs的miRNA结合能力,发现1401个circRNA中有946个都具有一个或多个miRNA结合位点,涉及150个已知miRNA。
骨骼肌组织或肌细胞中circRNA通过竞争性结合来抑制miRNA下游通路。Wang等[77]预测发现上具有4个结合位点,在C2C12细胞中竞争性结合,从而解除后者对基因的抑制作用,抑制C2C12成肌分化。Ouyang等[76]发现在鸡()晚期胚胎骨骼肌发育中高表达,并验证发现鸡作为的分子海绵,使和基因表达上调,促进鸡成肌细胞增殖和分化,调控鸡晚期胚胎骨骼肌发育过程。另外,Li[81]和聂露[86]等在山羊胚胎中期骨骼肌中高表达,并验证了MRFs家族中基因作为转录因子对发挥正向调控作用,同时通过靶向结合,阻遏其对基因表达的抑制,从而揭示了促进山羊骨骼肌卫星细胞成肌分化的调控通路。

图2 circRNA在骨骼肌增殖分化中的作用
circRNA-miRNA-mRNA在骨骼肌发育过程中的调控网络图,circRNA参与调控骨骼肌卫星细胞活化、成肌细胞增殖以及成肌细胞分化为肌管的生理过程。
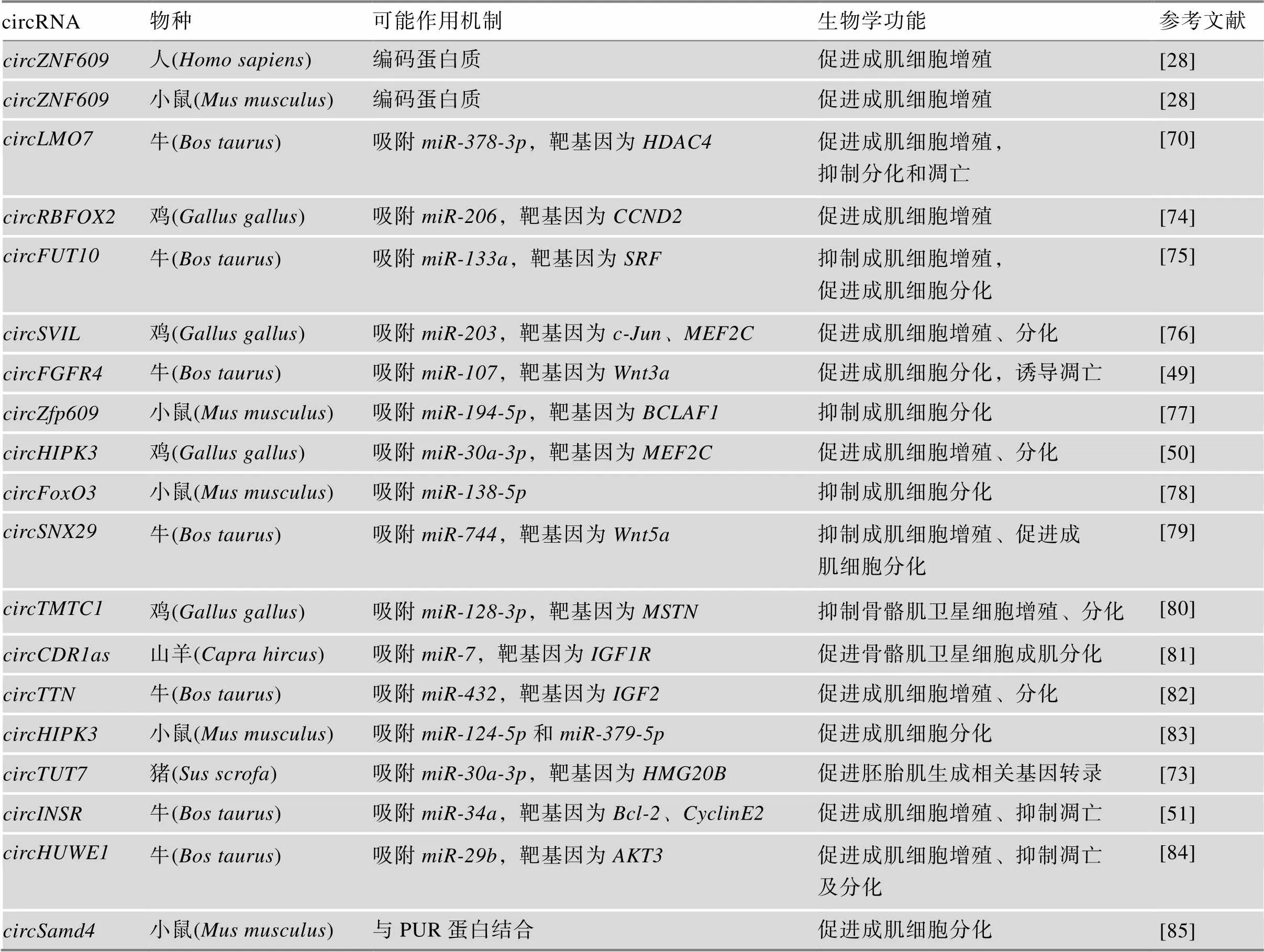
表1 circRNA参与不同动物骨骼肌发育的调控
除此之外,还有研究揭示了circRNA对骨骼肌生长发育的其他调控机制。2017年,Legnini等[28]首次利用测序数据筛选出通过参与蛋白质编码发挥调控作用的circRNA,将其命名为。在人和小鼠中有较高同源性,反向剪接时形成了一个开放阅读框,能够在应激条件下被特殊翻译成蛋白质发挥作用,可能促进成肌细胞增殖。此外,Pandey等[85]发现与PUR蛋白相结合,阻遏PUR蛋白对(myosin heavy chain)基因转录的拮抗作用,从而促进成肌细胞分化,加快骨骼肌生长发育进程。这是首次发现circRNA通过与RBPs协同作用发挥对骨骼肌生长发育的调控作用。
3.1.3 circRNA参与骨骼肌纤维类型转换过程调控
不同类型骨骼肌纤维具有不同的生理学特性,在动物骨骼肌发育过程中,成熟分化的肌管将逐渐转变为不同功能类型的肌纤维,包括氧化型和酵解型肌纤维,又称慢肌和快肌纤维。目前有研究表明,circRNA可能参与骨骼肌纤维生理功能调控及肌纤维类型转换调控。Shen等[87]选取处于生长拐点青峪猪的腰大肌和背最长肌,测序获得这两种类型肌肉的mRNA、lncRNA和circRNA差异表达谱,发现氧化型和酵解型肌肉不同的生理特性与其中差异表达的circRNA存在相关,这些circRNA的宿主基因被发现参与ATP代谢、快慢肌纤维转换等生理过程。值得关注的是,其中3个circRNA是由肌纤维分型相关的、和基因转录形成。研究提示circRNA在不同类型骨骼肌发育过程中起到特异的调控功能,并且circRNA还可能参与骨骼肌纤维类型转换过程的调控,其分子机制有待深入研究。
Li等[88]通过测序技术从猪快肌(肱二头肌)和慢肌(比目鱼肌)中分析circRNA差异表达谱,找到242个差异表达circRNAs,通过GO和KEGG分析发现这些差异表达circRNA宿主基因也与骨骼肌纤维类型形成的相关通路和生理过程有关,包括cGMP- PKG和AMPK信号通路,肌肉收缩、肌肉结构发育等生理学过程。此外,该研究还发现其中许多差异表达circRNA可能通过竞争性结合,参与调控肌纤维类型相关的骨骼肌疾病发生过程。circRNA在动物骨骼肌纤维发育形成及发挥生理功能上具有潜在作用,有助于进一步理解动物骨骼肌生长发育的表观遗传机制。
3.2 circRNA参与骨骼肌病理变化过程的调控
circRNA可变剪接在不同组织和发育阶段受到严格的调控[89],其降解调控可能影响增殖信号转导、细胞凋亡、血管生成等生物学过程,从而导致广泛的人类疾病[90]。在动物正常的骨骼肌中通常可检测到高表达的circRNA[70,91],而在肌肉疾病病例中存在circRNA表达失调[92]。研究发现circRNA表达水平在肌营养不良症及心肌病等疾病中发生改变[93,94],这暗示了circRNA的表达失调可能与肌肉病理状态相关(表2)。
杜氏肌营养不良(Duchenne muscular dystrophy, DMD)是一种由于基因发生移码突变,主要是外显子缺失引起肌营养不良蛋白缺失,从而产生肌无力等症状的疾病[95]。由基因转录产物剪接生成的circRNA是最早在骨骼肌中被识别的环状RNA之一[96]。有研究表明,由基因45~55号外显子区域剪接产生的circRNA,可能使基因外显子缺失的患者症状减轻[92,97]。也有研究分析了来自DMD患者和正常人成肌细胞的RNA测序结果,发现DMD患者来源的成肌细胞在circRNA表达水平方面确实具有独特的差异特征[28]。有趣的是,在正常成肌细胞体外分化过程中上调的和在DMD患者来源的成肌细胞中表达下调[98];相反,在正常成肌细胞肌生成过程中下调的编码蛋白的,在DMD成肌细胞分化过程中表达水平上调[28]。另外,Song等[99]从DMD模型小鼠的肌肉中鉴定出197个与对照组差异表达的circRNAs;Weng等[100]在坐骨神经损伤后的萎缩肌肉中鉴定出236个差异表达circRNAs,这些研究都表明circRNA与肌肉疾病病理过程有密切联系。
另一种肌营养不良症为1型肌强直性营养不良(myotonic dystrophy type 1, DM1)是一种由肌强直性营养不良蛋白激酶(myotonic dystrophy protein kinase,)基因3′UTR的CTG重复扩增导致mRNA剪接异常的多系统疾病[101]。与在DMD患者来源成肌细胞中的结果相似,Czubak等[102]发现DM1肌肉组织或细胞中,circRNA也呈现特异性表达,还初步发现DM1疾病程度与circRNA可变剪接变化之间的关联,并且circRNA整体表达水平呈上调趋势,在疾病个体中、、和的环状/线性比均升高[28,103](表2)。目前需要对circRNA可变剪接相关疾病中大量表达的circRNA进行识别与鉴定,以突出其在发病机制中的作用,并开发未来的疾病治疗方法。
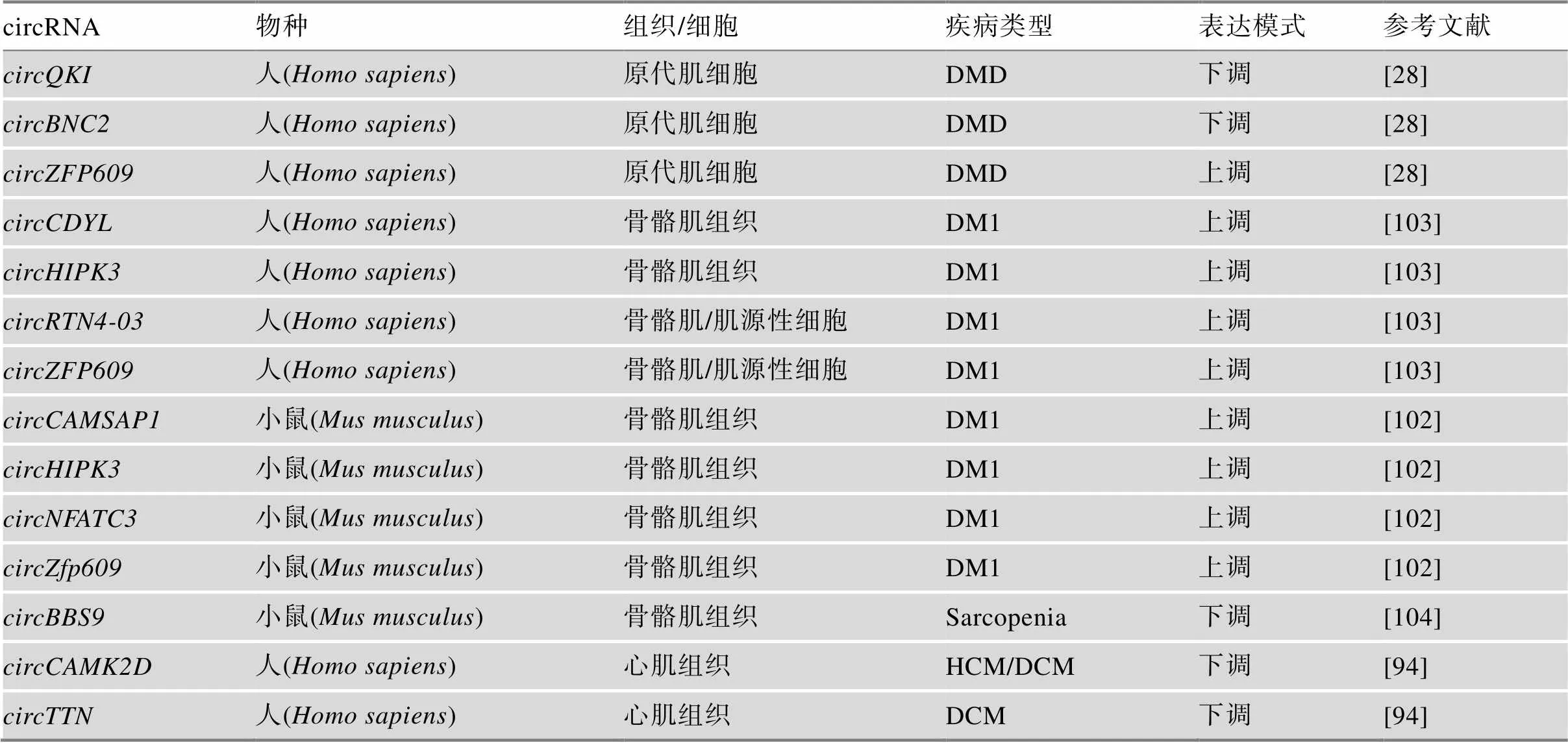
表2 circRNA在肌肉病理过程的表达变化
DMD:杜氏肌营养不良;DM1:I型肌强直性营养不良;Sarcopenia:老年性肌肉衰减症;HCM:肥厚型心肌病;DCM:扩张型心肌病。
4 结语与展望
circRNA的动态表达模式、复杂的调控机制和在不同细胞水平上扮演的新角色共同表明,它们并不是非正常剪接的“噪声”,而是在生物体发挥重要作用的新型调控分子。尽管大部分circRNA尚未被解析验证,但已有数千个circRNAs在不同物种、不同组织中被识别。
目前针对动物骨骼肌circRNA的大量研究,展现了circRNA在调控骨骼肌正常生长发育和骨骼肌相关疾病发生过程中的重要性及其分子机制的复杂性,仍存在许多科学问题有待深入探究。已经发现相当数量的circRNAs作为ceRNA在骨骼肌细胞增殖、分化等过程发挥调控作用,也有愈来愈多的研究者开始关注circRNA与骨骼肌纤维分型以及骨骼肌异常代谢发育的关系,其在骨骼肌相关疾病诊断、监测过程中有重要意义。目前circRNA形成机制与circRNA转录后调控机制均未被完全阐明,circRNA与蛋白质互作、调控亲本基因转录、参与编码蛋白质等一系列复杂的作用机制仍有待探索。
[1] Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ., 2012, 8(8): 457–465.
[2] Chal J, Pourquié O. Making muscle: skeletal myogenesisand., 2017, 144(12): 2104– 2122.
[3] Chargé SBP, Rudnicki MA. Cellular and molecular regulation of muscle regeneration., 2004, 84(1): 209–238.
[4] Epstein HF, Fischman DA. Molecular analysis of protein assembly in muscle development., 1991, 251(4997): 1039–1044.
[5] Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD., 1989, 56(4): 607–617.
[6] Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development., 1992, 71(3): 383– 390.
[7] Wang YN. The roles of MEF2A in the regulation of skeletal muscle myoblasts proliferation and differentiation in Qinchuan beef cattle[Dissertation]. Northwest A&F University, 2019.王亚宁. MEF2A对秦川牛骨骼肌成肌细胞增殖和分化的调控作用及机理研究[学位论文]. 西北农林科技大学, 2019.
[8] Whittemore LA, Song K, Li XP, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, Maylor R, O'hara D, Pearson A, Quazi A, Ryerson S, Tan XY, Tomkinson KN, Veldman GM, Widom A, Wright JF, Wudyka S, Zhao L, Wolfman NM. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength., 2003, 300(4): 965–971.
[9] Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions., 2007, 23: 645–673.
[10] Li XY , Fu LL , Cheng HJ , Zhao SH. Advances on microRNA in regulating mammalian skeletal muscle development., 2017, 39(11): 1046– 1053.李新云, 付亮亮, 程会军, 赵书红. MicroRNA调控哺乳动物骨骼肌发育. 遗传, 2017, 39(11): 1046–1053.
[11] Zhou R, Wang YX, Long KR, Jiang AA, Jin L. Regulatory mechanism for lncRNAs in skeletal muscle development and progress on its research in domestic animals., 2018, 40(4): 292–304.周瑞, 王以鑫, 龙科任, 蒋岸岸, 金龙. LncRNA调控骨骼肌发育的分子机制及其在家养动物中的研究进展. 遗传, 2018, 40(4): 292–304.
[12] Zhang PP, Chao Z, Zhang R, Ding RQ, Wang YL, Wu W, Han Q, Li CC, Xu HX, Wang L, Xu YJ. Circular RNA regulation of myogenesis., 2019, 8(8): 885.
[13] Gao Y, Wang JF, Zheng Y, Zhang JY, Chen S, Zhao FQ. Comprehensive identification of internal structure and alternative splicing events in circular RNAs., 2016, 7: 12060.
[14] Shen T, Han M, Wei G, Ni T. An intriguing RNA species—perspectives of circularized RNA.,2015, 6(12): 871–880.
[15] Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu SS, Yang L, Chen LL. Circular intronic long noncoding RNAs., 2013, 51(6): 792–806.
[16] Li ZY, Huang C, Bao C, Chen L, Lin M, Wang XL, Zhong GL, Yu B, Hu WC, Dai LM, Zhu PF, Chang ZX, Wu QF, Zhao Y, Jia Y, Xu P, Liu HJ, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus., 2015, 22(3): 256–264.
[17] Liang GM, Yang YL, Niu GL, Tang ZL, Li K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages.,2017, 24(5): 523–535.
[18] Shuai MX, Hong JW, Huang DH, Zhang X, Tian YQ. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity., 2018, 16(5): 6495–6501.
[19] Bi W, Huang JY, Nie CL, Liu B, He GQ, Han JH, Pang R, Ding ZM, Xu J, Zhang JW. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway., 2018, 37(1): 275.
[20] Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Kjems J. The biogenesis, biology and characterization of circular RNAs., 2019, 20(11): 675–691.
[21] Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs., 2020, 21(8): 475–490.
[22] Das A, Das A, Das D, Abdelmohsen K, Panda AC. Circular RNAs in myogenesis.,2020, 1863(4): 194372.
[23] Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges., 2013, 495(7441): 384–388.
[24] Yang F, Hu AP, Li D, Wang JQ, Guo YH, Liu Y, Li HJ, Chen YJ, Wang XJ, Huang K, Zheng LD, Tong QS. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation.,2019, 18(1): 158.
[25] Piwecka M, GlažAr P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function., 2017, 357(6357): eaam8526.
[26] Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing., 2014, 56(1): 55–66.
[27] Yang YB, Gao XY, Zhang ML, Yan S, Sun CJ, Xiao FZ, Huang NN, Yang XS, Zhao K, Zhou HK, Huang SY, Xie B, Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis., 2018, 110(3): 304–315.
[28] Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis.,2017, 66(1): 22–37.e29.
[29] Zeng Y, Du WW, Wu YY, Yang ZG, Awan FM, Li XM, Yang WN, Zhang C, Yang Q, Yee A, Chen Y, Yang FH, Sun H, Huang R, Yee AJ, Li RK, Wu ZK, Backx PH, Yang BB. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair.,2017, 7(16): 3842–3855.
[30] Du WW, Fang L, Yang WN, Wu N, Awan FM, Yang ZG, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity., 2017, 24(2): 357–370.
[31] Van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology., 2014, 30(9): 418–426.
[32] Wang J, Ren QL, Hua LS, Chen JF, Zhang JQ, Bai HJ, Li HL, Xu B, Shi ZH, Cao H, Xing BS, Bai XX. Comprehensive analysis of differentially expressed mRNA, lncRNA and circRNA and their ceRNA networks in the longissimus dorsi muscle of two different pig breeds.,2019, 20(5): 1107.
[33] Dahl M, Daugaard I, Andersen MS, Hansen TB, Grønbæk K, Kjems J, Kristensen LS. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies.,2018, 98(12): 1657–1669.
[34] Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges.,2016, 17(11): 679–692.
[35] Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs.,2016, 26(9): 1277–1287.
[36] Sekar S, Cuyugan L, Adkins J, Geiger P, Liang WS. Circular RNA expression and regulatory network prediction in posterior cingulate astrocytes in elderly subjects., 2018, 19(1): 340.
[37] Zhang QL, Ji XY, Li HW, Guo J, Wang F, Deng XY, Chen JY, Lin LB. Identification of circular RNAs and their altered expression under poly(I:C) challenge in key antiviral immune pathways in amphioxus, 2019, 86: 1053–1057.
[38] Sekar S, Geiger P, Cuyugan L, Boyle A, Serrano G, Beach TG, Liang WS. Identification of circular RNAs using RNA sequencing., 2019, (153).
[39] Hansen TB, Venø MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools., 2016, 44(6): e58.
[40] Chuang TJ, Chen YJ, Chen CY, Mai TL, Wang YD, Yeh CS, Yang MY, Hsiao YT, Chang TH, Kuo TC, Cho HH, Shen CN, Kuo HC, Lu MY, Chen YH, Hsieh SC, Chiang TW. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells., 2018, 46(7): 3671–3691.
[41] Wang LY, Long HY, Zheng QH, Bo XT, Xiao XH, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression., 2019, 18(1): 119.
[42] Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, Dudekula DB, Abdelmohsen K, Gorospe M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs., 2017, 45(12): e116.
[43] Łabaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling.,2011, 27(13): i383–i391.
[44] Li SS, Teng SS, Xu JQ, Su GN, Zhang Y, Zhao JQ, Zhang SW, Wang HY, Qin WY, Lu ZJ, Guo Y, Zhu QY, Wang D. Microarray is an efficient tool for circRNA profiling., 2019, 20(4): 1420–1433.
[45] López-Jiménez E, Rojas AM, Andrés-León E. RNA sequencing and prediction tools for circular RNAs analysis., 2018, 1087: 17–33.
[46] Guan YJ, Ma JY, Song W. Identification of circRNA- miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data., 2019, 19: 183.
[47] Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ, Feng ZB, Chen G. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma.,2018, 16(1): 220.
[48] Zhai ZS, Fu Q, Liu CJ, Zhang X, Jia PC, Xia P, Liu P, Liao SX, Qin T, Zhang HW. Emerging roles of hsa-circ- 0046600 targeting the miR-640/HIF-1α signalling pathway in the progression of HCC.,2019, 12: 9291–9302.
[49] Li H, Wei XF, Yang JM, Dong D, Hao D, Huang YZ, Lan XY, Plath M, Lei CZ, Ma Y, Lin FP, Bai YY, Chen H. CircFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a., 2018, 11: 272–283.
[50] Chen B, Yu J, Guo LJ, Byers MS, Wang ZJ, Chen XL, Xu HP, Nie QH. Circular RNA circHIPK3 promotes the proliferation and differentiation of chicken myoblast cells by sponging miR-30a-3p., 2019, 8(2): 177.
[51] Shen XM, Zhang XY, Ru WX, Huang YZ, Lan XY, Lei CZ, Chen H. circINSR promotes proliferation and reduces apoptosis of embryonic myoblasts by sponging miR-34a., 2020, 19: 986–999.
[52] Huang SL, Li XZ, Zheng H, Si XY, Li B, Wei GQ, Li CL, Chen YJ, Chen YM, Liao WJ, Liao YL, Bin JP. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice., 2019, 139(25): 2857–2876.
[53] Du WW, Zhang C, Yang WN, Yong TQ, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction., 2017, 7(17): 4183–4191.
[54] Barra J, Leucci E. Probing long non-coding RNA- protein interactions., 2017, 4: 45.
[55] Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1., 2017, 14(3): 361–369.
[56] Du WW, Yang WN, Liu E, Yang ZG, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progressionforming ternary complexes with p21 and CDK2., 2016, 44(6): 2846–2858.
[57] Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110infection., 2017, 67(2): 214–227.e217.
[58] Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, Liu H, Yang F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production., 2019, 45: 155–167.
[59] Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs., 2019, 38(16): e100836.
[60] Kong S, Tao M, Shen XJ, Ju SQ. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products., 2020, 483: 59– 65.
[61] Yang Y, Fan XJ, Mao MW, Song XW, Wu P, Zhang Y, Jin YF, Yang Y, Chen LL, Wang Y, Wong CC, Xiao XS, Wang ZF. Extensive translation of circular RNAs driven by N6-methyladenosine.,2017, 27(5): 626– 641.
[62] Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez- Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of circRNAs., 2017, 66(1): 9–21.e27.
[63] Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences., 2000, 28(6): 1102, 1104.
[64] Zhao J, Wu J, Xu TY, Yang QC, He JH, Song XF. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features., 2018, 45(7): 403–406.
[65] Wei LY, Chen HR, Su R. M6APred-EL: a sequence- based predictor for identifying N6-methyladenosine sites using ensemble learning., 2018, 12: 635–644.
[66] Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysisof translation with nucleotide resolution using ribosome profiling., 2009, 324(5924): 218–223.
[67] Zhang ML, Zhao K, Xu XP, Yang YB, Yan S, Wei P, Liu H, Xu JB, Xiao FZ, Zhou HK, Yang XS, Huang NN, Liu JL, He KJ, Xie KP, Zhang G, Huang SY, Zhang N. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma., 2018, 9(1): 4475.
[68] Zhang ML, Huang NN, Yang XS, Luo JY, Yan S, Xiao FZ, Chen WP, Gao XY, Zhao K, Zhou HK, Li ZQ, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of thegene suppresses glioma tumorigenesis., 2018, 37(13): 1805–1814.
[69] Ling YH, Zheng Q, Zhu L, Xu LN, Sui MH, Zhang YH, Liu Y, Fang FG, Chu MX, Ma YH, Zhang XR. Trend analysis of the role of circular RNA in goat skeletal muscle development., 2020, 21(1): 220.
[70] Wei XF, Li H, Yang JM, Hao D, Dong D, Huang YZ, Lan XY, Plath M, Lei CZ, Lin FP, Bai YY, Chen H. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p., 2017, 8(10): e3153.
[71] Xie YQ, Chen T, Luo JY, Xi QY, Zhang YL, Sun JJ. Mechanism of circRNA and its effect on development of animal muscles., 2018, 45(8): 2270–2275.谢月琴, 陈婷, 罗君谊, 习欠云, 张永亮, 孙加节. circRNA作用机制及其对动物肌肉发育的影响. 中国畜牧兽医, 2018, 45(8): 2270–2275.
[72] Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, De Cabo R, Gorospe M. Circular RNAs in monkey muscle: age-dependent changes., 2015, 7(11): 903–910.
[73] Hong LJ, Gu T, He YJ, Zhou C, Hu Q, Wang XW, Zheng EQ, Huang SX, Xu Z, Yang J, Yang HQ, Li ZC, Liu DW, Cai GY, Wu ZF. Genome-wide analysis of circular RNAs mediated ceRNA regulation in porcine embryonic muscle development.,2019, 7: 289.
[74] Ouyang HJ, Chen XL, Wang ZJ, Yu J, Jia XZ, Li ZH, Luo W, Abdalla BA, Jebessa E, Nie QH, Zhang XQ. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens., 2018, 25(1): 71–86.
[75] Li H, Yang JM, Wei XF, Song CC, Dong D, Huang YZ, Lan XY, Plath M, Lei CZ, Ma Y, Qi XL, Bai YY, Chen H. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a., 2018, 233(6): 4643–4651.
[76] Ouyang HJ, Chen XL, Li WM, Li ZH, Nie QH, Zhang XQ. Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken., 2018, 9: 172.
[77] Wang YH, Li ML, Wang YH, Jia L, Zhang M, Fang XT, Chen H, Zhang CL. A Zfp609 circular RNA regulates myoblast differentiation by sponging miR-194-5p.,2019, 121: 1308–1313.
[78] Li XY, Li C,Y Liu ZJ, Ni W, Yao R, Xu YR, Quan RZ, Zhang MD, Li HX, Liu L, Hu SW. Circular RNA circ-FoxO3 inhibits myoblast cells differentiation., 2019, 8(6): 616.
[79] Peng SJ, Song CC, Li H, Cao XK, Ma YL, Wang XG, Huang YZ, Lan XY, Lei CZ, Chaogetu B, Chen H. Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+signaling pathway., 2019, 16: 481–493.
[80] Shen XX, Liu ZH, Cao XN, He HR, Han SS, Chen YQ, Cui C, Zhao J, Li DY, Wang Y, Zhu Q, Yin HD. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p., 2019, 15(10): 2265–2281.
[81] Li L, Chen Y, Nie L, Ding X, Zhang X, Zhao W, Xu XL, Kyei B, Dai DH, Zhan SY, Guo JZ, Zhong T, Wang LJ, Zhang HP. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells.,2019, 1862(8): 807–821.
[82] Wang XG, Cao XK, Dong D, Shen XM, Cheng J, Jiang R, Yang ZX, Peng SJ, Huang YZ, Lan XY, Elnour IE, Lei CZ, Chen H. Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway., 2019, 18: 966–980.
[83] Yao R, Yao Y, Li CY, Li XY, Ni W, Quan RZ, Liu L, Li HX, Xu YR, Zhang MD, Ullah Y, Hu SW. Circ-HIPK3 plays an active role in regulating myoblast differentiation., 2019, 155: 1432–1439.
[84] Yue BL, Wang J, Ru WX, Wu JY, Cao XK, Yang HY, Huang YZ, Lan XY, Lei CZ, Huang BZ, Chen H. The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development., 2020, 19: 1086–1097.
[85] Pandey PR, Yang JH, Tsitsipatis D, Panda AC, Noh JH, Kim KM, Munk R, Nicholson T, Hanniford D, Argibay D, Yang XL, Martindale JL, Chang MW, Jones SW, Hernando E, Sen P, De S, Abdelmohsen K, Gorospe M. circSamd4 represses myogenic transcriptional activity of PUR proteins., 2020, 48(7): 3789–3805.
[86] Nie L. Mechanism of circ-CDR1as regulating goat skeletal muscle satellite cells differentiation[Dissertation]. Sichuan Agricultural University, 2018.聂露. 环状RNA CDR1as调控山羊骨骼肌卫星细胞分化的机制研究[学位论文]. 四川农业大学, 2018.
[87] Shen LY, Gan ML, Tang QZ, Tang GQ, Jiang YZ, Li MZ, Chen L, Bai L, Shuai SR, Wang JY, Li XW, Liao K, Zhang SH, Zhu L. Comprehensive analysis of lncRNAs and circRNAs reveals the metabolic specialization in oxidative and glycolytic skeletal muscles., 2019, 20(12): 2855.
[88] Li BJ, Yin D, Li PH, Zhang ZK, Zhang XY, Li HQ, Li RY, Hou LM, Liu HL, Wu WJ. Profiling and functional analysis of circular RNAs in porcine fast and slow muscles., 2020, 8: 322.
[89] Feng J, Chen K, Dong X, Xu XL, Jin YX, Zhang YX, Chen WB, Han YJ, Shao L, Gao Y, He CJ. Genome- wide identification of cancer-specific alternative splicing in circRNA., 2019, 18(1): 35.
[90] Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field., 2018, 37(5): 555–565.
[91] Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu JZ, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats., 2013, 19(2): 141–157.
[92] Suzuki H, Aoki Y, Kameyama T, Saito T, Masuda S, Tanihata J, Nagata T, Mayeda A, Takeda SI, Tsukahara T. Endogenous multiple exon skipping and back-splicing at the DMD mutation hotspot., 2016, 17(10): 1722.
[93] Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis., 2017, 66(1): 22–37.e9.
[94] Khan MaF, Reckman YJ, Aufiero S, Van Den Hoogenhof MMG, Van Der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, Van Der Velden J, Creemers EE, Pinto YM. RBM20 regulates circular RNA production from the titin gene., 2016, 119(9): 996–1003.
[95] Shieh PB. Emerging strategies in the treatment of duchenne muscular dystrophy., 2018, 15(4): 840–848.
[96] Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing., 1999, 8(3): 493–500.
[97] Aoki Y, Yokota T, Nagata T, Nakamura A, Tanihata J, Saito T, Duguez SMR, Nagaraju K, Hoffman EP, Partridge T, Takeda SI. Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery., 2012, 109(34): 13763–13768.
[98] Cazzella V, Martone J, Pinnarò C, Santini T, Twayana SS, Sthandier O, D'amico A, Ricotti V, Bertini E, Muntoni F, Bozzoni I. Exon 45 skipping through U1-snRNA antisense molecules recovers the Dys-nNOS pathway and muscle differentiation in human DMD myoblasts., 2012, 20(11): 2134–2142.
[99] Song ZB, Liu YM, Fang XB, Xie MS, Ma ZY, Zhong ZG, Feng XL, Zhang WX. Comprehensive analysis of the expression profile of circRNAs and their predicted protein-coding ability in the muscle of mdx mice., 2020, 20(3): 397–407.
[100] Weng J, Zhang PX, Yin XF, Jiang BG. The whole transcriptome involved in denervated muscle atrophy following peripheral nerve injury., 2018, 11: 69.
[101] Fu YH, Pizzuti A, Fenwick RG, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, De Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy., 1992, 255(5049): 1256– 1258.
[102] Czubak K, Taylor K, Piasecka A, Sobczak K, Kozlowska K, Philips A, Sedehizadeh S, Brook JD, Wojciechowska M, Kozlowski P. Global increase in circular RNA levels in myotonic dystrophy., 2019, 10: 649.
[103] Voellenkle C, Perfetti A, Carrara M, Fuschi P, Renna LV, Longo M, Sain SB, Cardani R, Valaperta R, Silvestri G, Legnini I, Bozzoni I, Furling D, Gaetano C, Falcone G, Meola G, Martelli F. Dysregulation of circular RNAs in myotonic dystrophy type 1., 2019, 20(8): 1938.
[104] Guo MW, Qiu J, Shen F, Wang SN, Yu J, Zuo H, Yao J, Xu SN, Hu TH, Wang DM, Zhao Y, Hu YP, Shen FX, Ma XR, Lu J, Gu XJ, Xu LY. Comprehensive analysis of circular RNA profiles in skeletal muscles of aging mice and after aerobic exercise intervention., 2020, 12(6): 5071–5090.
circRNA on animal skeletal muscle development regulation
Ting Zheng1, Mailin Gan1, Linyuan Shen1, Lili Niu1, Zongyi Guo2, Jinyong Wang2, Shunhua Zhang1, Li Zhu1
Circular RNA (circRNA) is a type of closed circular RNA molecules formed by reverse splicing, which exists widely in organisms and has become a research hotspot in non-coding RNAs in recent years. Skeletal muscle plays the role of coordinating movement and maintaining normal metabolism and endocrine in organisms. With the development of sequencing and bioinformatics analysis technology, the functions and regulation mechanisms of circRNAs in skeletal muscle development have been gradually revealed. In this review, we summarize the types of molecular regulatory mechanisms, the classical research ideas and the functional research methods of circRNAs, and the research progress of circRNAs involved in normal development of skeletal muscle and regulation of skeletal muscle disease, in order to provide a reference to further study of the genetic mechanisms of circRNAs in the regulation of skeletal muscle development.
circRNA; skeletal muscle development; skeletal muscle diseases; RNA-seq
2020-07-06;
2020-10-13
国家自然科学基金项目(编号:31972524),国家现代农业产业技术体系四川生猪创新团队项目(编号:SCSZTD-3-008)和四川省科技支撑计划项目(编号:2016NYZ0050)资助[Supported by the National Natural Science Foundation of China (No. 31972524), Sichuan Pig Innovation Team Project of National Modern Agricultural Industrial Technology System (No. SCSZTD-3-008), and the Science and Technology Program of Sichuan Province (No. 2016NYZ0050)]
郑婷,在读硕士研究生,专业方向:动物遗传育种。E-mail: 741377392@qq.com
张顺华,博士,助理研究员,硕士生导师,研究方向:动物遗传育种。E-mail: 363445986@qq.com
朱砺,博士,教授,博士生导师,研究方向:动物遗传育种。E-mail: zhuli7508@163.com
10.16288/j.yczz.20-207
2020/12/2 15:43:23
URI: https://kns.cnki.net/kcms/detail/11.1913.R.20201202.1335.002.html
(责任编委: 蒋思文)
