Role of non-coding RNAs in pathogenesis of gastrointestinal stromal tumors
2020-12-16IoannisStefanouMariaGazouliGeorgiosZografosKonstantinosToutouzas
Ioannis K Stefanou,Maria Gazouli,Georgios C Zografos, Konstantinos G Toutouzas
Ioannis K Stefanou, Department of Surgery, Hippocration Hospital Athens, Athens 11527, Greece
Maria Gazouli, Department of Basic Medical Sciences, Laboratory of Biology, National and Kapodistrian University of Athens, Athens 11527, Greece
Georgios C Zografos, Konstantinos G Toutouzas, 1st Propaedeutic Department of Surgery, National and Kapodistrian University of Athens, Athens 11527, Greece
Abstract
Key words: Gastrointestinal stromal tumors; Non-coding RNA; MicroRNA; Transcriptomics; Biomarker; Long non-coding RNAs
INTRODUCTION
Non-coding RNAs
The discovery of transfer RNA (tRNA), and ribosomal RNA (rRNA), in the 1950s is the beginning of the history of the non-coding RNAs (ncRNAs) that play functional roles in the eukaryotic cells[1].James Watson imagined the one gene, one ribosome and one protein hypothesis (central dogma).Therefore, RNA changed from being a just information carrying molecule, to having three flavors.rRNA, tRNA and everything else was assumed to be mRNA[2].Later on, in the 70s Starket al[3]published the existence of other functional RNAs like ribonuclease P and snRNAs[4].One of the prominent examples about how huge was the surprise at this period of time, was the eventual renaming of signal recognition protein to signal recognition particle (SRPRNA).That happened after the discovery, that it contains a 7S RNA (by Walteret al[5]).In the early 90s, other long intergenic non coding RNAs were discovered, like XIST, by Brockdorffet al[6]Nowadays, it is generally known, according to the encyclopedia of DNA elements (published by the ENCODE Project[7]) consortium that the 80% of the human genome is transcripted for RNA molecules that have no protein coding capacity[8].In the past, it was believed that this huge amount of RNA molecules was a transcriptional noise.Contrariwise, they appear to have direct function as regulators in several endocytic molecular paths.They seem to play crucial role in differentiation, development, and apoptosis of normal cells[9], so even in the era of complete genome sequences, non-coding RNAs gene have been eventually invisible.These features of non-coding RNAs have turned them into one of the most promising fields of scientific research.
ncRNAs are classified into two big subgroups according to their size[10].
Short ncRNAs, with < 200 nucleotides (nts) in length and include:MicroRNAs (miRNAs) usually bind to a specific molecular locum at the mRNA to induce degradation or block the prosses of translation.In addition, this may be done in the context of a feedback mechanism that involves chromosome methylation.
Small interfering RNAs (siRNAs) have a similar function as miRNAs with the additional feature of inducing heterochromatin formation through RNA transcriptional silencing complex which, when bound to siRNA, promotes H3K9 methylation and chromatin condensation.
Piwi-interacting RNAs seem to interact with the piwi family proteins.They involve in chromatin regulation and suppression of transposon activity in germline and somatic cells[11].
Long ncRNAs (lncRNAs) are longer than 200nt and may comprise thousands ofnucleotides[12]:This group includes the long intergenic ncRNAs (lincRNAs), the natural antisense transcript, the transcripted ultraconserved regions and non-coding pseudogenes[13].It seems to be transcribed mostly by RNA polymerase 2 as the mRNA does but they do not undergo the standard processing steps[14].The mechanism of their function is generally unknown, but it is suggesting that it is similar to that of HOX antisense intergenic RNA (HOTAIR) which is the most studied lncRNA.It regulates chromatin methylation of the HOXD locus through polycomb repressive complex 2.HOTAIR was recently reported to play a crucial role in metastatic disease and may be a good prognostic marker in patients with breast cancer[15].
Post-transcriptional modifications that occur in RNA molecules started being explored at the recent years and therefore led to a new field of research called epitranscriptomics.Equivalent to epigenetics, which analyzes the post-transcriptional events occurring in DNA, epitranscriptomics investigates modifications resulting from all RNA processing events, such as RNA splicing, RNA editing, or methylation[16].
Gastrointestinal stromal tumors
Gastrointestinal stromal tumors (GISTs) are specific, generally c-Kit (CD117)-positive, mesenchymal tumors of the gastrointestinal tract, encompassing a majority of tumors previously considered gastrointestinal smooth muscle tumors[17].They are believed to originate from interstitial cells of Cajal or related stem cells.Interstitial cells of Cajal and GIST cells express the hematopoietic progenitor cell marker CD34 and the growth factor receptor c-Kit.Expression of the c-Kit gene protein product, CD117, has emerged as an important defining feature of GISTs[18,19].Using these criteria, the incidence of GISTs has been estimated to be 6 to 15 cases per million individuals per year[20].They constitute a significant percentage ranging from 1%-2% of all the gastrointestinal neoplasms.The most common genetic alterations found in GISTs include mutations of growth factors genes such as c-Kit (70–80%) and PDGFRA (platelet-derived growth factor A, 5%-8%).Several features of GISTs have been postulated in the past to predict their clinical behavior.Nowadays, much is known about the histological, immunohistochemical and molecular aspects of GISTs especially in diagnostic purposes[21,22].However, little is known about the clinicopathological features that can predict the biological behavior of these tumors.
At the recent years, plenty of studies have revealed the specific molecular characteristics of GISTs.Nowadays, these tumors are considered among the best genetically understood human cancers[23].
Especially after the discovery of their sensitivity to tyrosine kinase inhibitors, GISTs tend to be referred as ideal tumor for novel molecular targeted therapies.Apart from that, the fact that many studies have been published specific chromosomal changes (e.g.loss of 14q), genetic mutations (e.g.KIT,PDGFRA), gene expression profiles (e.g.ETV1,fascin1) and miRNA expression profiles, have contributed to make them one of the well-recognized tumors[24].It is important to mention thatKITandPDGFRAmutations are almost exclusive in GISTs, which makes them specific biomarkers of these tumors.The gold standard therapy in primary localized GISTs is a R0 surgical resection[25].First line therapy for the advanced disease is Imatinib that offers a dramatic response, in most of the cases, for about 2-3 years[26].After long term treatment, resistance is quite common.Sunitinib and regorafenib are the second line agents in imatinib resistant GISTs with also unsatisfactory outcomes in progressive disease[27].Therefore, further fundamental clinical studies are being conducted in order to provide improved diagnostic modalities to increase the possibility for the patients to be diagnosed in early disease, and furthermore provide novel therapeutic options for the advanced disease cases.
NcRNAs in GISTs
At the present, a clear relationship with GISTs has been reported for only a few ncRNA classes, especially miRNAs and some lncRNAs such as the ultra-conserved genes,HOTAIR,H19,MALAT1andCCDC26[28,29].The other types of ncRNAs it seems to participate in the genetic puzzle that gives rise to carcinogenic phenotype[13].
miRNAs are the most widely studied class of ncRNAs in GISTs and generally in human cancer.These small ncRNAs of approximately 22 nucleotides, mediate posttranscriptional gene silencing by controlling the translation of mRNA into proteins.miRNAs are estimated to regulate the translation of more than 60% of protein-coding genes[30].They are involved in regulating many processes, including proliferation, development, differentiation, and apoptosis.Alterations of miRNAs expression profile has been reported in GISTs, and is associated with tumor location, mutation status, tumor risk, and chromosomal changes[31].Two excellent reviews by Nanniniet al[32], and Kupcinskaset al[33]have perfectly analyzed relevant miRNA profiling studies.Since then several papers came out concerning ncRNA and GISTs.
STUDIES SELECTION
This review included all studies published in PubMed database related to the role of ncRNAs in GIST published from 2008 to 2020.The keywords we used to retrieve the papers were GIST, ncRNAs, miRNAs and lncRNAs.82 papers selected using these keywords.According the selection criteria, only 52 of them were relevant to the topic, 32 profiling studies, 9 reviews, 11 other studies (Figure 1).
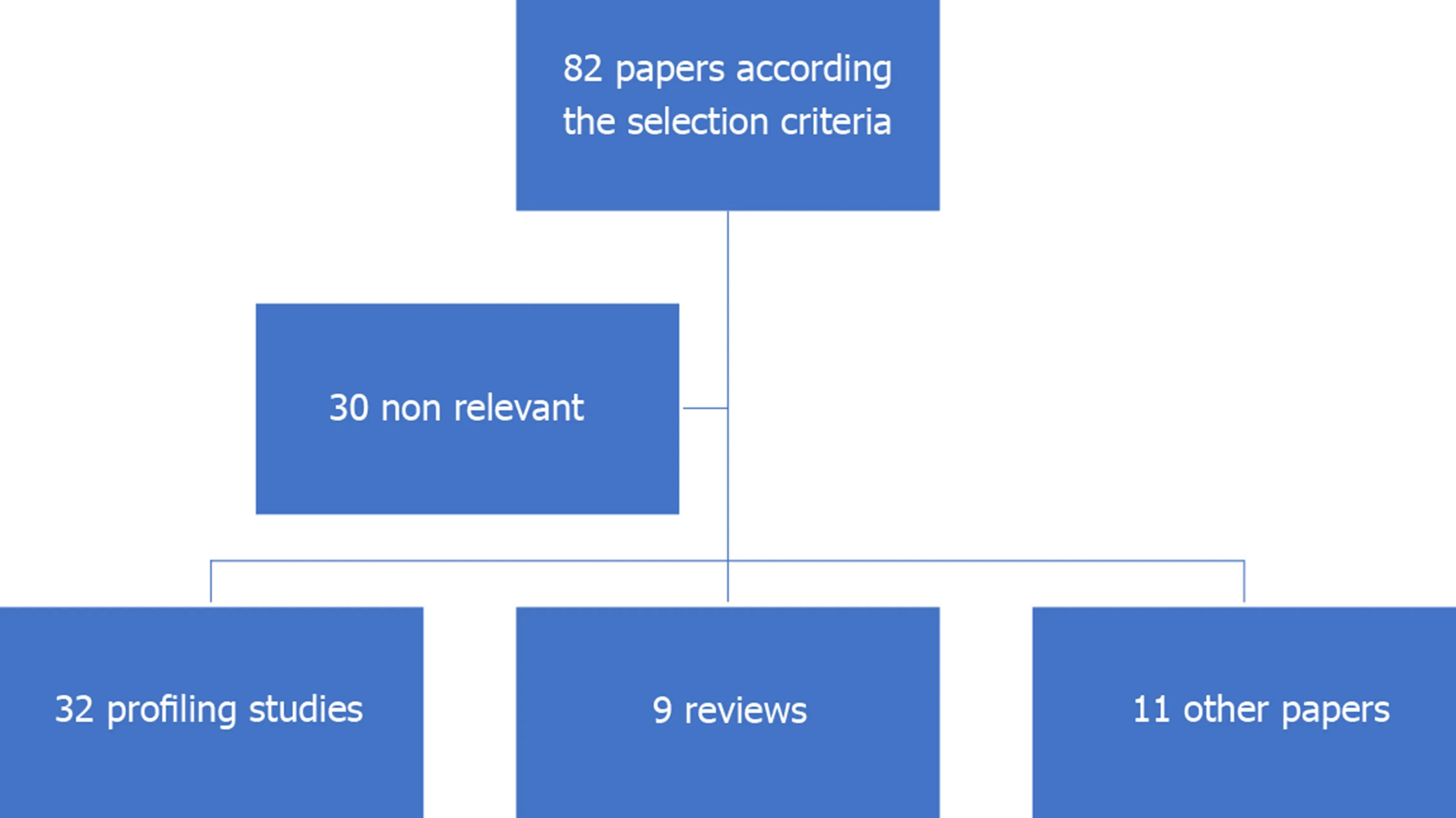
Figure 1 Studies selection.
CHROMOSOMAL LOSS OF 14q AND MIRNA EXPRESSION
Chromosomal deletions have been reported as frequent and characteristic aberrations and are related to the carcinogenesis of the GISTs[34].The most common described are in 14q, 22q, and 1p.Among them, partial or entire chromosomal loss of 14q is the most frequently found (60%–70%) and represents the majority of gastric GISTs, while 1p loss is usually present in small bowel GISTs[35]and its characterized by poor clinical outcome[36].None of the other common chromosome eliminations[37](22q, 1p) seems to affect the miRNAs expression profile.Table 1 summarizes the studies related to chromosomal loss of 14q and miRNAs expression.miRNAs seem to form two distinct clusters on the 14q chromosome.A study by Choiet al[38]published in 2010, identified a clear correlation between the 14q loss and deregulation of miRNA expression profile in 20 tumors.They noticed that, 6 GISTs that did not have 14q loss, formed a separate cluster.Furthermore, they found 73 deregulated miRNAs at a significant level according to 14q loss status.Among the 73 miRNAs, 38 were encoded on 14q.Kellyet al[39]studied a cluster of miRNAs on 14q32 region and revealed similar downregulated miRNAs according to 14q loss statue, in both adult and pediatric patients, but distinguish miRNA expression pattern between the adult and pediatric GISTs.They suggest that this happens due to the different methylation state of the maternal and paternal allele during the aging.Another study by Halleret al[40]identified 44 miRNAs located at 14q32.31 chromosomal region.Moreover, in a qRTPCR analysis of additional 49 GIST, the authors observed a significant lower expression of miRNA-134 and miRNA-370 in GIST with 14q loss.As mentioned above these miRNAs found to affect the mutational status ofKITandPDGRFA, and some of them including miRNA-494 are experimentally confirmed to targetKITorPDGFRA.Deregulation of these miRNAs were associated with tumor progression and shorter disease-free survival, suggesting that GIST with low expression of miRNAs located at the 14q32.31 chromosomal loss might represent o group with higher risk of tumor progression[36].
POTENTIAL DIAGNOSTIC AND PROGNOSTIC BIOMARKERS
GISTs are considered among the best recognized tumors, regarding their specific phenotypic and molecular characteristics.The diagnosis relies on the specific morphology and the unique immunohistochemistry (CD117,CD34and/orDOG1).Although, despite the high specific value of these biomarkers, in many cases, the diagnosis may be difficult.Table 2 summarizes the studies concerning ncRNAs as new emerging novel biomarkers, highly specific to GISTs.First of all, Subramanianet al[41]founded 16 upregulated and 10 downregulated miRNAs specifically in GISTs.In this study, they compared 84 miRNAs (that met the filtering criteria) expression status of 27 mesenchymal tumors (including GISTs), 5 normal smooth muscle and 2 normal skeletal muscle.Remarkably, the miRNA expression patterns suggested that two of the mesenchymal tumors had been misdiagnosed and this was confirmed by reevaluation of the tumors using immunohistology and molecular analyses.These findings demonstrated that miRNA expression profiling is unique for each tumor type, suggesting the potential use of miRNAs as diagnostic biomarkers.
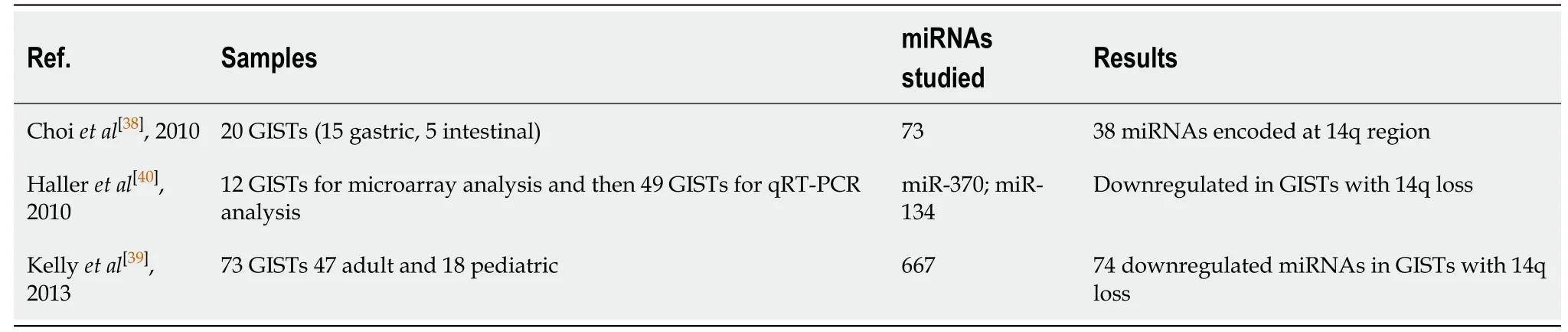
Table 1 Chromosomal loss of 14q and miRNA expression studies
Koelzet al[42]were the first who found significant depressed the 220/221 miRNAs compared to peripheral healthy tissue and blood samples.Niinumaet al[43], after the examination of 56 GISTs founded that, overexpression of miRNA-196a andHOTAIRwas associated with high-risk grade, metastasis, and poor survival among GISTs.Yamamotoet al[44]later in 2013 published a clear correlation between fachin-1 overexpression and miRNA-133b downregulation in the progression of gastrointestinal stromal tumor, making fascin-1 as a useful potential biomarker to predict the aggressive behavior.Another two studies by Halleret al[40]and Gitset al[45]are coming to confirm the downregulation of these two miRNAs 220/221 specific in GIST.However, according to the findings of all the previously mentioned studies the 220/221 miRNAs may not have had any impact on routine diagnostics because KITpositive and KIT-negative GIST exhibited a completely inverse expression pattern.One recent study by Gyvyteet al[46]2017, the first one which used the next generation sequencing kit in order to reveal deregulated miRNAs in GISTs and their possible associations with oncogenes.They found 19 deregulated miRNAs, 13 of which were not previously reported.They also proposed miRNA-215-5p to be negatively correlated with the risk grade, while miRNA-509-3p to be associated with epithelioid and mixed histological subtypes.The same research team, one year later (2018)[47], found a significant correlation between a lincRNA H19 and GIST oncogene ETV1, and between H19 and miRNA-455-3p.A Polish study, by Kosela-Paterczyket al[48], aimed to identify the miRNA expression profiles in four common soft tissue tumors.They also founded different miRNA signatures in serum samples in each soft tissue tumor, included GISTs.At the recent years, many studies came out regarding the lncRNAs and their task in GIST progression.A Chinese study by Huet al[49], questioned for the first time about the role of lncRNA AOC4P in GIST development.They identified that AOC4P regulate the epithelial mesenchymal transition (EMT) related proteins, which is important step for the metastatic ability of the tumor cells.One year later Badalamentiet al[50], questioned about the role of H19 and MALAT1 in GISTs.They found high expression levels of both lncRNAs in tumor samples which could be associated with prognosis and clinical response to IM.Yanet al[51]in a latest study through a microarray analysis, compared 3 metastatic GISTs with 3 normal tissue and 3 low grade GISTs and found significant expression of certain lncRNAs, including lnc-DNAJC6-2 in high risk tumors.
THE ROLE OF NON-CODING RNAS IN IMATINIB RESISTANCE
Numerus studies (Table 3) have been released about the imatinib resistance GISTs and their potential prognostic biomarkers.Overexpression of miRNA-196a in GIST tissues was associated with high-risk grade, metastasis, and poor survival.Akçakayaet al[52]highlighted a novel functional role of miRNA-125a-5p on imatinib response.They experimentally showed that overexpression of miRNA-125a-5p suppressedPTPN18expression and furthermore this eventually increased the GIST cells viability upon imatinib treatment.Almost the same research team (Huanget al[53]2018) evaluated phosphorylated FAK (pFAK) as a candidate target of PTPN18.They revealed a downstream regulation of pFAK and direct association with imatinib resistance.Fanet al[54]explored the role of miRNA-218 on imatinib resistance GIST cells and they found a clear correlation between the downregulation of miRNA-218 and imatinib resistance.They also proposed that, miRNA-218 overexpression can improve the sensitivity of GIST cells to imatinib mesylate, with PI3K/AKT signaling pathway possibly involved mechanism.Leeet al[55]revealed that HOTAIR is upregulated is GISTs and can promote GIST cell metastatic statusin vitro.HOTAIR found to regulate promoter methylation of protocadherin 10 (PCDH10) and promote tumor invasion status.Bureet al[56]come to confirm the correlation of HOTAIR and tumor aggressiveness and propose specific methylation patterns caused by the upregulation of HOTAIR during the progression of carcinogenesis.Zhanget al[57]proposed HsamiRNA-28-5p and hsa-miRNA-125a-5p to be involved in the development and progression of GIST and therefore may be able to serve as prognostic markers for imatinib-response in GIST patients.Yanet al[58]In their study they found that lncRNA CCDC26 induced imatinib resistance and decreased imatinib induced apoptosis.These results introduced lncRNA CCDC26 to be a possible target to reverse IM resistance.The same author[51]also proposed lnc-DNAJC6-2 to be associated with the HIF-1 pathway.HIF-1 is responsive for the modulation of over 200 genes that are associated with proliferation, cycle arrest, apoptosis, and drug efflux.Therefore, investigating molecules that target the HIF-1 pathway may identify a novel treatment strategy.
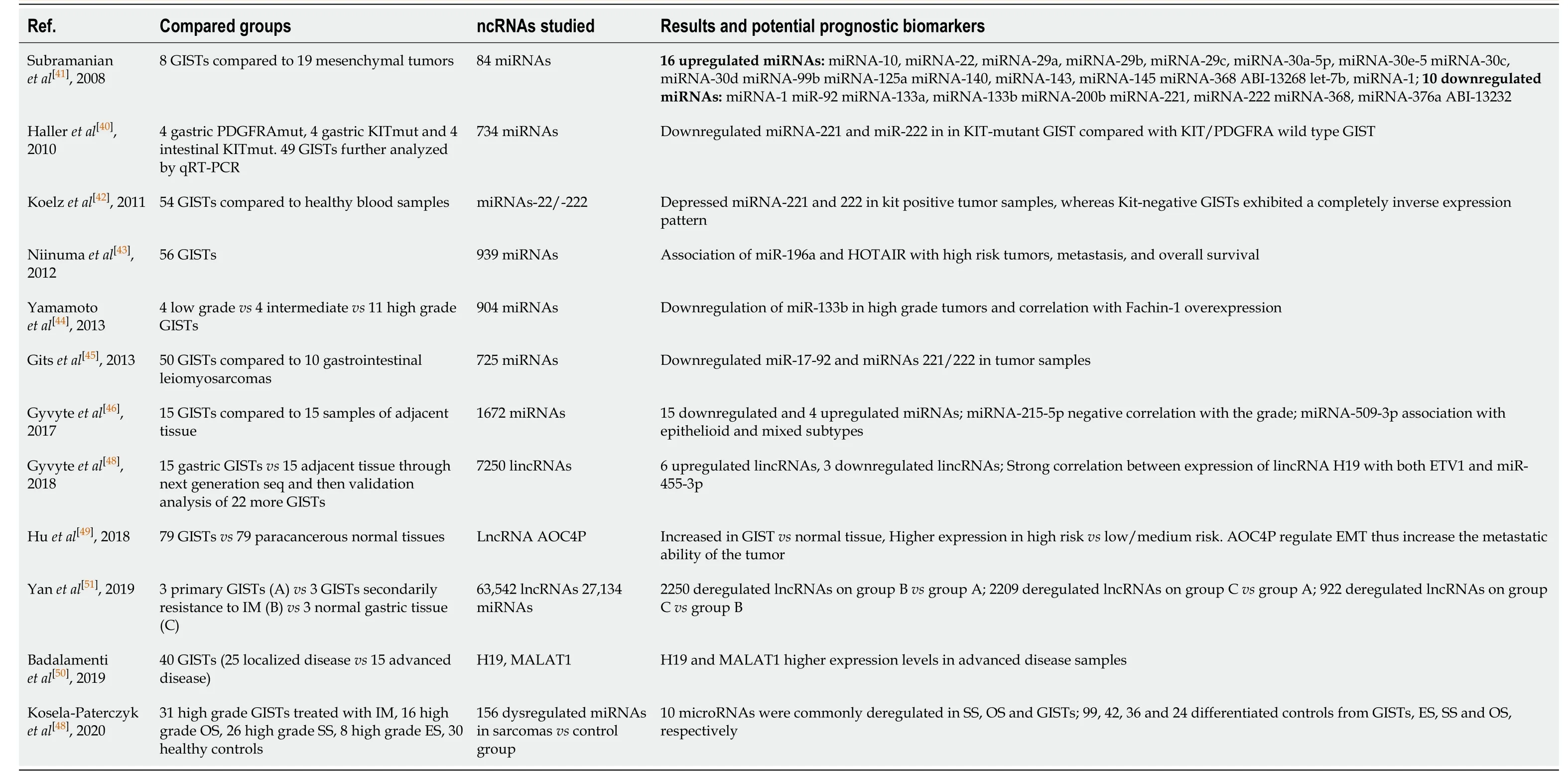
Table 2 Non-coding RNAs as potential prognostic biomarkers of gastrointestinal stromal tumors
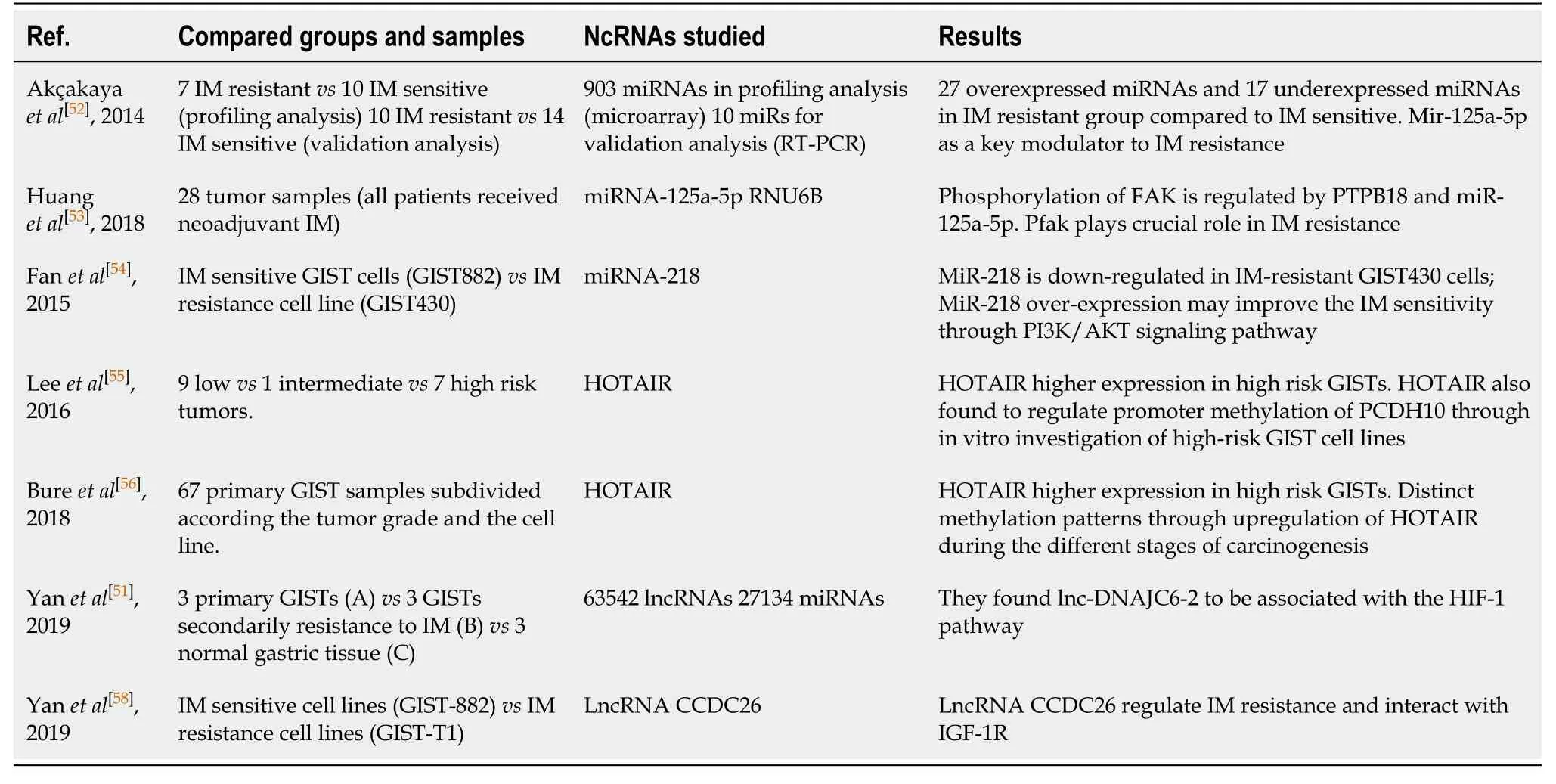
Table 3 Studies about the role of non-coding RNAs expression profile and imatinib resistance
There have been observations that miRNAs constantly export from cells and circulate in body fluids as a part of a lipoprotein complexes called exosomes, containing miRNAs and proteins[59].Furthermore, to date, there is no study looking at the role of circulating miRNAs in GIST patients, which is essential for the potential clinical use.
GENE REGULATING NON-CODING RNAS AND THEIR ROLE IN GIST CARCINOGENESIS
miRNAs are thought to act as regulators in gene expression.AlthoughKITgene mutations and KIT protein overexpression are the main genetic characteristics of GISTs, little is known about the mechanism of KIT overexpression.It is essential to identify molecules that regulate c-KIT and other relative genes as they could be excellent candidates for future clinical trials on GIST treatment.Plenty of recent studies suggesting that miRNAs directly regulate KIT protein expression levels and inhibit cell proliferation in GISTs.Felliet al[60]reported, in 2005, the downregulation of KIT receptor by miRNA-221/miRNA-222 in erythroleukemic cells.MiRNA-221 and miRNA-222 are highly homologous miRNAs, whose upregulation has been recently described in several types of human tumors.Later studies have been proposed them as oncomirs, acting by targeting tumor suppressor genes such as PTEN, TIMP3 p57, p27Kip1and BIM[61].MiRNA-221/222 overexpression induces cell proliferation through the activation of cell cycle and the Akt pathway and blocks TRAIL-induced apoptosis.Koelzet al[42]was the first to show that miRNA-221 and miRNA 222 act as regulators of Kit protein expression in GISTs and hence reveals a new aspect in the molecular pathogenesis of these tumors.They found a completely inverse expression among KIT positive and KIT negative tumors.Further studies came to correspond this by the observation that miRNA-222 and miRNA -17/20a directly target KIT and ETV1 in GISTs.MiRNA-494 is proposed as a potential KIT targeting miRNA by Kimet al[62].This study showed that miRNA-494 is a negative regulator of KIT in GISTs and an overexpressing miRNA-494 may be a promising approach to GIST treatment.Gitset al[45]published that miRNA-17, miRNA-20a directly target KIT.They also showed that overexpression of these two miRNAs induced apoptosis and significantly inhibited cell proliferation.Interestingly they did not found correlation of miRNA494 and KIT expression like Kimet al[62]did! Luet al[63]founded at their study, that miRNA-152 induced cell apoptosis, prevents cell proliferation and migration by repressing cathepsin L, suggesting miRNA-152 an attractive anti-tumor agent.In a latest study, Badalamentiet al[50]founded that the expression levels ofMALAT1lncRNA seem to affect the c-KIT mutational status.A recent Chinese study by Longet al[64], indicated that miRNA-374b inhibits apoptosis promotes viability of GIST cells by targetingPTENgene through the PI3K/Akt signaling pathway.Another similar study[65]focused on the effects of neferine, an alkaloid derivative of lotus plant, in GIST development.They interestingly founded that neferine possibly upregulate miRNA-449a and then inactivate the PI3K/AKT and Notch pathways and by this mean suppress growth and migration of GIST cells.A latest paper came out from Chenet al[66].Their results suggested that miR-4510 downregulation could promote GIST development, including growth, metastasis and invasion, through increasingAPOC2expression.Needless to say that much more scientific effort is needed in order to clarify the exact role of non-coding RNAs in GIST carcinogenesis and their interaction with tumor related genes and the respectively molecular endocytic paths.
POTENTIAL ROLE OF NON-CODING RNAS IN GIST TREATMENT
The potential role of ncRNAs as treatment tools against cancer has been explored through many studies during the recent years.The main treatment strategies aim to inhibit cell proliferation by importing exogenous ncRNAs through viral vectors (adenoviral, lentiviral and rectoviral vectors), which are mainly tumor suppressor miRNAs[67].A recent study by Tuet al[68]suggested miR-218 loaded nanoparticle as tumor suppressor miRNA in GIST.Another study by Dursoet al[69]proposed modified miRNAs 221/222 as effective inhibitors of KIT.Nowadays it is generally accepted that miRNAs can act as oncogenes or tumor suppressor genes.For this reason, it seems reasonable to manipulate those molecules against the carcinogenetic process.For example, synthesized miRNAs mimics imports into the cells and enhance endogenous miRNA function (antagomirs)[70].Another strategy is proposed for the inhibition of over-expressed oncogenic miRNAs (oncomirs), by the use of antisense oligonucleotides[71].This strategy includes inhibition or replacement of miRNAs through anti-miRNA oligonucleotides, antagomirs, miRNA sponges and nanoparticles.Only a few of the investigated miRNAs are currently in phase 2 stage[72].But it must be pointed out that, up to now, although they have been shown remarkable success inin vitromodels, none of these particles have been tested in GIST clinical trials.
CONCLUSION
A huge amount of preclinical data introduces non-coding RNAs as a new weapon against cancer in biomedical sciences armamentarium, although many efforts need to be done in order to understand the role of epitranscriptomics in GISTs.Especially for GISTs, numerus studies identified association patterns among specific ncRNAs with subsequent phenotypic characteristics.NcRNAs related to the tumor progression, grade, site, chromosomal eliminations, and imatinib sensitivity could probably be of importance as diagnostic or prognostic tumor biomarkers.In vitrostudies revealed some of the mechanisms of action of these molecules.The endocytic paths could be served as guidance for future targeted drugs, acting as interfering or enhancing molecules.In addition, published data concerning GISTs and ncRNAs is based mainly onin vitrocell lines and fresh frozen paraffin-embedded tumor tissue blocks, thus necessitating high quality, randomized, multicentric clinical studies at a large scale of patients.
ACKNOWLEDGEMENTS
This study was supported by a non-profit organization of Greek Society of Cancer Biomarkers and Targeted Therapy.
杂志排行
World Journal of Meta-Analysis的其它文章
- COVID-19: Off-label therapies based on mechanism of action while waiting for evidence-based medicine recommendations
- Learning and competence development via clinical cases – what elements should be investigated to best train good medical doctors?
- Immunotherapy in hepatocellular carcinoma: Combination strategies
- Combined endoscopy/laparoscopy/percutaneous transhepatic biliary drainage, hybrid techniques in gastrointestinal and biliary diseases
- Thrombopoietin-receptor agonists in perioperative treatment of patients with chronic liver disease
- Exclusive cigar smoking in the United States and smoking-related diseases: A systematic review
