A Novel Method for the Prenatal Diagnosis of Cleft Palate Based on Amniotic Fluid Metabolites
2020-08-29WancongZHANGLewenJIANGZhiweiSHENJiashengCHENXiaopingZHONGWeipingZENGJiandaZHOUZhihaoHEShijieTANG
Wancong ZHANG ,Lewen JIANG ,Zhiwei SHEN ,Jiasheng CHEN ,Xiaoping ZHONG ,Weiping ZENG ,Jianda ZHOU,Zhihao HE,Shijie TANG*
1 Department of Plastic Surgery and Burn Center,Second Affiliated Hospital,Shantou University Medical College,Shantou,Guangdong,China
2 Department of Plastic Surgery,Shenzhen Longgang District Maternity& Child Health care Hospital,Shenzhen,Guangdong,China
3 Department of Medical Imaging,Second Affiliated Hospital of Shantou University Medical College,Shantou,Guangdong,China
4 Department of Plastic and Reconstructive Surgery,Central South University Third Xiangya Hospital,Changsha,Hunan,China
ABSTRACT Background and purpose The prenatal diagnosis of cleft palate is an important component of sequential therapy,but the relevant diagnostic methods are still limited.We aimed here,to explore the possibility of an early prenatal diagnosis of cleft palate by assessing metabolites in pregnant mice.Methods Twenty-four inseminated females were randomly divided into retinoic acid(RA)-treated (treated with retinoic acid at 10.5 gestation days) and control groups.The metabolites of the embryonic palatal tissue,maternal amniotic fluid,and serum were characterized using 9.4T magnetic resonance spectroscopy in vitro.Then,a predictive model was established through the principal component analysis (PCA),and the correlations between the metabolites of amniotic fluid and palatal tissue were explored using orthogonal-2 partial least squares (O2-PLS).Results The incidences of cleft palate were 100% and 0% in the RA-treated and control groups,respectively.A predictive PCA model with a high specificity and sensitivity was established for the early prenatal diagnosis of isolated cleft palate using amniotic fluid metabolic data.Between RA-treated and control mice,we found that two metabolites in the amniotic fluid and palatal tissue were correlated.Creatinine showed the same trend in the palatal tissue and amniotic fluid,while choline showed opposite trends in the two tissues.However,the data for serum metabolites could not be used to establish a prediction model.Conclusion This study indicates that assessing the metabolites of amniotic fluid is a potential approach for the prenatal diagnosis of isolated cleft palate.
KEY WORDS
INTRODUCTION
The orofacial cleft is one of the most common forms of congenital malformations and displays a substantial phenotypic variation[1,2].Based on the position where the oral cavity fails to close during embryogenesis,the orofacial cleft is classified into three categories:cleft lip only (CLO),cleft palate only (CPO),cleft lip and palate(CLP)[3].There is evidence indicating that the CPO is the most common form of orofacial cleft[4].
The prenatal diagnosis of cleft palate can lead to the discovery of congenital abnormalities.This helps parents to prepare for the birth of a baby with this deformity and reassures them regarding subsequent management[5,6].Prenatal diagnosis of cleft palate will motivate parents to meet the treatment team before birth and ensure family preparations for the child's birth[7].Moreover,informing parents beforehand about the care and feeding of a baby with a cleft palate can reduce perinatal anxiety and morbidity[8].Davalbhakta et al.reported 89% favorable results of prenatally diagnosed children with cleft palate[9].
Ultrasonography has been the main technique in the prenatal diagnosis of CLO and CLP for over 35 years[10,11].Despite considerable advances in ultrasonography,successful imaging of the palate has eluded this investigation technique[12-14].However,the detection rate of CPO varies from about 0% to 22% using this technique[15].To avoid exposing the fetus to radiations,many imaging tests are not performed during pregnancy.Although a wide cleft lip is often associated with a complete cleft through the entire anterior and posterior palate,variation in phenotype is sufficiently common to render such associations valueless for predictive counseling[16].Ultrasonography has a low diagnosis rate mainly due to the location of the palate.Other influencing factors can be oligohydramnios,maternal obesity,complex fetal anomalies,overlying limbs,advanced gestational age,and the ability of the operators[17].Most clinical centers have reached a consensus on the limitations of ultrasonography in CPO diagnosis.A study analyzed the performance of two-dimensional (2D)ultrasonography for the detection of isolated cleft palate with a 0% prenatal detection rate[16,18].
Recently,magnetic resonance imaging (MRI) has been widely used to evaluate fetal anomalies[17,19].MRI has excellent soft-tissue resolution and a large field of view[20].However,MRI has not been popularized as a prenatal examination in developing countries and regions because of its cost.
Magnetic resonance spectroscopy (MRS) is used to obtain information about metabolites in tissues[21].Recently,it has been used in the investigation of many diseases,including cancers[22]and neurodegenerative diseases[23,24].In many diseases,MRS technology has identified biomarkers,which play a positive role in the diagnosis,treatment,and prognosis of diseases.The nuclear magnetic resonance (NMR)-based metabolomics platform was widely used in identifying the metabolites involved in organ developmental processes[25].However,there is little literature showing the value of MRS in the diagnosis of cleft palate by the detection of metabolites in body fluids.
In this study,we explored the possibility of prenatal diagnosis of cleft palate by metabonomics of body fluids.We detected the metabolic changes in the amniotic fluid and plasma of maternal mice with cleft palate induced by RA using 9.4T NMR metabolomic platforms.Subsequently,we compared the accuracy in the diagnosis of cleft palate between amniotic fluid and plasma metabolites.
RESULTS
Embryos exposed to retinoic acid (RA) displayed 100% incidence of cleft palate.
Among RA-exposed embryos,136 out of 143 exhibited cleft palate and seven died before analysis.While 100%of the RA-treated embryos exhibited a cleft palate,none of the 155 embryos in the control group exhibited a cleft palate (Figure 1).
Amniotic fluid and serum metabolites,as well as palatal tissue were detected by 9.4T MRS.
To explore the relationship between amniotic fluid(Figure 2),serum,and cleft palate,the metabolites in different organs,including the embryo palatal tissue and maternal amniotic fluid and serum,were characterized using 9.4T MRSin vitro(Figure 3).Then principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA)modeling were used to detect abnormal metabolites(Figures 4 and 5).
In the embryo palatal tissue,the water-soluble and fatsoluble metabolites were analyzed separately.The result for water-soluble metabolites is shown in (Figure 4).The data for the control and RA-treated group samples were separated obviously in the score plot (Figures 4a and 4b,Figures 5a and 5d),indicating that several metabolites in the RA-treated group were significantly different compared with those in the control group.The satisfactory stability of the OPLS-DA model was proved by its 200-times permutation (Figure 4c,Figures 5b and 5e).The volcano plot (Figure 4d,Figure 5c and 5f) revealed a significant difference (Students' t-test;P<0.05) in the levels of several metabolites,with a fold change >2.Significant differences in metabolite expression between the groups were confirmed through the direct inspection of VIP and p-value (VIP>1 andP<0.05).A similar analysis was carried out for the lipidsoluble metabolites.The tendency of several metabolites,such as phosphocholine,citrate,betaine,glutamine,and cholesterol,are shown in Table 1.Cholesterol,lipid,and choline were identified as the key metabolites closely related to RA-treated cleft palate.
In the maternal amniotic fluid,the differentially-expressed metabolites identified between the RA-treated and control groups included alanine,creatinine,choline,glycine,hippurate,phosphocholine,and valine (Table 1).
In the maternal serum lower expression levels of choline,citrate,creatinine,glutamine,and cholesterol were observed in the RA-treated group compared to those in the control group (Table 1).
A novel method for the Prenatal Diagnosis of Cleft Palate using Amniotic Fluid Choline.
Through 1H-NMR detection and multivariate statistical analysis of several metabolites,differentially-expressed metabolites were identified between the RA-treated and control groups,including alanine,creatinine,choline,glycine,hippurate,phosphocholine,and valine (Table 1).Based on the inner connections,as earlier mentioned,we established an O2-PLS model to explore the reasons for the strong predictive ability of prenatal diagnosis of cleftpalate by amniotic fluid analysis.Between the RA-treated and control mice,we found that two metabolites were correlated in amniotic fluid and palatal tissue.Creatinine showed the same trend in the palatal tissue and amniotic fluid (Pearson correlation coefficient:0.56),while choline showed opposite trends in the two tissues (Pearson correlation coefficient:0.42) (Table 2).Moreover,choline was the only metabolite with the same trend in both the palatal tissue and the amniotic fluid.Therefore,choline may be an important biomarker in the amniotic fluid for the diagnosis of cleft palate.
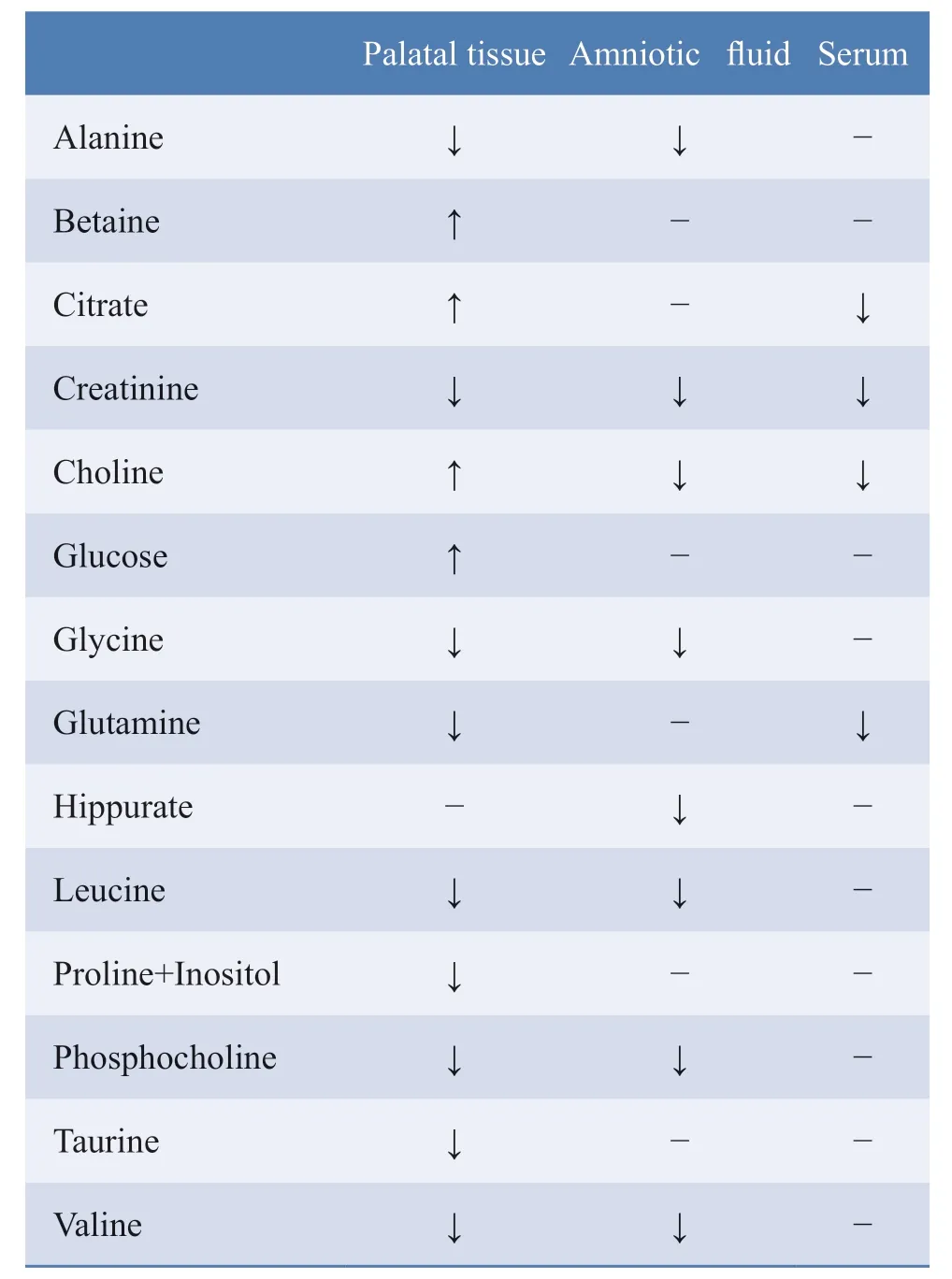
Table.1 Metabolic changes in multiple organs (including embryonic palatal tissue and maternal amniotic fluid and serum) in pregnant mice treated with or without RA
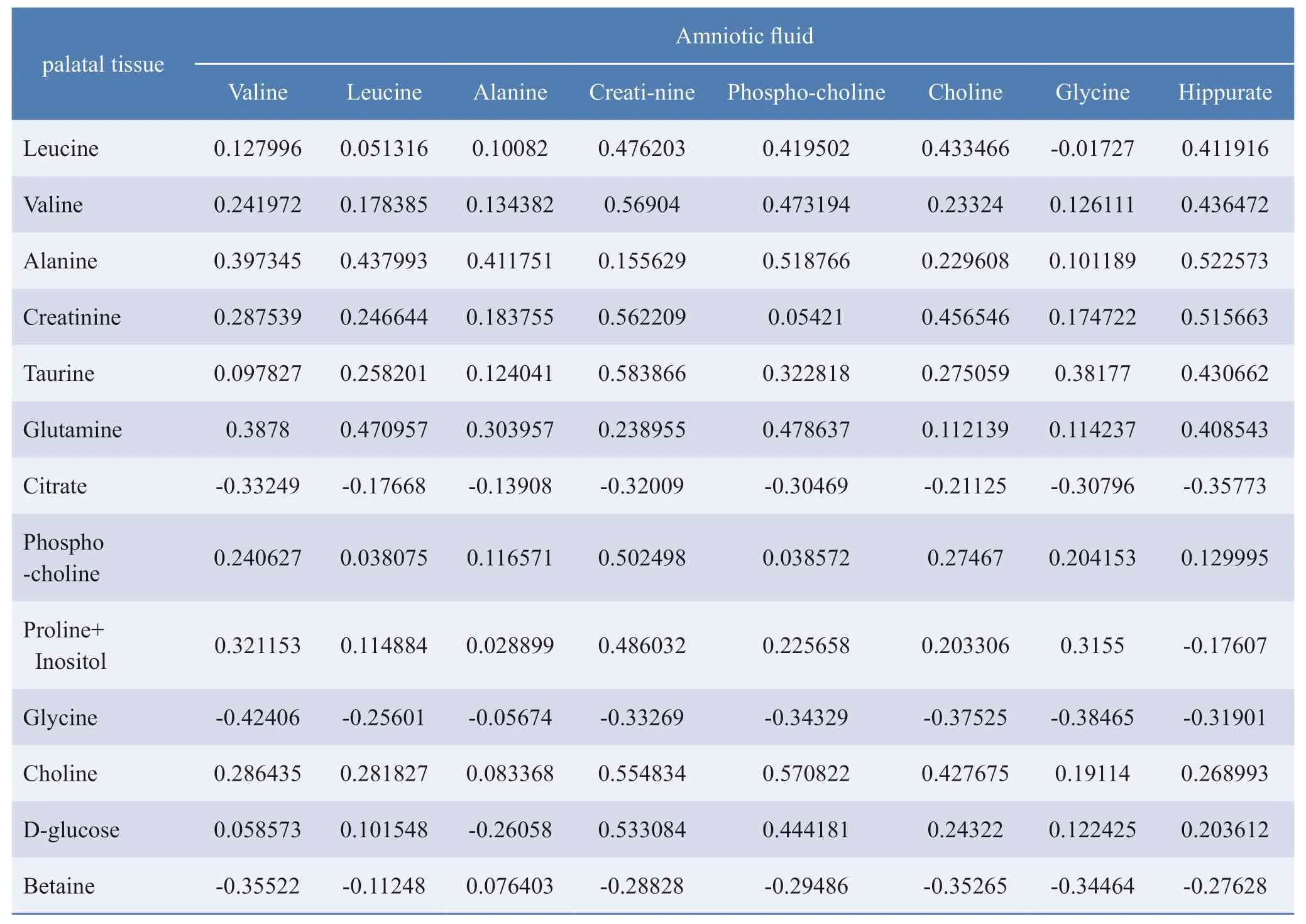
Table.2 Correlation between metabolites of amniotic fluid and palatal tissues
To further verify the stable abnormality of amniotic fluid metabolomics in the RA-treated group,a PCA-class model was established,and the discriminative abilities for the RA-treated and control groups were analyzed.Using the PCA-class model of the control group,all samples of the control group could be correctly identified,and all samples of the RA-treated group could be completely excluded (Figure 6a).The area under curve (AUC) of this model was 1 (Figure 6b).That means the specificity and sensitivity were 100% in the PCA-class model based on the 12 control samples.In the PCA-class model of the RA-treated group,10 samples of the RA-treated group could be correctly identified,and all samples of the control group could be completely excluded (Figure 6c).The AUC of this model was 1 (Figure 6d).That means sensitivity was 83% and specificity was 100%.These results indicated the potential of the amniotic fluid model in distinguishing cleft from normal palate.
As a further validation,six new samples (three RAtreated,and three controls),added to the original 24,were introduced into the model and the discriminative ability of the model was analyzed.In the PCA-class model of the control group,all new samples could be correctly identified (Figure 6e and 6f).In the PCA-class model of the RA-treated group,93.3% of the RA-treated samples were correctly identified,and 100% of the control samples were excluded.These results further verified the ability of amniotic fluid metabolites for the diagnosis of cleft palate.
Prenatal diagnosis of cleft palate cannot be established using serum metabolites
In the maternal serum,lower levels of expression of choline,citrate,creatinine,glutamine,and cholesterol were found in the RA-treated group compared with those in the control group.
To further verify the stable abnormality of serum metabolites in the RA-treated group,a PCA-class model was established and its ability to discriminate between the RA-treated and control groups was analyzed.
In the PCA-class model of the control group,11 samples of the control group could be correctly identified,but one could not be identified (Figure 7a).The AUC of this model was 0.79 (Figure 7b).That means the sensitivity was about 92%.However,eight samples (67%) of the RA-treated group were mistakenly identified as belonging to the control group using this model,and only four samples of RA-treated group (33%) could be excluded(Figure 7c).That means the specificity was 33%,which is poor (Figure 7d).
These findings indicate that a prediction model of metabolites of serum for cleft palate cannot be effectively established.
DISCUSSION
In this study,metabolites in different organs of the cleft palate mouse model were detected and analyzed by 9.4T MRS,including embryo palatal tissue,maternal amniotic fluid,and serum.We successfully established a model with high specificity and sensitivity for the early prenatal diagnosis of cleft palate using amniotic fluid metabolic data.In addition,we validated the reliability of the predictive model and found that creatinine and choline in palatal tissue and amniotic fluid were the key factors for the success of this model.Contrarily,the analysis of serum metabolites could not be used to establish a specific prediction model with good specificity.
Currently,ultrasonography plays a significant role in determining the structural malformations of CLP,of which cleft lip with or without palate accounted for approximately 86% to 90%[26].However,the detection rate of isolated cleft palate is about 0% to 22% using this technique[27].Actually,ultrasonography examination is not an effective method for the prenatal diagnosis of isolated cleft palate.Recently,chromosomal microarray analysis(CMA) was applied in the early diagnosis of orofacial clefts.However,CMA is only effective for syndromic CLP with chromosomal changes but not in isolated cleft palate[3].
In this study,the cleft palate samples were accurately distinguished using the predictive model employing amniotic fluid.The PCA-class models of both the control and RA-treated groups had 100% specificity and sensitivity.Therefore,this method is likely to be reliable for the prenatal diagnosis of isolated cleft palate.Importantly,we investigated the prediction models by analyzing the correlation of metabolites between amniotic fluid and palatal tissue.We found that choline in amniotic fluid could be used as a potential biomarker for diagnosis of cleft palate.In the organ analysis of the O2-PLS model,we found that two metabolites were correlated in amniotic fluid and palatal tissue.Creatinine showed the same trend in palatal tissue and amniotic fluid,while choline showed opposite trends in the two tissues.In other disease models,amniotic fluid metabolites,such as the glycosaminoglycan,are also useful biomarkers for prenatal diagnosis[28]and can be valuable for early clinical diagnosis.Thus,it is worth investigating whether a combination of these potential metabolite biomarkers could be used as a reliable diagnostic tool.
It is convenient to establish predictive models by detecting blood metabolites,because blood tests are routine in early prenatal diagnosis.Prenatal diagnosis through plasma has been carried out in many genetic diseases[29-32].Wu et al.conducted metabolite analyses of plasma between a dexamethasone-treated and control group and suggested that the two groups can be distinguished[33].In our study,we also tried to set up a prediction model using serum metabolites for the early prenatal diagnosis of cleft palate.In the PCA score scatter plot,the two groups were distributed in different regions but overlap,similarly to Wu's study.However,compared with amniotic fluid prediction models,serum prediction models were much less reliable,mainly because of the 33% specificity.This suggested that the placental barrier reduces the association of metabolites between blood and palatal tissues.
Although,the result of abnormal metabolite levels in the amniotic fluid of the RA-treated group could be further developed into a novel diagnostic approach for cleft palate,we need to remember that no clinical trial has been performed along these lines.It remains a challenge to obtain amniotic fluid from pregnant women during early pregnancy in developing countries.Due to the birth rate of CPO,a huge number of amniotic fluid samples is required during early pregnancy.All of these temporarily limit clinical trials.
In conclusion,the results of abnormal metabolite levels in amniotic fluid could be further developed into a novel diagnostic approach for cleft palate.The PCA-class model developed herein indicated that the cleft palate group could be distinguished from the control group by analyzing the metabolite data obtained using NMR.The predictive model had a high specificity and sensitivity for cleft palate.However,sufficient data cannot be obtained for serum metabolites to build a prediction model.
EXPERIMENTAL PROCEDURES
Animal Experiments
This study was approved by the Animal Ethics Committee of Shantou University Medical College.Eight-week-old Kun Ming mice were raised in a room at 22-25 °C with 45% humidity.Female mice were mated with male mice overnight (2 females:1 male,in the Center Laboratory Animal Sciences of Shantou Medical University).The timing was determined as embryonic gestation day 0.5(E0.5) when vaginal plugs were observed the next day.Twenty-four inseminated females were randomly divided into two groups.At E10.5,the treatment group was administered RA (Sigma,MO,USA) dissolved in sesame oil by gavage,at 70 mg/kg.
In Vitro Metabolite Measurements
In VitroPreparation of Samples
I have a daughter, said the old woman, who is so beautiful that she has not her equal in the world, and is well fitted to be your wife; if you will make her lady-queen I will show you the way out of the wood
Collected samples included bio-fluids (serum and amniotic fluid) and tissue (palatal tissue of the embryos).Palatal tissue of approximately 30-40 mg was separated and homogenized in a plastic centrifuge tube; then,1.5 ml of the mixture was extracted with methanol:chloroform(2:1 v:v).Then,the mixture was centrifuged to separate the water-soluble and lipid-soluble layers.The solvents were removed by lyophilization or evaporated under nitrogen gas to extract hydrophilic and lipid metabolites,respectively.Finally,the water-soluble metabolites were dissolved in 600 µL of D2O with 0.5 mm 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP),while the lipid-soluble metabolites were dissolved in 600 µL of CDCl3 (0.03% v/v TMS).
Bio-fluids (amniotic fluid and serum) were centrifuged for 10 min at 4°C to remove particulate contaminants.Then,300 µL of each sample was blended with 250 mL of D2O.Serum or amniotic fluid was mixed with 200 µL phosphate buffer solution (0.2 M Na2HPO4/NaH2PO4,pH 7.4,99.9% D2O) to reduce pH variation across samples.In addition,0.3 mm TSP was used as an internal reference standard at 7.0T MRS.
In VitroMetabolomic Measurements by 9.4T NMR
The samples were analyzed using a 9.4T NMR spectrometer (Bruker Avance,Switzerland).After autoshimming,a zgpr pulse (ns 64,TM 100 us,SW 13.9 ppm,TD 16K,d11.5s) sequence was used.A weak irradiation on water signal was applied to suppress the solvent.Chemical shifts in all samples were referenced to TSP.All spectra detected using the 9.4T NMR spectrometer were processed using MestReNova9.0.1 (Mestrelab Research SL,CA).For data analysis,metabolites in all 1H-NMR spectra were assigned with reference to published data and the HMDB database.
Multivariate Statistical Analysis by SIMCA
Data from thein vitroexperiment were managed using the SIMCA-P 14.1 software (Umetrics,Umea,Sweden) for multivariate statistical analysis.After an overview of the NMR data using PCA,OPLS-DA was used for supervised analysis.By applying PLS-DA,a 20-fold cross-validation was applied to obtain Q2 and R2 values,which represent the predictive ability of a model and the explain variance,respectively.To further validate the quality of the OPLSDA model,permutation tests with 200 iterations were performed.In the organ analysis,the O2-PLS model was used to determine the correlation of metabolites between the two organs.Moreover,predictive PCA-classes 1 and 2 were set up in the amniotic fluid.
HISTOLOGICAL SECTIONS
The palates of embryos were harvested at E14.5 and processed in paraffin wax.Histological sections of embryonic palates were cut and stained with hematoxylin and eosin (HE).
ACKNOWLEDGEMENTS
Not applicable.
AUTHORS' CONTRIBUTIONS
ST and WZ were involved in study design,performed the analyses,and wrote the manuscript.WZ,and LJ designed and conducted the study.JC,XZ,HZ,ZH,and SZ collected the data.JZ was involved in study design,data analysis,and the writing of the manuscript.JZ and WZ were involved in data analysis and the writing of the manuscript.All authors were involved in the writing of this manuscript and approved of the final,submitted version.
FUNDING
This study was funded by the Guangdong Basic and Applied Basic Research Foundation (2019A1515011857),the Guangdong Medical Research Foundation Project(A2019108,A2020099,A2020538),the Guangdong Science and Technology Innovation Strategy Special Fund (Vertical Collaborative Management Direction)Project ([2018] 157-45),the Guangdong Higher Education Teaching Reform Project (No.246),the Shantou University Chuangqiang Provincial Special Fund Construction Project (925-38230120),the Shantou University Special Support for In-school Research of the School of Arts (STURCS201813),and the Shantou Science and Technology Project ([2019]10602).It was also supported by the Department of Education of Guangdong Province under the Top-tier University Development Scheme for Research and Control of Infectious Diseases and the grant for Key Disciplinary Project of Clinical Medicine under the Guangdong Highlevel University Development Program,and supported by 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG18B,2020LKSFG02E).
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of interest:The authors declare that they have no competing interests.
Ethical approval:This study was approved by the Animal Ethics Committee of Shantou University Medical College(Shantou,China).
杂志排行
Chinese Journal of Plastic and Reconstructive Surgery的其它文章
- Diagnosis and Treatment of Axillary Web Syndrome:An Overview
- Regenerative Therapeutic Applications of Mechanized Lipoaspirate Derivatives
- Biomaterial Scaffolds for Improving Vascularization During Skin Flap Regeneration
- A Case of Coexistence of Aplasia Cutis Congenita and Giant Congenital Melanocytic Nevus:Coexistence of Two Rare Skin Diseases
- Pedicled or Free Flap from Contralateral Breast for Autologous Breast Reconstruction
- Comprehensive Strategy for Keloid Treatment:Experience at Shanghai Ninth People's Hospital
