Genome of a thermophilic bacterium Geobacillus sp. TFV3 from Deception Island, Antarctica
2020-12-18XinJieCHINGChuiPengTEOHDexterLEEMarceloGONZLEZARAVENANazalanNAJIMUDINYokeKqueenCHEAHParisLAVINClementeMichaelVuiLingWONG
Xin Jie CHING, Chui Peng TEOH, Dexter J. H. LEE, Marcelo GONZÁLEZ-ARAVENA, Nazalan NAJIMUDIN, Yoke Kqueen CHEAH,Paris LAVIN & Clemente Michael Vui Ling WONG,6*
1 Biotechnology Research Institute, Universiti Malaysia Sabah, Jalan UMS, 88400 Kota Kinabalu, Sabah, Malaysia;
2 Instituto Antártico Chileno, Plaza Muñoz Gamero 1055, Punta Arenas, Chile;
3 School of Biological Science, Universiti Sains Malaysia, Persiaran Bukit Jambul 11900 Bayan Lepas, Penang, Malaysia;
4 Department of Biomedical Science, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor Darul Ehsan, Malaysia;
5 Universidad de Antofagasta, Facultad de Ciencias del Mar y Recursos Biologicos, Departamento de Biotecnologia; Laboratorio de Complejidad Microbiana y Ecología Funcional, Instituto Antofagasta, Chile;
6 National Antarctic Research Centre, University of Malaya, 50603 Kuala Lumpur, Malaysia
Abstract Thermophilic microorganisms have always been an important part of the ecosystem, particularly in a hot environment, as they play a key role in nutrient recycling at high temperatures where most microorganisms cannot cope. While most of the thermophiles are archaea, thermophiles can also be found among some species of bacteria. These bacteria are very useful in the fundamental study of heat adaptation, and they are also important as potential sources of thermostable enzymes and metabolites. Recently, we have isolated a Gram-positive thermophilic bacterium, Geobacillus sp. TFV3 from a volcanic soil sample from Deception Island, Antarctica. This project was undertaken to analyze the genes of this thermophilic Antarctic bacterium and to determine the presence of thermal-stress adaptation proteins in its genome. The genome of Geobacillus sp. TFV3 was first purified, sequenced, assembled, and annotated. The complete genome was found to harbor genes encoding for useful thermal-stress adaptation proteins. The majority of these proteins were categorized under the family of molecular chaperone and heat shock protein. This genomic information could eventually provide insights on how the bacterium adapts itself towards high growth temperatures.
Keywords temperature, 16S rDNA, genomes, Geobacillus, Deception Island
1 Intr oduction*
Geobacillusis a genus comprising of rod-shaped, Gram- positive thermophilic bacteria. Previously categorized within the Bacillaceae, these strains were subsequently placed into the new genusGeobacillus, as there was a significant difference in their genotype as well as phenotype (Nazina et al., 2001).Geobacillusis quite abundant and has been detected in a wide range of habitats, from the sediment in the Mariana Trench, which is 10897 m below sea level (Takami et al., 2004), to the upper troposphere at an altitude above 10000 m (DeLeon-Rodriguez et al., 2013). Most of the members can grow at a temperature ranging from 45℃ to 75℃ ,whereas some of them can extend this to as low as 35℃ or as high as 80℃ (Zeigler, 2014). Despite having high growth temperature, these thermophiles can be found not only in hot environments, but they can also be easily isolated from cool soils and cold ocean sediments (Zeigler, 2014; Studholme, 2015). Currently, there are around 14 known species in this genus as depicted in National Center for Biotechnology Information (NCBI, part of the United States National Library of Medicine (NLM)) database, the most extensively studied ones beingG.stearothermophilusandG.kaustophilus. Their property of growing at high temperature allows them to be one of the great interests in the field of extremophile researches, as they can produce thermostable hydrolytic extracellular enzymes that can function optimally at a temperature higher than the majority of organisms can withstand (Weigand et al., 2013). Usually, bacteria would adopt more than one strategy to make up for their thermal adaptability. For instance, it has been found that the high thermal melting temperature of dihydrofolate reductase fromGeobacillus stearothermophiluswas attributed to several aspects, which includes additional proline residues, removal of water-accessible thermolabile residues, ligand binding, and also the presence of salt or cosolvents (Guo et al., 2014).
Even though more and moreGeobacilli have been discovered recently, not much is known about the thermal adaptation strategies adopted by the bacteria at elevated temperature. Genome analysis is an important step in providing information on how the bacteria respond to high growth temperatures since everything gets down to the molecular level nowadays. Therefore, the objective of the project was to analyze the genome of a thermophilic Antarctic bacteriumGeobacillussp. TFV3 isolated from Deception Island, Antarctica, and to determine the presence of thermal-stress adaptation genes in its genome.
2 Material and methods
Strain TFV3 was isolated from the volcanic soil of Deception Island (62°58'57.6"S, 60°39'52.3"W). It was grown and routinely sub-cultured in Trypticase Soy Medium at 65℃.High-quality genomic DNA was extracted using an optimized protocol for QIAGEN DNeasy Blood and Tissue kit. PCR was carried out using primer BSF-8/20 (5’-AGAGTTTGATCCTGGCTCAG-3’) and BSR-1541/20 (5’-AAGGAGGTGATCCAGCCGCA-3’) to amplify the 16S rDNA. The amplicon was sequenced and aligned against sequences in NCBI GenBank using Basic Local Alignment Search Tool (BLAST) to identify the strain.
The extracted genomic DNA was then sequenced using Pacific Biosciences Sequencer RSII. The polymerase read statistics and sub-read statistics are shown in Tables 1 and 2 respectively. The raw sequencing data were then corrected, trimmed, and assembled using Canu (Koren et al., 2017), followed by circularizing the genome assembly using Circlator (Hunt et al., 2015). The circular genome was then annotated using RAST (Rapid Annotation using Subsystem Technology) (Aziz et al., 2008).

Table 1 Polymerase read statistics

Table 2 Subread statistics
A phylogenetic tree was constructed using the multiple reference genes present in the whole genome sequence of strain TFV3 instead of just using one highly conservative gene like 16S rDNA for a more reliable phylogenetic analysis. To achieve this, software PhyloSift (Darling et al., 2014) was used to compare the genome of strain TFV3 against the genome of 17 otherGeobacilliand sevenParageobacilliobtained from the NCBI database. From the phylogenetic tree, a few closest related species were selected, and the thermal-stress related genes in their genomes were compared against those of strain TFV3.
The complete genome sequence ofGeobacillussp. TFV3 has been deposited to the NCBI database, under the accession number MPSP01000000.
3 Results and discussion
The DNA alignment result of the 16S rDNA sequence showed that strain TFV3 has the highest similarity to the partial sequence 16S ribosomal RNA gene ofGeobacillussp. strain Et7/4 (99.42%), followed byGeobacillussp. strain Et2/3 (99.41%), both with 99% query coverage and 0 E-value. The identification was further supported by the phylogenetic analysis using PhyloSift. As shown in Figure 1, strain TFV3 formed a distinctive cluster withG.jurassicusNBRC 107829,G.kaustophilusEt7/4, andG.kaustophilusEt2/3. This suggested that these 3Geobacillihave the closest phylogenetic relationship with strain TFV3.
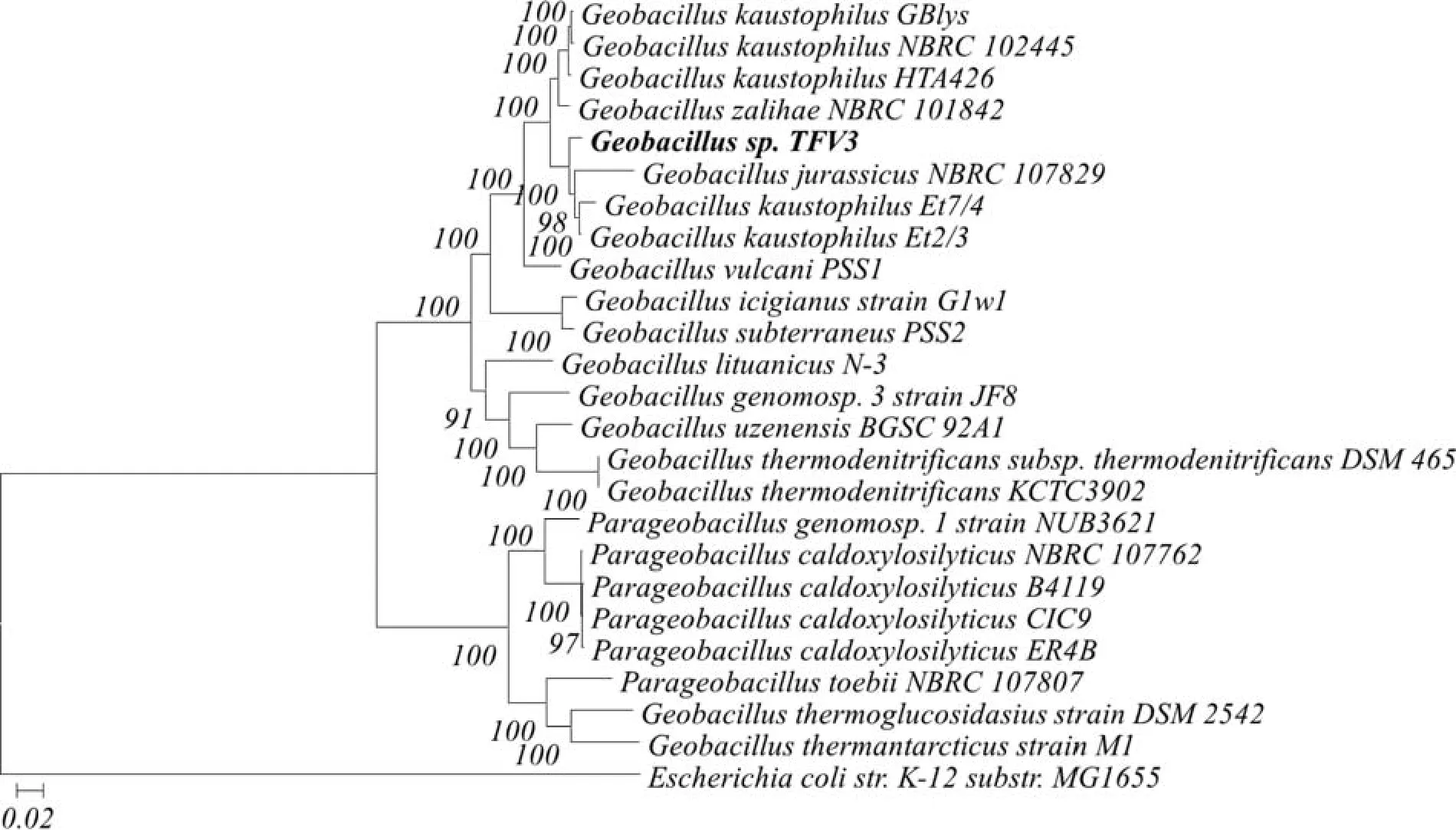
Figure 1 Whole genome phylogenetic tree constructed by PhyloSift, using the Maximum Likelihood method based on a Generalised Time-Reversible (GTR) model. The tree shows the close relationship between TFV3 related species, while E. coli K-12 substr. MG1655 is included to serve as outlier.
The genome properties of strain TFV3 after assembly and annotation are listed in Table 3. The genome of strain TFV3 was assembled into a single contig, suggesting that there is no plasmid in this bacterium. The genome was 3408572 bp in size, and with a G + C content of 51.9%. This falls into the 51% to 53% G + C content range of most of theGeobacilli. The genome has 3903 genes, including 3787 putative protein-coding sequences (CDS), 27 rRNA, 88 tRNA, and 1 tmRNA. A genome map was constructed using DNAPlotter (Carver et al., 2009) for better visualization of the position of each element in the genome (Figure 2). All the annotated genes were classified into orthologous groups based on their function using eggNOG (Jensen et al., 2008). Figure 3 shows that most of the genes fall into group S, suggesting that all the hypothetical genes and some of the genes encoding for proteins with unknown function occupied 32.11% of the total predicted genes in the genome. Aside from the hypothetical genes, the group that has the second most genes is group L (10.22%), which is responsible for DNA replication, recombination, and repair. This makes sense as strain TFV3 lived in a rather extreme environment, it is important that they protect and constantly evolve their DNA so that they adapt to the harsh living conditions. An even distribution was observed between most of the categories related to cellular metabolisms, energy production, transport, and signal transduction (~3%–6%), except for nucleotide transport and metabolism (2.31%). These genes are equally important in maintaining the routine cellular process and vitality of strain TFV3. Most of the genes encoding for thermal-stress proteins were categorized under group O (post-translational modification, protein turnover, and chaperones) and group K (transcriptional).
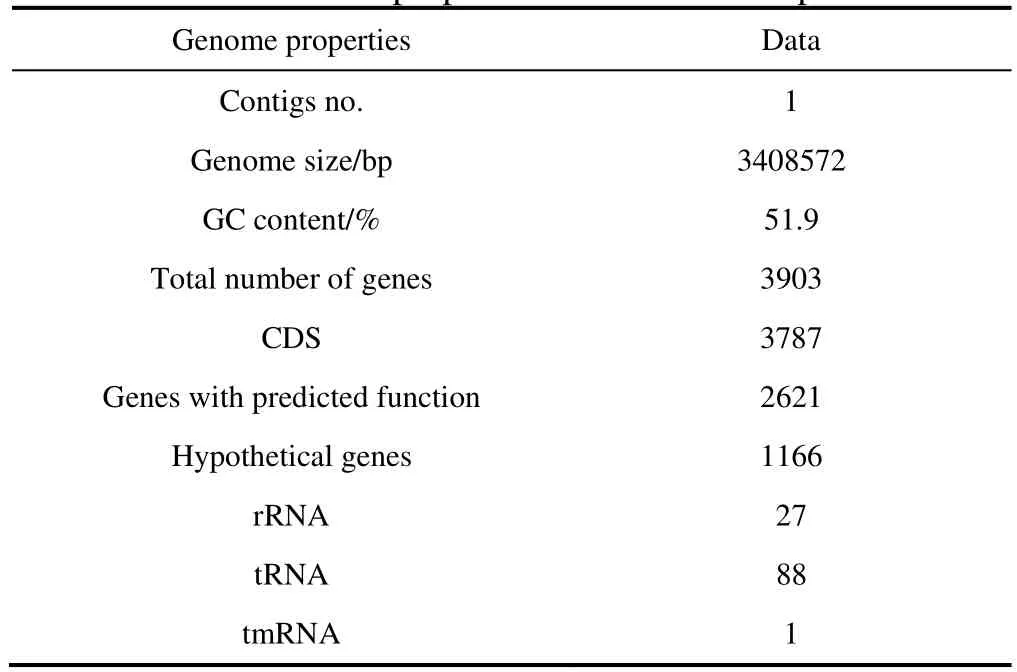
Table 3 Genome properties of Geobacillus sp. TFV3
Several common thermal-stress related genes were found in the genome of strain TFV3, including genes encoding for cold-shock protein (CSP), heat-shock protein (HSP), and molecular chaperone (Table 4). These genes are very important as they have a major contribution to maintaining cell viability in response to temperature fluctuations. Cold-related protein such as cold-shock protein CspD is thought to facilitate the initiation of translation as they serve as RNA chaperones to prevent the formation of secondary structure in mRNA when the temperature drops (Phadtare, 2004). Meanwhile, the other heat-related proteins such as HSPs and chaperones are useful in the event of temperature rise, as they are able to prevent protein aggregation (Whitley et al., 1999), repair damaged proteins (De Maio, 1999), and promote degradation of damaged proteins (Raboy et al., 1991).
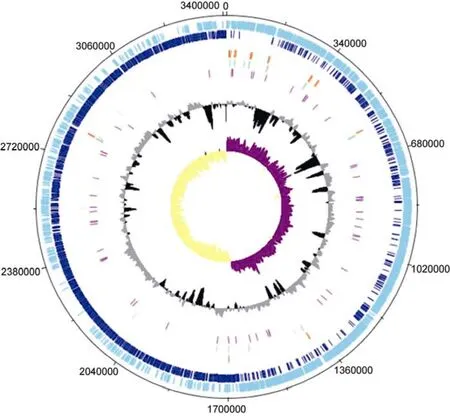
Figure 2 Genome map of Geobacillus sp. TFV3 as viewed DNAplotter. The outermost track represents forward CDS, followed by reverse CDS, rRNA, tRNA, miscRNA, tmRNA, GC plot, and GC skew.
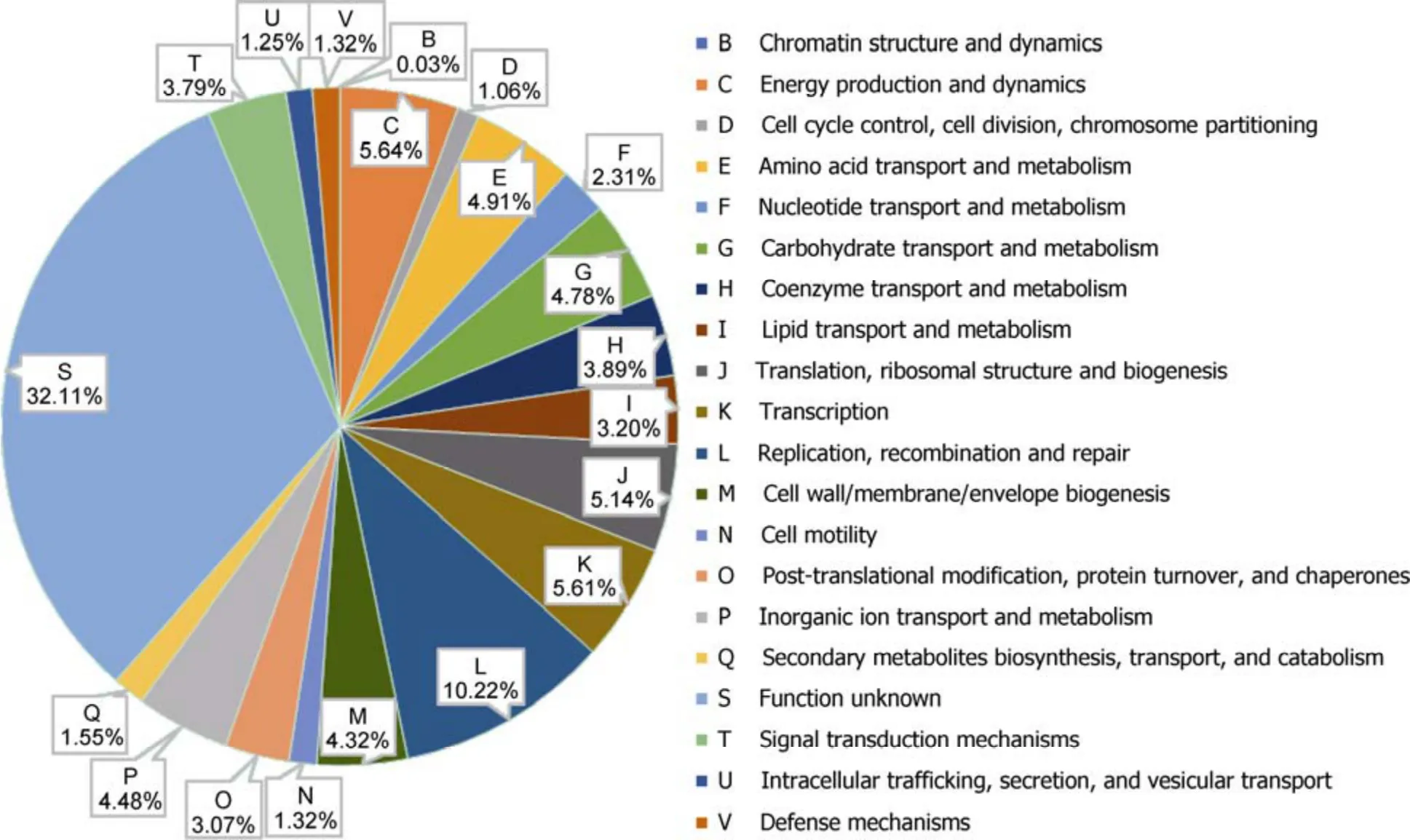
Figure 3 Functional distribution of genes within Geobacillus sp. TFV3 genome classified by clusters of orthologous groups (COG) using eggNOG.
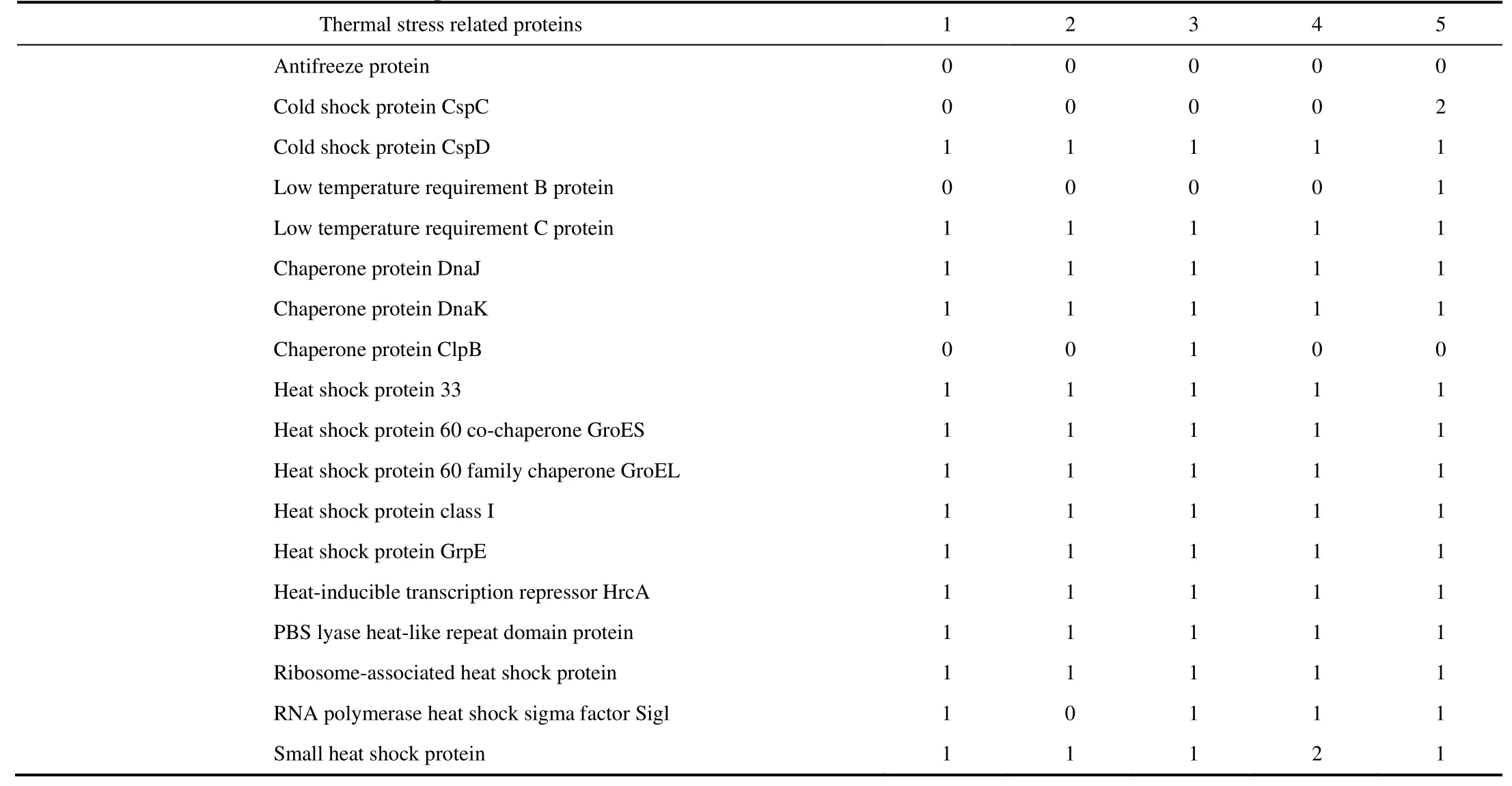
Table 4 Number of gene copies for thermal stress related proteins found in each bacterial genome. (1) Geobacillus sp. TFV3, (2) G. jurassicus NBRC 107829, (3) G. kaustophilus Et2/3, (4) G. kaustophilus Et7/4, and (5) Parageobacillus caldoxylosilyticus ER4B as an outlier for comparison
Some thermal-stress related genes found in strain TFV3 were also compared to the three closest species and aParageobacillusspecies (Table 4). It was found that the presence of thermal-stress related genes in the genomes of all five bacterial strains are highly similar, withP.caldoxylosilyticusER4B exhibiting a relatively different pattern in terms of gene presence. Other than antifreeze- related proteins, which are absent in all the strains, cold-shock protein, CspC and low-temperature requirement B protein which were only present in the genome of strain ER4B, was absent in all four Geobacilli. As well, chaperone protein ClpB was also absent in strain TFV3.
The presence of proteins such as chaperone protein DnaJ, chaperone protein DnaK, heat-shock protein GrpE, heat-shock protein 60 family chaperone GroEL, and heat-shock protein 60 co-chaperone GroES suggested that these proteins are crucial for the Geobacilli to be able to survive at high temperature. These molecular chaperones are primarily responsible for maintaining the integrity of cellular proteins and preventing cellular aggregation in response to environmental stress (Ranford et al., 2000; Harrison, 2003). They do not work independently but work in conjunction with one another. For instance, GroEL requires lid-like GroES to function properly. Binding of GroES to the apical domain binding site of GroEL would induce several changes which would lead to easier protein folding (Samali et al., 1999; Lee et al., 2002; Horwich et al., 2007). The same applies to DnaJ and GrpE which not only help to stimulate the ATPase activity of DnaK (Cyr et al., 1994; Hennessy et al., 2005), but also form a ternary complex with DnaK and polypeptide chains to prevent the aggregation of the polypeptide chains (Han and Christen, 2004). Although these heat shock proteins exist in almost every bacterium, the thermos-stability of the proteins is different in each bacterium and this influences the ability of the bacteria to grow at elevated temperature (Endo et al., 2006; Kumwenda et al., 2013).
The genome information of strain TFV3 provides insights into the genetic makeup of a thermophilic bacterium that lives in a hot environment. It harbors many genes encoding thermal-stress related proteins that might be significant for the bacterium in response to the high yet ever-changing temperature around the volcanic region in Antarctica. The genome data is important for the subsequent analysis to determine the key genes required for this bacterium to grow and survive under hot conditions. For instance, by performing a systematic gene knock-out or gene silencing study, functions of a gene related to thermal adaption can be determined. The complete genome data is also useful in comparative genomics among thermophilic bacteria as it could provide gene functional annotation by comparing homologous thermal-stress related proteins from different bacteria. In addition, the genome of strain TFV3 also contains genes encoding for enzymes such as lipases, proteases, amylases, glycosyltransferases, RNA polymerases, DNA polymerases, and ligases with industrial applications which are most likely to be thermostable. Lastly, with the genome data, this bacterium can be manipulated to be a host to express genes at high temperatures. In conclusion, the genome ofGeobacillussp. TFV3 data is not only an important source of information for fundamental study on thermal-adaptation of this bacterium and potentially as a host for gene expression study but also serves as a source of thermostable enzymes.
AcknowledgmentsThe funding support from the Ministry of Science, Technology, and Innovation (MOSTI), Malaysia, under the Antarctica Flagship Programme (Sub-Project 1: Grant no. FP1213E036) is gratefully acknowledged. We appreciate very much two anonymous reviewers, and guest editor for their helpful and constructive comments on the manuscript of this paper.
杂志排行
Advances in Polar Science的其它文章
- Foreword
- Information for authors
- Arctic envir onmental change r esearch and Antar ctic studies have mutual benefits
- Structure and function of a novel cold r egulated cold shock domain containing pr otein fr om an obligate psychrophilic yeast, Glaciozyma antarctica
- The growth of the Rhodococcus sp. on diesel fuel under the effect of heavy metals and differ ent concentrations of zinc
- Effects of elevated temperatur es on gr owth and photosynthetic performance of polar Chlorella
